Abstract
Recently, two crystal structures of the Thermus thermophilus 70S ribosome in the same functional state were determined at 2.8 and 3.7 Å resolution but were different throughout. The most functionally significant structural differences are in the conformation of the peptidyl-transferase center (PTC) and the interface between the PTC and the CCA end of the P-site tRNA. Likewise, the 3.7 Å PTC differed from the functionally equivalent structure of the Haloarcula marismortui 50S subunit. To ascertain whether the 3.7 Å model does indeed differ from the other two, we performed cross-crystal averaging of the two 70S data sets. The unbiased maps suggest that the conformation of the PTC–CCA in the two 70S crystal forms is identical to that of the 2.8 Å 70S model as well as that of the H. marismortui 50S subunit. We conclude that the structure of the PTC is the same in the functionally equivalent 70S ribosome and the 50S subunit.
The ribosome catalyzes the final step in the flow of genetic information from DNA to proteins using the decoding machinery situated in the small ribosomal subunit and the peptidyl-transferase center (PTC) located in the large ribosomal subunit. Insights into the structural basis of mRNA decoding have come largely from structures of the Thermus thermophilus 30S ribosomal subunit and its substrate complexes (1–4), whereas understanding of the structural basis of peptide bond formation has been derived from structures of the Haloarcula marismortui 50S subunit (Hma50) complexed with substrate, intermediate, and product analogues (5–9). The structures of the Hma50 complexed with a peptidyl-CCA substrate or with an analogue of the intermediate show that the CCA bound in the P-site interacts with the P-loop, nucleotides 2246–2258 of the 23S rRNA, and that the attacking α-NH2 group of a bound CC-puromycin substrate analogue is hydrogen-bonded to both the 2′-OH of the terminal A76 of the peptidyl-CCA P-site substrate and to the N3 of the ribosomal base A2451 (Escherichia coli numbering). The 2′-OH is essential for catalysis (8, 10–18) and is thought to provide a proton shuttle from the attacking α-NH2 group to the P-site 3′-OH of A76, whereas A2451 assists in the proper positioning of the attacking α-NH2 group. Currently, almost all structural, biochemical, genetic, and kinetic data are consistent with the proton shuttle mechanism (19) suggested initially by Dorner et al. (13).
Two crystal structures of the 70S ribosome from T. thermophilus (Tth70) at 2.8 and 3.7 Å resolution have recently been published. They represent the same functional state of the 70S particle with the P- and E-sites fully occupied albeit with different tRNAs (20, 21). The P-site is occupied by tRNAPhe in the 3.7 Å crystal and by tRNAfMet in the 2.8 Å crystal. The A-site is unoccupied in the 3.7 Å crystal but is partially occupied in the 2.8 Å crystal, with only the anticodon stem–loop region of tRNAPhe ordered and the antibiotic paromomycin bound to the decoding A-site. Moreover, whereas an endogenous mixture of tRNAs is bound in the E-site of the 3.7 Å crystal form, the 2.8 Å crystal has tRNAPhe in the corresponding position. Finally, the 2.8 Å crystal contains a 24-mer mRNA with a strong Shine-Dalgarno sequence, whereas the 3.7 Å crystal form contains a 10-mer mRNA.
Although the general characteristics of the two crystal forms render the ribosome in the same functional state, the atomic models determined from the two crystals differ throughout. The most functionally significant differences are in the conformations of the PTC and the hydrogen-bonding network between the PTC and the CCA end of the P-site tRNA (which we shall call the PTC–CCA interface). The structure of the PTC–CCA interface of the 3.7 Å-resolution Tth70 (21) deviates significantly from that of the Hma50 (7) as well. Most importantly, these structural variations present in the 3.7 Å Tth70 model are not consistent with the mechanistic results derived from the studies of Hma50 in isolation. The proposed differences in the conformation of the P-site CCA, in the position of A2451, and in the closer orientation of A76 to an adjacent noncanonical A2450–C2063 base pair, are inconsistent with the proton shuttle mechanism of peptide bond formation.
To reevaluate the data leading to the structures of the 70S ribosome PTC, we performed cross-crystal averaging of the two Tth70 data sets. Cross-crystal averaging is very powerful in improving phases and the resulting electron density maps while removing the model bias that is present in the initial phase set by applying a combination of solvent-flattening, histogram matching, and density averaging (22). It has several applications ranging from improving the density of poorly ordered parts of a structure to complete phasing of one crystal form with the phase set from another crystal form (23–26).
Here we present greatly improved electron density-averaged maps of the two Tth70 crystal forms using their corresponding models as the source for the initial phase sets. Our results suggest that the conformation of the PTC–CCA interface in the two Tth70 crystal forms is identical to that of the current 2.8 Å 70S model (20) and to that observed earlier in the Hma50 structure (7). Therefore, we conclude that the PTC–CCA interface adopts the same conformation in the functionally equivalent 70S ribosome as in the 50S subunit. The structure of the PTC–CCA interface we establish herein is in agreement with the previous structural, biochemical, and kinetic consensus on the mechanism of peptide bond catalysis by the ribosome.
Results
The Cross-Crystal Averaged Map of the 3.7 Å Crystal Form Fits the 2.8 Å Model Better than the 3.7 Å Model.
We used cross-crystal averaging of the two Tth70 data sets to assess the basis for the structural discrepancies between the 3.7 Å and the 2.8 Å models, particularly in the PTC–CCA interface. In this case, the two copies of 70S ribosome in the 2.8 Å crystal and one in the 3.7 Å crystal allow for 3-fold averaging. To further eliminate any phase bias from the starting model in the resulting maps, we intentionally removed portions of the 2.8 Å model that either differed or were in question before the initial phase set calculation (see Methods). On the other hand, the entire 3.7 Å model was used for calculating phases for the 3.7 Å data set, thereby introducing model bias toward the low-resolution model. Independent of the choice of the averaging reference model and the initial phase set, the results of cross-crystal averaging suggested that the 2.8 Å Tth70 model agreed better with the resulting 3.7 Å density-averaged maps than the 3.7 Å Tth70 model.
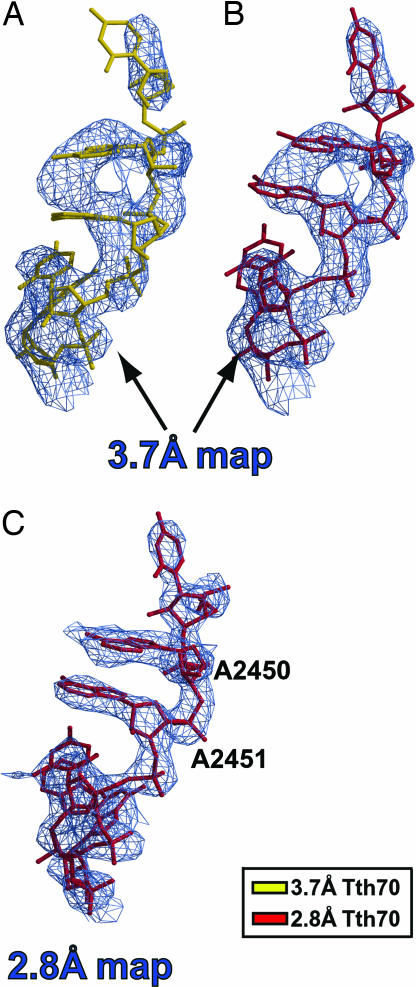
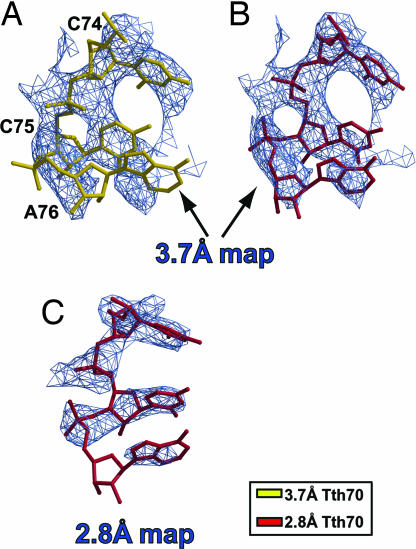
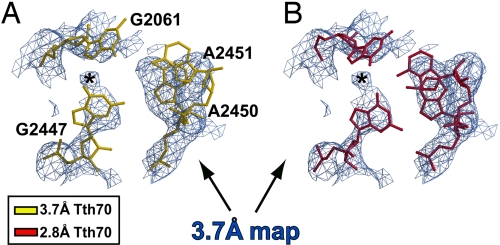
After superimposing the final 3.7 Å density-modified map separately on PTC residues 2449–2453 from both Tth70 models, it was immediately clear that the unadjusted 2.8 Å model fit the resulting map well (Fig. 1B), including the critical residue A2451, whereas the 3.7 Å model did not (Fig. 1A). The 2.8 Å Tth70 model also agreed well with the 2.8 Å density-modified map in the same region even after omitting the PTC–CCA domain from averaging (Fig. 1C). Furthermore, the CCA end of the 2.8 Å model agreed with the 3.7 Å modified map better than the CCA end of the 3.7 Å model (Fig. 2). The map unambiguously revealed the tRNA backbone density, and the smeared density at the CCA end indicated base stacking between the bases of C75 and A76. This stacking was not present in the 3.7 Å Tth70 model, and the alternative path for the backbone resulted in steric clashes with the 23S rRNA [see supporting information (SI) Text] (21). All of these steric clashes disappeared once the 2.8 Å model was used instead. Additionally, the CCA end of the 2.8 Å Tth70 agreed completely with the 2.8 Å density-modified map even when the PTC–CCA domain was not averaged (Fig. 2C).
Fig. 1.
Superposition of the 3.7 Å and 2.8 Å models on cross-crystal averaged maps of the PTC. (A) The 3.7 Å model (gold) does not fit the 3.7 Å density-modified map contoured at 1.5σ. (B) Conversely, the 2.8 Å model (crimson) agrees with the resulting map. (C) The 2.8 Å cross-crystal averaged map after omitting the PTC–CCA from averaging, with the corresponding model in crimson superimposed. The map is contoured at 1.2σ.
Fig. 2.
Superposition of the 3.7 Å and 2.8 Å models on cross-crystal averaged maps of the CCA end of the P-site tRNA. (A) The 3.7 Å cross-crystal averaged map with the 3.7 Å Tth70 model (gold) superimposed. (B) The 2.8 Å model (crimson) fits the cross-crystal averaged map of the low-resolution crystal form better than the corresponding 3.7 Å model (gold). (C) The density-modified map of the 2.8 Å crystal form after omitting the PTC–CCA from averaging. The maps are contoured at 1.5σ.
The unadjusted PTC of the 2.8 Å Tth70 fit the 3.7 Å modified map better than the 3.7 Å PTC even when the 3.7 Å Tth70 was used as the averaging reference model (SI Fig. 7 A and E). Although the quality of the 3.7 Å map was significantly worse when the PTC–CCA was omitted from the averaging, the backbone of the unadjusted 2.8 Å PTC again agreed with the resulting map better than the 3.7 Å PTC (SI Fig. 7B). Finally, the 2.8 Å Tth70 devoid of the PTC–CCA, P-site tRNA, A-site tRNA, and mRNA was used as the source for the initial phase set for both crystal forms. The truncated 2.8 Å Tth70 was superimposed onto the 3.7 Å model and then refined against the 3.7 Å data set. Thus, neither of the initial phase sets contained information about the PTC–CCA conformation. The quality of the modified maps improved significantly with the altered phase set (SI Fig. 7 C, D, and F). The density for the backbone was continuous even when the PTC–CCA was excluded from the averaging (compare SI Fig. 7 B and D), whereas density for the entire residue U2449 became visible when the domain was 3-fold averaged (compare SI Fig. 7 A and F).
Phasing of the 3.7 Å Crystal with the 2.8 Å Tth70 Model Further Established That the Conformation of the PTC Is Identical to That of the 2.8 Å Tth70.
In a separate experiments the two Tth70 models, devoid of the parts of the PTC–CCA, were refined against the data set from the other crystal form and the omit maps were analyzed. All of the maps obtained by this procedure confirmed that the conformation of the PTC–CCA is identical to that of the 2.8 Å Tth70.
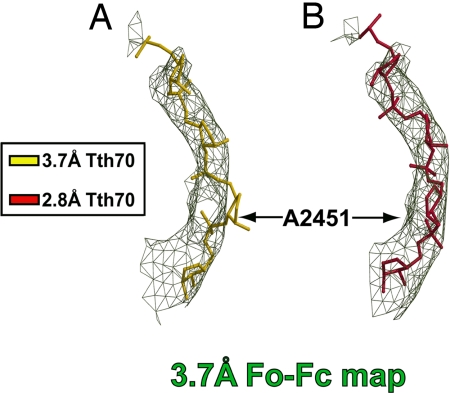
The 2.8 Å Tth70 model that did not include residues 2448–2456 of the PTC and the CCA end of the P-site tRNA (nucleotides 73–76) was superimposed onto the 3.7 Å model and then rigid-body-refined against the 3.7 Å data set. After superimposing the resulting 3.7 Å omit map onto PTC residues 2448–2456 from both Tth70 models, it was clear that the 2.8 Å Tth70 fit the 3.7 Å omit map better than the 3.7 Å Tth70 model (Fig. 3). For instance, the backbone of the residue A2451 from the 3.7 Å Tth70 model was completely out of the density (Fig. 3A), whereas the 2.8 Å Tth70 model fit the map well (Fig. 3B).
Fig. 3.
Superposition of the 3.7 Å and 2.8 Å models on the 3.7 Å omit map of the PTC. (A) The 3.7 Å model (gold) does not fit the omit map (green). (B) Conversely, the backbone of the 2.8 Å model (crimson) fits the same map well. The model is rotated ≈90° counterclockwise around the vertical axis relative to Fig. 1. Only the RNA backbone atoms of residues 2448–2456 are shown, and the map is contoured at 1.4σ.
Finally, the entire 3.7 Å Tth70 model was refined against the 2.8 Å data set, and we observed the following: (i) the density for the bases was completely missing in the 2Fo–Fc map; (ii) strong positive density in the Fo–Fc map appeared for the backbone of PTC residues 2449–2453, bases A2450 and A2451, and bases C75 and A76 of the P-site tRNA; and (iii) the PTC–CCA of the 2.8 Å Tth70 fit the resulting Fo–Fc map better than the PTC from the 3.7 Å model (data not shown).
Evidence in Maps for the Lack of Model Bias.
The appropriate density-modified maps confirm that the method of cross-crystal averaging used did not result in maps that exhibited bias toward either the starting model or the averaged electron density from the stronger diffracting 2.8 Å crystal. Perhaps the strongest positive control showing the lack of bias toward the starting model is the appearance in the averaged electron density maps of unique structural features that were omitted from both the initial phase calculation and the averaging procedure. Only the 2.8 Å Tth70 model contained the partially ordered and occupied A-site tRNA, the antibiotic paromomycin, and the large-subunit proteins L31 and L33, whereas the 3.7 Å Tth70 model included the large subunit protein L36. The L1 stalk orientation was also proposed to be different in the two crystal forms. The L1 stalk in the 3.7 Å model was tilted by 11° toward the E-site tRNA. Finally, the 3.7 Å crystal contained the 10-mer mRNA, whereas the mRNA was a 24-mer in the 2.8 Å crystal form. Consequently, to confirm the lack of electron density bias we examined the reappearance of these unique structural features in both density-modified maps.
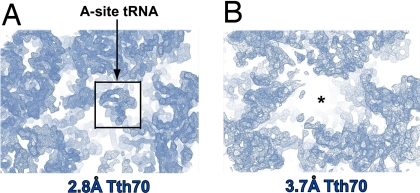
Electron density corresponding to the antibiotic moiety paromomycin and a partially ordered A-site tRNA appeared in the 2.8 Å cross-crystal averaged map but not in the 3.7 Å map (Fig. 4). Electron density for the large subunit proteins L31 and L33 also reappeared exclusively in the 2.8 Å map, although with density of significantly lower quality (data not shown). The somewhat lower-quality density in these regions is probably a consequence of these proteins being omitted both from the initial phase calculation and cross-crystal averaging while being protected from density flattening only with the solvent mask. Furthermore, the features reappeared exclusively in the 2.8 Å map even when the whole 2.8 Å Tth70 model was included and 3-fold averaging of these regions was used. Finally, although the proteins L31 and L33 and the A-site tRNA were modeled into the 3.7 Å Tth70 and 3-fold averaging was used, the density reappeared only in the 2.8 Å map.
Fig. 4.
Unbiased electron density maps produced by cross-crystal averaging. (A) The averaged electron density for the anticodon stem loop of the A-site tRNA reappeared only in the 2.8 Å map. (B) On the other hand, the 3.7 Å map did not contain any density in the same area. The black box demarcates the tRNA electron density. The maps are contoured at 1.5σ.
Furthermore, the averaged 2.8 Å electron density map in the region proposed to contain protein L36 in the 3.7 Å model did not reveal density for the L36 protein, thus confirming the original conclusions (20). However, surprisingly the density corresponding to L36 was also missing completely from the 3.7 Å map, although the protein had been included in the initial phase calculation and was protected by the averaging mask.
Also of interest is the large ribosomal protein L28, which was built differently in the two Tth70 models. For this reason the entire L28 protein was removed from the 2.8 Å Tth70 model used here for the initial phasing of the 2.8 Å data, whereas the one from the 3.7 Å Tth70 was included in the initial phasing. The 2.8 Å modified map showed clear density for the protein backbone, although density for many side chains was missing, which may have resulted in placing the incorrect protein sequence register into the density in these regions (V. Ramakrishnan, personal communication). On the other hand, the density for the same protein in the 3.7 Å map was of extremely poor quality, and it was not improved even when L28 was introduced in the initial phasing of the 2.8 Å crystal and subsequent averaging.
The density-modified maps confirmed the original conclusions that the L1 stalk is positioned differently in the two crystal forms (20, 21). A series of tests with different levels of averaging and with alternating reference and target models was performed. The same results were obtained independent of the experimental conditions. The density for the L1 stalk was well defined in the 3.7 Å map except for residues 2130–2160. The map for this region was of poor quality even when 3-fold averaging was used (data not shown). Furthermore, the unadjusted L1 stalk and the E-site tRNA from the 3.7 Å Tth70 model fit the 3.7 Å map well (data not shown). Similarly, the L1 stalk of the 2.8 Å model agreed with the 2.8 Å modified map (data not shown), hence confirming the proposed different orientations of the L1 stalk in the two crystals. In addition, the cross-crystal averaged maps confirmed that the two crystal forms contained different P-site tRNA and mRNA molecules (data not shown). The quality of the 3.7 Å map was superior when the 3-fold averaging of the P-tRNA anticodon stem loop was applied, yet the 3.7 Å Tth70 model fit the map (data not shown), confirming the original findings (20, 21).
The Density-Modified Maps Reveal the Presence of the PTC Triple Base Pair in both Tth70 Crystal Forms.
The 3.7 Å-resolution map obtained by cross-crystal averaging confirmed the existence of the triple base pair at the heart of the PTC and also contained density for a metal ion coordinated by these bases. The PTC triple base pair is formed among the bases G2447, A2451, and G2061, and it has been previously seen in the Hma50 model (5, 7, 9). In that model the hydrogen-bonding network was stabilized by a metal ion positioned between the bases G2447 and G2061. This ion had been identified as potassium from anomalous scattering difference Fourier maps calculated by using data from crystals of the Hma50 subunit soaked in solutions containing rubidium (5). The triple base pair was present in the 2.8 Å Tth70 model with a Mg2+ ion modeled at the position of the density corresponding to the metal ion (SI Fig. 8B), whereas these interactions among the bases were completely missing in the 3.7 Å model (SI Fig. 8A). However, the averaged 3.7 Å map showed unambiguously that the triple base pair is present in this crystal form as well (Fig. 5). The positions of residues G2447 and A2451 in the 3.7 Å Tth70 model did not fit in the resulting averaged electron density map (Fig. 5A), whereas the 2.8 Å Tth70 model was in good agreement with the same map (Fig. 5B). Moreover, the electron density adjacent to the base of the G2447 residue exhibited an extension that became spherical at higher contour levels. The sphere represents the density for metal ion, presumably potassium, which assists in stabilizing the pair. Observing the metal ion at this relatively low resolution confirms the phasing power of the method used. The triple base pair constructed in the published 2.8 Å Tth70 model fit the 2.8 Å cross-crystal averaged map very well (data not shown), although the density corresponding to the metal ion was somewhat shifted. However, the ion is likely to be potassium rather than magnesium for two reasons: (i) the coordination is planar rather than octahedral, and (ii) the distances between the ion and the heavy atoms of the surrounding bases are somewhat longer than those observed in high-resolution RNA structures containing Mg2+ ions.
Fig. 5.
Superposition of the two Tth70 models on unbiased cross-crystal averaged maps of the PTC triple base pair. (A) The cross-crystal averaged map of the 3.7 Å crystal form in the triple base pair region with the 3.7 Å Tth70 model (gold) superimposed. (B) The same map as in A with the 2.8 Å model (crimson) superimposed. The putative metal ion density is labeled with an asterisk.
The Structure of the PTC–CCA Interface in the 3.7 Å Tth70 Model Differs Significantly from That of the Hma50.
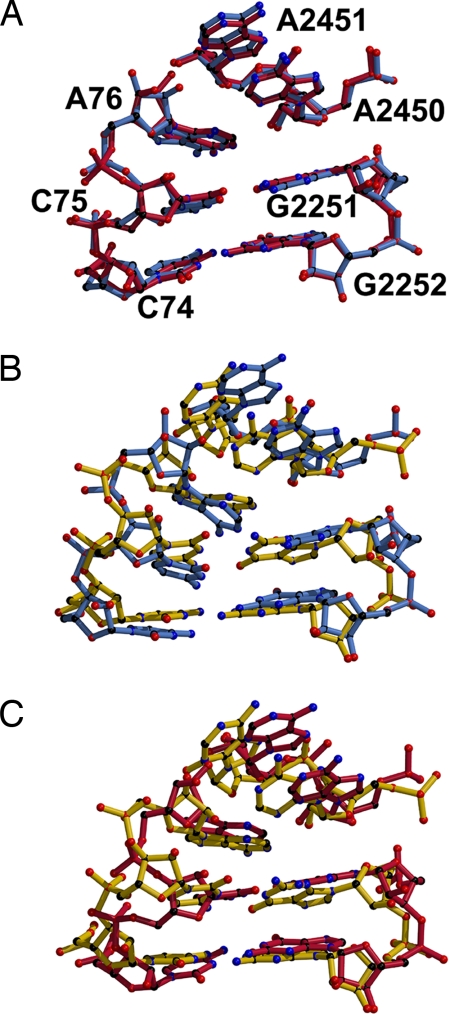
Least-squares superposition of the 50S subunits of the two Tth70 models onto that of the Hma50 revealed major differences between the PTC model of the 3.7 Å Tth70 and the other two, which agree with each other. First, the two crystal structures of the T. thermophilus 70S (20, 21) were compared with that of the Hma50 (7), and then both Tth70 models were compared with each other (see Methods). The backbone atoms of the PTC residues and the CCA end of the P-site tRNA from Hma50 superimposed with an rmsd of 0.75 Å on the corresponding atoms of the 2.8 Å Tth70 despite sequence differences elsewhere (Fig. 6A). The rmsd value increased to 1.03 Å when the atoms of the bases were included in the calculation as well. In contrast, the PTC of the 3.7 Å Tth70 model differed significantly from both models (Fig. 6 B and C). An rmsd of 1.58 Å was obtained when the backbone atoms of the PTC–CCA of the 3.7 Å Tth70 were superimposed onto that of the Hma50, whereas the value increased to 1.78 Å when the atoms of the bases were incorporated in the estimate. Similarly, the 3.7 Å PTC–CCA superimposed onto the 2.8 Å PTC–CCA with rmsd of 1.41 Å (for the backbone atoms only) and 1.61 Å (for all of the atoms). Major disparities were noticed in the positions of the A2451 residue of the 23S rRNA and of C75 of the P-site tRNA.
Fig. 6.
Comparison of the two Tth70 and the Hma50 models in the region of the PTC–CCA interface. (A) Superposition of the Hma50 PTC–CCA (blue) onto the 2.8 Å Tth70 structure (crimson). (B) Superposition of the Hma50 (blue) onto the 3.7 Å Tth70 structure (gold). (C) Comparison of the 2.8 Å Tth70 (crimson) and the 3.7 Å Tth70 (gold) models.
Discussion
The recently reported structure of the 70S ribosome from T. thermophilus at 3.7 Å resolution differs significantly both from a functionally equivalent structure of Tth70 at 2.8 Å resolution and from the structure of the H. marismortui 50S subunit. Importantly, the structural differences in the modeled structures of the interface between the PTC and the CCA end of the P-site tRNA would have serious implications for our understanding of the ribosome-catalyzed peptidyl-transferase reaction because the 3.7 Å Tth70 model is inconsistent with the proton shuttle mechanism of catalysis of peptide bond synthesis. The remarkable differences in the modeled conformations of the active site suggested the need for an additional analysis of the diffraction data available from the two crystal forms to provide improved and unbiased electron density maps of the PTC–CCA in both Tth70 crystals.
An improved electron density map of the 3.7 Å Tth70, unbiased by the initial phasing model, was achieved through cross-crystal averaging of the two Tth70 data sets. Although the initial model used in the first round of phasing introduced bias toward the 3.7 Å Tth70 model (see Results and Methods), the procedure of averaging the corresponding densities of the three separate 70S ribosome particles resulted in improved phase sets and electron density maps that were unbiased by the starting model. The resulting averaged maps revealed that the 2.8 Å Tth70 model agreed well with the 3.7 Å density-modified map, whereas the corresponding 3.7 Å model did not. The current 2.8 Å model was in good agreement with the 2.8 Å density-modified map even when the PTC–CCA was omitted both from the refinement and averaging. Similar results were obtained when the 2.8 Å Tth70 model devoid of the PTC–CCA interface was refined against the 3.7 Å data set. A series of the maps in which the PTC was omitted from the phase calculation fit the 2.8 Å Tth70 model better than the 3.7 Å Tth70 model. These results suggest that the structure of the PTC is indeed the same in the two crystals and is identical to the 2.8 Å Tth70 model.
The differences between the final averaged electron density maps of the 3.7 Å and 2.8 Å Tth70 crystals that stem from the original experimental differences can be used to establish the validity of the averaging procedure used. Although unique structural features, such as the antibiotic moiety paromomycin, the stem–loop of the A-site tRNA, and the large ribosomal proteins L31 and L33, were not included in the starting model and subjected to 3-fold averaging, these features were seen exclusively in the 2.8 Å map and not the 3.7 Å map. We further confirmed these results by modeling proteins L31 and L33 and the A-site tRNA into the 3.7 Å Tth70 model. Furthermore, the final 2.8 Å map revealed that the backbone of L28 was built correctly, whereas the 3.7 Å modified map corresponding to the equivalent region was of poor quality, implying that an alternative conformation of L28, as it was suggested in the published work (21), was probably a consequence of map misinterpretation. Moreover, the averaged maps revealed that neither of the crystal forms contained the L36 protein, consistent with the 2.8 Å model (20) but not the 3.7 Å model (21). On the other hand, our results validate the different proposed orientations of the L1 stalk in the two crystal forms (20, 21). Finally, the density-modified maps obtained after 3-fold averaging and with the 2.8 Å Tth70 as the averaging reference model confirmed that two crystals indeed contained mRNA molecules of different lengths. The aforementioned observations provide further evidence that the averaging method used here converged on the correct phases and resultant electron density for both crystal forms and was unbiased by the starting model.
If the maps of the 3.7 Å Tth70 obtained by Korostelev et al. (21) allowed them to correctly discover several model features of the 70S ribosome complex such as the orientation of the L1 stalk and the positions and interactions made by the P-site and E-site tRNAs, why were detailed features of the structure inaccurate? The existence of a significant number of disallowed steric clashes in the published 3.7 Å model (see SI Text) suggests that insufficient steric and conformational constraints were imposed during refinement to prevent overfitting of the model to the weak 3.7 Å-resolution data set. The starting model used for phasing the 3.7 Å crystal form (27) contained only 138 close contacts (defined here as distances <2.5 Å), whereas the number of steric clashes in the final 3.7 Å Tth70 model (21) was a staggering 6,167, of which 2,405 steric clashes were found in the 23S rRNA alone. Because the 〈I〉/σI at the highest-resolution shell was 1.5 and the overall 〈I〉/σI was only 3, the number of significant reflections measured with an intensity greater than two σ is not as large as implied by the 3.7 Å-resolution description (only 47% of the entire data set), and, hence, it was not sufficient to constrain the model for the type of the refinement used (21). Because the bias introduced into the phasing by the model is even more pronounced at lower resolution of this study, errors introduced into the model would be difficult to correct subsequently. Furthermore, the theoretically built model of Tth70 (27) that was used for phasing the 3.7 Å crystal form is almost identical to that of the 2.8 Å Tth70 (20), consistent with our conclusion that the subsequent errors in the 3.7 Å Tth70 model resulted from an insufficiently constrained coordinate refinement. We conclude that the PTC of the 3.7 Å Tth70 as modeled would be a highly unstable structure. Also, the number of steric clashes observed within the A2450–C2063–A76 triple pair invalidates the proposal by Korostelev et al. (21) about its possible involvement in the peptide bond synthesis reaction.
Finally, major differences in the structure of the PTC–CCA domain were seen between the 3.7 Å Tth70 model (21) and that of Hma50 (7), whereas the Hma50 and 2.8 Å Tth70 (20) models were identical in this region. Korostelev et al. (21) attributed the observed differences between the 3.7 Å Tth70 and the Hma50 models to the species-dependent sequence divergence or to differences in crystal contacts. Although the rRNA sequences and proteins of the archaeal and eubacterial ribosomes are divergent, the residues forming the PTC are highly conserved. Crystal contacts can affect the conformation of the solvent-exposed domains and rarely, if ever, have any effect on the structure of the active site that is hidden in the interior of the molecule. Strikingly, the same degree of structural difference is seen between the two Tth70 models despite the sequence identity and the similar crystallization conditions. In contrast, the PTC–CCA interfaces from the Hma50 model and the 2.8 Å Tth70 were completely superimposable despite the sequence divergence and large differences in crystallization conditions.
Our results provide an answer to the longstanding question of whether the structures of the PTC and its complexes with substrates are the same in the isolated 50S ribosomal subunit as in the complete 70S ribosome (21, 28, 29). The comparison of the cross-crystal averaged Tth70 maps with the Hma50 structure prompts us to conclude that they are indeed identical in the same functional state. The position and interactions of the CCA end of the tRNA bound in the P-site of the PTC in Hma50 and the 2.8 Å Tth70 represent the genuine structure of the ribosome active site. Consequently, the conclusions drawn from earlier studies of the structural basis of peptide bond synthesis using the Homaris mortui 50S subunit are based on relevant structures (5–9). We finally conclude that the 50S ribosomal subunit is, indeed, a valid model system for studying the mechanism of the peptide bond synthesis.
Methods
The initial phase and figure-of-merit sets were calculated from the two Th70 models that had been refined against the corresponding data sets by using positional refinement with bulk solvent correction in Refmac 5 (30). The entire 3.7 Å model (21) was used for the 3.7 Å phase calculation, whereas in the case of the 2.8 Å phase calculation (20) the following parts of the model were omitted from the refinement and subsequent 2.8 Å phase calculation: the PTC (comprising the following 23S rRNA residues: 2050–2075, 2240–2260, 2430–2460, 2500–2510, 2565–2575, and 2598–2604), the CCA end of the P-site tRNA (residues 1–16 and 69–76), the anticodon stem loop of the P-site tRNA (residues 31–38), the A-site tRNA, mRNA, the antibiotic moiety (paromomycin), proteins L28, L31, and L33, and the metal ions (Mg2+ and Zn2+). The solvent masks covering the entire asymmetric unit in both crystal forms were calculated in MAMA (31). Each experiment was performed twice alternating the reference and target models. The averaging reference model (defined as the model used for the mask calculation used in the averaging) was divided into the following six domains: 50S, PTC–CCA (PTC and CCA end), L1 stalk (23S rRNA residues 2104–2186 and L1 protein), 30S, E-tRNA, and P-tRNA (residues 17–68). The rotation–translation matrices to be used for the density averaging were calculated by using Lsqman (31); the domain from the reference molecule was placed onto the position of the corresponding domain from the target model using all of the phosphate and sugar backbone atoms. The quality of superposition was analyzed in Coot (32) by visual inspection. For the sake of simplicity we used the matrices derived from the phosphate–sugar backbone and applied them also to the appropriate ribosomal proteins. Several proteins that differed significantly between the two models (L28, L29, and THX) and those that were absent from one or the other model (L31 and L33, but not L36) were excluded from the cross-crystal averaging procedure. Averaging masks covering the domains of the reference molecule were calculated, and any overlaps between adjacent masks were removed by using the program MAMA (31). The quality of all masks was visually inspected in O (33). Density averaging and solvent flattening followed by cycles of phase extension were performed by using the program DMMULTI (34). All calculations were performed with three and five cycles of averaging. The final maps with different models superimposed were examined by using the program Coot (32). For the 3.7 Å omit map calculation the PTC residues (nucleotides 2448–2456), the CCA end of the P-site tRNA (nucleotides 73–76), the anticodon stem loop of the P-site tRNA (nucleotides 31–38), and the mRNA were omitted from both Tth70 models before refinement. The coordinates and structure factors deposited under Protein Data Bank ID codes 2I1C and 1VS9 were used for the 3.7 Å Tth70 model refinement and averaging, whereas for the 2.8 Å Tth70 model those under Protein Data Bank ID codes 2J00, 2J01, 2J02, and 2J03 were used. The same results were obtained by using the updated set of coordinates for the 3.7 Å Tth70 model.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Yong Xiong, Peter Moore, and C. Axel Innis for suggestions. This work was supported by National Institutes of Health Grant GM22778 (to T.A.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711076105/DC1.
References
- 1.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 4.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 5.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 6.Schmeing TM, et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 7.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 10.Das GK, Bhattacharyya D, Burma DP. A possible mechanism of peptide bond formation on ribosome without mediation of peptidyl transferase. J Theor Biol. 1999;200:193–205. doi: 10.1006/jtbi.1999.0987. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlin SI, Merino EJ, Weeks KM. Catalysis of amide synthesis by RNA phosphodiester and hydroxyl groups. Proc Natl Acad Sci USA. 2002;99:14688–14693. doi: 10.1073/pnas.212527799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen JL, Schmeing TM, Moore PB, Steitz TA. Structural insights into peptide bond formation. Proc Natl Acad Sci USA. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorner S, Panuschka C, Schmid W, Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31:6536–6542. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 15.Sharma PK, Xiang Y, Kato M, Warshel A. What are the roles of substrate-assisted catalysis and proximity effects in peptide bond formation by the ribosome? Biochemistry. 2005;44:11307–11314. doi: 10.1021/bi0509806. [DOI] [PubMed] [Google Scholar]
- 16.Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci USA. 2005;102:12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trobro S, Aqvist J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry. 2006;45:7049–7056. doi: 10.1021/bi0605383. [DOI] [PubMed] [Google Scholar]
- 18.Rangelov MA, Vayssilov GN, Yomtova VM, Petkov DD. The syn-oriented 2′-OH provides a favorable proton transfer geometry in 1,2-diol monoester aminolysis: Implications for the ribosome mechanism. J Am Chem Soc. 2006;128:4964–4965. doi: 10.1021/ja060648x. [DOI] [PubMed] [Google Scholar]
- 19.Rodnina MV, Beringer M, Wintermeyer W. How ribosomes make peptide bonds. Trends Biochem Sci. 2007;32:20–26. doi: 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 21.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Cowtan K. Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr D. 1999;55:1555–1567. doi: 10.1107/s0907444999007416. [DOI] [PubMed] [Google Scholar]
- 23.Elkins PA, et al. Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor. Nat Struct Biol. 2000;7:808–815. doi: 10.1038/79047. [DOI] [PubMed] [Google Scholar]
- 24.Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 26.Lomakin IB, Xiong Y, Steitz TA. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129:319–332. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Tung CS, Sanbonmatsu KY. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys J. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 29.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Kleywegt GJ, Jones TA. Software for handling macromolecular envelopes. Acta Crystallogr D. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.