Abstract
Brh2, the ortholog of the BRCA2 tumor suppressor in Ustilago maydis, works hand in hand with Rad51 to promote repair of DNA by homologous recombination. Previous studies established that Brh2 can stimulate DNA strand exchange by enabling Rad51 nucleoprotein filament formation on replication protein A-coated ssDNA. But, more recently, it was noted that Brh2 has an inherent DNA annealing activity, raising the notion that it might have roles in recombination in addition to or beyond the mediator function. Here, we found that Brh2 can autonomously promote the formation of D-loops in reactions with plasmid DNA and homologous single-stranded oligonucleotides. The reaction differs from that catalyzed by Rad51 in having no requirement for cofactors or preloading phase on ssDNA. D-loop formation was most effective when Brh2 was mixed with plasmid DNA before addition of single-stranded oligomer. D-loop formation catalyzed by Rad51 was also enhanced when Brh2 was premixed with plasmid DNA. Brh2 rendered defective in Rad51 interaction by mutation in the BRC element was still capable of promoting D-loop formation. However, the mutant protein was unable to enhance the Rad51-catalyzed reaction. The results suggest a model in which Brh2 binding to plasmid DNA attracts and helps capture Rad51-coated ssDNA.
Keywords: BRCA2, BRC, Dss1, homologous pairing, Rad51
In eukaryotes, repair of DNA by recombination requires Rad51 for homologous pairing and strand exchange. BRCA2 plays a fundamental role in the process by governing formation of the Rad51 nucleoprotein filament, the polymerized form of Rad51 that catalyzes pairing and DNA strand invasion (1, 2). BRCA2's interplay with Rad51 is mediated by BRC motifs located medially and the unrelated C-terminal interaction element. Productive recombination apparently results from a functional balance between the two different binding regions (3, 4).
Brh2, the conserved but smaller BRCA2 ortholog in Ustilago maydis (5), has provided a useful model system for understanding mechanistic details of BRCA2 function. Cooperation between its single BRC motif and C-terminal Rad51 binding element is mediated by Dss1 (6), a small polypeptide that is essential for Brh2 function (7, 8) and forms a tight complex with the medial region corresponding to part of the helical domain and OB1 fold in the BRCA2 crystal structure (9). Brh2 was previously shown to nucleate Rad51 filament formation on replication protein A (RPA)-coated DNA at junctions of ssDNA and dsDNA (10). Investigation of its DNA binding properties by using oligonucleotides revealed that D-loop DNA was a preferred substrate (11). Given this binding preference, we set out to investigate whether the protein could augment Rad51-promoted D-loop formation, not necessarily by promoting Rad51-filament formation, but possibly by stabilizing the nascent DNA joint molecules. During the course of our studies, as reported here, we found that Brh2 itself could promote D-loop formation.
Results
Brh2 Makes D-Loops.
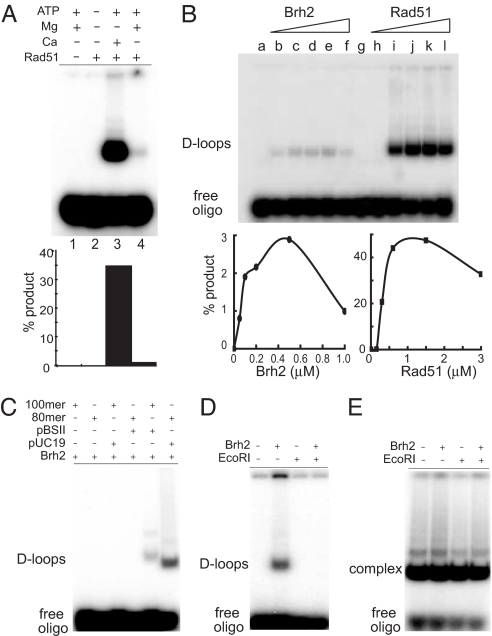
We studied D-loop formation by using superhelical plasmid DNA and radiolabled synthetic oligonucleotide substrates, and we used U. maydis Rad51and/or Brh2 to promote reactions. The Brh2 was purified as a complex with Dss1; in all of the experiments below the complex is referred to simply as Brh2 (11). D-loop formation was monitored by using gel electrophoresis to track the uptake of a single-stranded (ss) oligonucleotide by superhelical DNA. Standard assays used pBluescript II plasmid DNA and a homologous 32P-labeled ss100-mer oligonucleotide. D-loop formation was evident in autoradiograph images by the appearance of a 32P-labeled band with mobility slower than free oligonucleotide but corresponding to that of the superhelical DNA in primarily the monomer form but also to a lesser extent dimer form. D-loop formation catalyzed by Rad51 depended on ATP and a divalent cation, and in accord with findings on the human protein (12) it was strongly stimulated by Ca2+ as compared with Mg2+ (Fig. 1A). Product formation expressed as the percentage of D-loops was taken as the fraction of radiolabel present in the slow migrating complexes.
Fig. 1.
Making and unmaking D-loops (A) D-loop formation by Rad51. Reaction mixtures contained 6 nM 32-labeled ss100-mer, 18 nM plasmid, 750 nM Rad51, and cofactors as indicated (4 mM MgCl2, 4 mM CaCl2, 2 mM ATP). Rad51 was preincubated with 32-labeled ss100-mer, and reactions were processed as described in Materials and Methods. (B) D-loop formation by Brh2 compared with Rad51. Standard reaction mixtures contained 4 mM CaCl2, 2 mM ATP, 6 nM 32P-labeled ss100-mer, 18 nM plasmid DNA, and variable concentrations of Brh2 or Rad51. Reactions were processed as above with protein preincubated with ss100-mer. Brh2, lanes a–f: a, no protein; b, 50 nM; c, 100 nM; d, 200 nM; e, 500 nM; f, 1 μM. Rad51, lanes g–l: g, no protein; h, 100 nM; i, 300 nM; j, 600 nM; k, 1.5 μM; l, 3 μM. (C) Homology requirement in D-loop formation. Reactions contained 4 mM CaCl2, 2 mM ATP, 500 nM Brh2, and the indicated combinations of ss100-mer and pBSII or ss80-mer and pUC19 DNA. (D) Verification of D-loop structure. Standard reactions were run as above except that MgCl2 was substituted for CaCl2. After an initial incubation period of 30 min, EcoRI (10 units) was added as indicated, and incubation continued for an additional 20 min before reactions were deproteinized with SDS and proteinase K. (E) Irreversibly denatured pBSII plasmid DNA was used in an identical set of reactions as in D.
During the course of studying its effects on the Rad51 reaction we discovered that Brh2 alone had an inherent ability to promote formation of DNA complexes that migrated with the same mobility as D-loops formed by Rad51 (Fig. 1B). In reactions containing plasmid DNA and ss100-mer oligonucleotide at a fixed ratio of three molecules of plasmid to one molecule of ss100-mer (a predetermined optimum for the Rad51 reactions), DNA complex formation promoted by Brh2 became apparent when about eight molecules of Brh2 were present per ss100-mer (Fig. 1B, lane b), and increased with additional protein until ≈80 Brh2 molecules per 100-mer were present (Fig. 1B, lane e). By comparison, addition of ≈17 Rad51 monomers per ss100-mer oligonucleotide was insufficient for D-loop formation (molar ratio of one Rad51 monomer per six nucleotides; Fig. 1B, lane h), whereas increasing that level to ≈100 (molar ratio of one Rad51 monomer per nucleotide; Fig. 1B, lane i) resulted in near maximal yield. In contrast to the Rad51-promoted reaction, D-loop formation by Brh2 did not require ATP or divalent cation (data not shown). However, in the subsequent analyses, 4 mM divalent cation (Ca2+ unless otherwise indicated) and 2 mM ATP were included in reactions so that conditions would be uniform by comparison with Rad51. It should be noted that in the well established yeast system Rad51 binding to ssDNA is optimal at a ratio of one monomer per four nucleotides (13, 14). The need for more Rad51 in D-loop formation as observed in the present study is probably caused by low efficiency of filament formation on the oligonucleotide substrate.
That the complexes formed in reactions with Brh2 were indeed joint molecules composed of plasmid DNA paired with 32P-labeled oligonucleotide in the form of D-loops was supported by three lines of evidence. First, the slowly migrating 32P-labeled complexes survived deproteinization by SDS and proteinase K, indicative of a nucleic acid composition rather than protein–nucleic acid aggregate (Fig. 1B). Second, complex formation depended on sequence homology between the 32P-labeled oligomer and cognate plasmid (Fig. 1C). No complexes were evident when plasmid DNA was omitted or when the heterologous plasmid pUC19 was substituted for pBluescript II DNA (pBSII). On the contrary, complexes were formed when an ss80-mer homologous to pUC19 was added to reactions with pUC19 plasmid DNA. These complexes migrated slightly faster than those formed in reactions with pBluescript II and its homologous ss100-mer, consistent with the difference in size of the plasmids. Third, complexes disappeared after a restriction endonuclease was added to cut the plasmid DNA at a site removed from the region homologous to the oligonucleotide sequence (Fig. 1D), a response consistent with the known instability of D-loops resulting from branch migration of a redundant strand in unconstrained DNA (15). In all of these determinations, we used plasmid DNA prepared without a denaturation–renaturation protocol to avoid any irreversible denaturation of the topologically constrained covalently closed circular DNA (16). Unlike the Brh2-dependent formation of complexes noted above in which plasmid DNA was prepared under neutral pH conditions, plasmid DNA deliberately irreversibly denatured by treatment with alkali spontaneously formed complexes with the 32P-labeled oligonucleotide in the absence of Brh2, and the latter did not dissociate upon digestion with restriction endonuclease (Fig. 1E). These results show that the joint molecules formed by Brh2 are in the form of authentic D-loops and are not complexes formed by annealing of the oligonucleotide to ssDNA inadvertently present in the plasmid DNA preparations.
DNA Binding by Brh2.
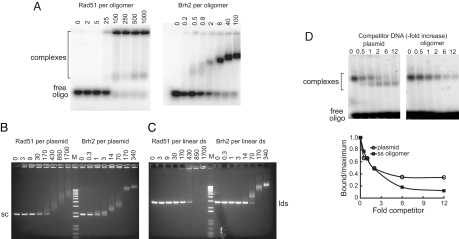
D-loop formation by Brh2 as noted above became apparent at levels of protein that nearly saturated the 100-mer oligonucleotide (eight Brh2 molecules per 100-mer) as determined in a gel mobility-shift assay (compare Figs. 1B and 2A). By visual inspection of an ethidium bromide-stained gel and interpolation from the Brh2 concentrations used, this level of protein also appeared sufficient to tie up almost all of the plasmid DNA molecules in complexes as well (Fig. 2B). About five times more Brh2 was required to generate complexes with the plasmid DNA in the linear double-strand (ds) form (Fig. 2C). With increasing protein there was a correspondingly greater mobility shift in DNA substrate, suggesting formation of protein–DNA complexes with larger and larger mass. For Rad51, at least 10 times more protein with respect to Brh2 was required to saturate each of the three different DNA substrates.
Fig. 2.
Protein–DNA binding. (A) 32P-labeled ss100-mer (6 nM) was incubated with Rad51 (Left) or Brh2 (Right) as indicated, and DNA binding was measured as described in Materials and Methods. (B) Similarly, supercoiled (sc) pBSII plasmid DNA (3.6 nM) was incubated with Rad51 or Brh2 as indicated. (C) Binding was performed as in B but pBSII DNA was precut with BamHI to the linear double-strand form (lds). (D) 32P-labeled ss100-mer (800 nM as nucleotide) was mixed with unlabeled pUC19 plasmid DNA or ss100-mer as competitor (expressed as fold increase of total nucleotide relative to the ss100-mer). Brh2 (≈ 2 molecules per 32P-labeled ss100-mer) was added to start reactions. The ratio of complexes remaining in the presence of competitor (bound) compared with no competitor (maximum) was determined from the intensity of the shifted bands.
Under these conditions (6 nM ss100-mer or 3.2 nM plasmid DNA) it seemed that Brh2 had a slight preference for binding to the superhelical plasmid DNA in comparison to the single-stranded oligomer. At a ratio of Brh2 to plasmid of 14:1, all of the DNA was bound in complexes, whereas with Brh2 to ss100-mer at a ratio of 40:1 some oligonucleotide remained unbound. However, as the DNAs in this analysis were compared on a per-molecule basis, there was a large excess of total nucleotide present in the case of the plasmid DNA. To standardize the comparison we used a competition assay to determine the binding of Brh2 to the 32P-labeled ss100-mer oligonucleotide with unlabeled plasmid or ss100-mer added as competitor on a per-nucleotide basis. By this assay it was evident that there was a slight preference of Brh2 for the ss100-mer over the plasmid DNA (Fig. 2D). Only one type of complex was evident with increasing levels of oligomer competitor, whereas a second complex presumably of lesser mass appeared with increasing plasmid DNA competitor. The basis for this difference is not known.
Order of Addition.
It has been well established that assembly of Rad51 on ssDNA to form an extended presynaptic filament is required for efficient homologous pairing (17). Similarly in the case of the Rad52-promoted D-loop reaction, it was reported that formation of a presynaptic complex with ssDNA was necessary for maximum product formation (18). Conversely, preincubating either Rad51 or Rad52 with duplex DNA effectively poisons any subsequent pairing reaction, presumably from sequestration of the protein on duplex DNA in a form inactive or unable to recycle (13, 18–20).
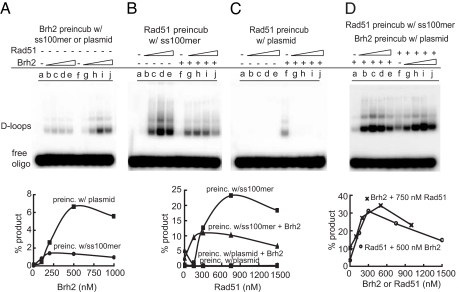
D-loop formation with Brh2 was several-fold more efficient when Brh2 was preincubated with plasmid DNA substrate compared with when it was preincubated with ss100-mer (Fig. 3A). In the Rad51 reaction D-loop formation was nonlinear with increasing Rad51 concentration, requiring a minimum threshold level of protein before any product was formed. At 150 nM Rad51 little D-loop formation was evident (<0.2% product), whereas at 300 nM the level achieved was about half (12% product) the maximal (Fig. 3B, lanes b and c). When Brh2 was present with 150 nM Rad51 during the preincubation period, D-loop formation was stimulated from an undetectable level (<0.2% product) to 10% product formation (Fig. 3B, compare lanes b and g). At higher levels of Rad51 where D-loop formation by Rad51 alone was maximal there was less product formed when Brh2 was present during the preincubation period. Thus, at the level of protein used in this experiment (500 nM Brh2), Brh2 appeared to act in either a stimulatory or inhibitory manner depending on the Rad51 concentration.
Fig. 3.
D-loop reaction parameters. (A) Brh2 was preincubated for 15 min with either 6 nM ss100-mer or 18 nM pBSII plasmid DNA. Reactions were then started by addition of the cognate DNA. Brh2 in lanes a-j: a and f, no protein; b and g, 100 nM; c and h, 200 nM; d and i, 500 nM; e and j, 1 μM. (B) Rad51 was preincubated for 15 min with 6 nM ss100-mer in the presence or absence of 500 nM Brh2 as indicated. Reactions were started by addition of plasmid DNA. Rad51 in lanes a-j: a and f, no protein; b and g, 150 nM; c and h, 300 nM; d and i, 750 nM; e and j, 1,500 nM. (C) Rad51 was preincubated for 15 min with18 nM plasmid DNA in the presence or absence of 500 nM Brh2 as indicated. Reactions were started by addition of ss100-mer. Rad51 in lanes a-j: a and f, no protein; b and g, 150 nM; c and h, 300 nM; d and i, 750 nM; e and j, 1,500 nM). (D) Rad51 at increasing concentrations [lanes a, no Rad51; b, 150 nM; c, 300 nM; d, 750 nM; e, 1,500 nM] was preincubated with 6 nM 32P-labeled ss100-mer for 15 min. At the same time Brh2 at a fixed concentration of 500 nM was preincubated separately for 15 min with 18 nM plasmid pBSII DNA. Reactions were started by combining the two mixes and allowing incubation to continue for 30 min. In lanes f—j, Rad51 at a fixed concentration of 750 nM was preincubated with 6 nM 32P-labeled ss100-mer for 15 min. At the same time Brh2 at increasing concentrations [lanes f, no Brh2; g, 100 nM; h, 200 nM; i, 500 nM; j, 1,000 nM] was preincubated separately for 15 min with 18 nM plasmid pBSII DNA. Reactions were started by combining the two mixes and incubation continued for 30 min.
Preincubation of Rad51 with plasmid DNA absolutely quenched the D-loop reaction (Fig. 3C). Preincubation of Brh2 with Rad51 on plasmid DNA was unable to mitigate the plasmid DNA-induced inactivation of Rad51. In addition, the Brh2-promoted reaction was effectively killed when Rad51 was present (Fig. 3C). The only D-loop formation evident when preincubation with plasmid DNA was performed was that promoted by Brh2 in the absence of any Rad51 (Fig. 3C, lane f). In contrast, D-loop formation was stimulated to maximal levels (≈35% product) when Rad51 was preincubated with ss100-mer and Brh2 was preincubated separately with plasmid DNA before combining all of the components (Fig. 3D). These results suggest that Rad51/ss100-mer filaments react better with plasmid DNA when the latter has independently formed complexes with Brh2. Perhaps Brh2 binding to the plasmid alters the DNA structure, making it more receptive for the incoming oligonucleotide, possibly by opening the duplex. Pairing of the invading oligomer with its complementary sequence within the plasmid DNA might then be enhanced as a consequence of the annealing activity inherent in Brh2.
Role of BRC Element in D-Loop Formation.
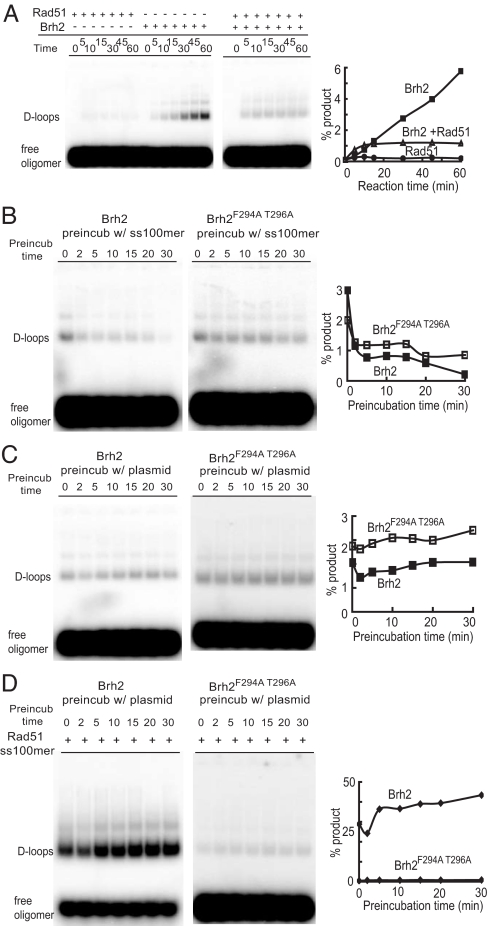
Rad51 added directly to reaction mixes containing both ss100mer and plasmid DNA substrates without a preincubation step was also completely ineffective in making D-loops (Fig. 4A). On the other hand, Brh2 added directly to mixes of ss100-mer and plasmid DNA promoted a slow, but significant, D-loop reaction with increasing product formed over time. When Rad51 and Brh2 were added together to such mixes the initial rate of D-loop formation was slightly faster than that by Brh2 alone, but the yield was reduced. In the absence of Rad51, Brh2 promoted a low level of D-loop formation regardless of whether it was first preincubated with ss100-mer or plasmid DNA (Fig. 4 B and C). The yield of product remained nearly constant over the entire period of preincubation of Brh2 with plasmid DNA, but diminished over time with ss100-mer. It is possible that Brh2 is slowly inactivated or becomes unstable in the presence of ssDNA.
Fig. 4.
Brh2 stimulation of Rad51 requires BRC element. (A) Rad51 (750 nM) or Brh2 (500 nM) or both was added without preincubation directly to standard reactions containing 6 nM ss100-mer and 18 nM pBSII plasmid DNA. Samples were removed at the indicated times. (B) Brh2 or Brh2F294A T296A mutant protein was preincubated with 6 nM 32P-labeled ss100-mer for the indicated times. Reactions were started by addition of 18 nM plasmid pBSII DNA and incubation continued for 30 min. (C) Brh2 or Brh2F294A T296A mutant protein was preincubated with 18 nM plasmid pBSII DNA for the indicated times. Reactions were started by addition of 6 nM 32P-labeled ss100-mer and incubation continued for 30 min. (D) Rad51 (750 nM) was preincubated with 6 nM 32P-labeled ss100-mer for 15 min. Brh2 protein or Brh2F294A T296A mutant protein was preincubated with 18 nM plasmid pBSII DNA for the indicated times (min). Reactions were started by combining the two mixtures and incubation continued for 30 min.
As noted above maximal D-loop reaction was observed when preformed Rad51/ss100-mer filaments were mixed with preformed Brh2/plasmid DNA complexes (Fig. 3D). When the Rad51 reaction was started by adding naked plasmid DNA to preformed Rad51/ss100-mer filaments, product formation was ≈10% (Fig. 3D, lane f), but when the Rad51 reaction was started by adding plasmid DNA premixed with Brh2 the yield of product jumped ≈4-fold (Fig. 3D, lane i). This competence of Brh2/plasmid DNA complexes in stimulating D-loop formation by preformed Rad51 filaments increased as the time of preincubating Brh2 with plasmid DNA was extended (Fig. 4D). When Brh2 was mixed with plasmid DNA and the mixture was added immediately to Rad51/ss100-mer filaments, product formation was ≈25%. However, when the period of preincubating Brh2 with plasmid DNA was extended over the course of 30 min the yield of product climbed to ≈40%. This increase in competence of the plasmid DNA to form D-loops when preincubated with Brh2 reinforces the notion that Brh2 induces a change in the plasmid DNA structure that makes it more favorable for pairing with the Rad51 filament.
Stimulation of the Rad51 reaction by Brh2 could also be caused in part by other reasons, including stabilization of the D-loop product through preferential binding by Brh2 (11) and/or enhancement of the reaction by physical interaction with Rad51. Because the BRC element mediates a well defined interaction with Rad51 via its polymerization interface (21, 22), we examined a mutant form of Brh2 with key residues in the BRC changed (F294A T296A) to abrogate interaction with Rad51 (6). Like the wild-type version, this Brh2F294A T296A mutant protein retained the autonomous D-loop forming activity regardless of whether it was preincubated on ss100-mer or plasmid DNA (Fig. 4 B and C). Thus, the inherent D-loop forming activity of Brh2 protein does not require a functional BRC element. However, the strong Rad51-dependent D-loop reaction observed when preformed Rad51 filaments were added to Brh2-plasmid DNA complexes was absent (Fig. 4D). Only the basal level of Brh2-promoted D-loops was evident. These results suggest that direct protein-mediated interaction between Rad51 filaments and Brh2-plasmid complexes is required for maximal D-loop formation when both proteins are present in reactions. It has been noted by others that BRC repeats from human BRCA2 can associate with Rad51 nucleoprotein filaments in both disruptive and nondisruptive modes, possibly providing some remodeling function (23–26). Our findings here suggest there could be an additional and more direct role of the BRC element in contributing to homologous pairing and joint molecule formation.
Discussion
There are three principal points from this study. First, Brh2 promotes uptake of a single-stranded oligonucleotide by a homologous superhelical plasmid DNA to form D-loops. This reaction differs in notable ways from that promoted by Rad51. There is no ATP requirement, nor is there any evidence to suggest that the D-loop reaction proceeds after formation of a nucleoprotein filament on ssDNA. Second, Brh2 is active in D-loop formation under conditions in which Rad51 is virtually dead. Indeed, the reaction proceeds quite well when Brh2 is mixed first with duplex DNA, a condition that effectually poisons Rad51. Third, Brh2 stimulates D-loop formation by acting in trans when Rad51 filaments formed on ssDNA are mixed with Brh2 bound to duplex DNA. Here, the stimulation depends on a functional BRC element, suggesting that Brh2 lures the Rad51 filament to the duplex through physical association. These findings suggest that the scope of Brh2's capabilities is wider than previously considered, but at the same time anchor Brh2 more firmly within the class of proteins that mediate nucleoprotein filament formation.
The emerging view of the initiating step in recombinational repair is recognition of sequence homology coupled with DNA strand exchange (27, 28). Rad51 in eukarya (29), RecA in eubacteria (30), and RadA in archaea (31) promote this process to achieve strand exchange over hundreds of base pairs. The nucleoprotein filament formed by the Rad51/RecA/RadA family proteins polymerizing on ssDNA becomes a molecular machine that catalyzes DNA strand exchange. The filament is conserved in structure and function across the domains of life, exemplifying the universal importance of its role in recombination (32). BRCA2 provides crucial activity for regulating the Rad51 filament of many, although not all, eukaryotes. As demonstrated with the human BRCA2, the medially located BRC elements exert a filament-destabilizing effect (23), whereas the unrelated C-terminal Rad51-interacting element protects the filament against disassembly through specific association with the polymerized Rad51 (3, 4). Balance between these opposing influences is required to ensure appropriate filament status and properly executed recombination. In prokaryotes RecX and DinI act as competing modulators to provide a functionally similar regulatory ability in governing dynamic equilibrium of the RecA filament (33). RecX promotes dissociation of the filament (34), whereas DinI stabilizes it (35).
Nucleation of the nascent Rad51 filament on RPA-coated ssDNA is another crucial function in the repertoire of regulatory activities provided by BRCA2 as demonstrated with Brh2 in a reconstituted in vitro system (10). The functionally equivalent activity in prokaryotes is provided by the RecFOR proteins, which act in concert to mediate RecA filament loading on (SSB)-coated DNA (36). The RecOR complex recognizes the ds/ssDNA junction on gapped DNA, loads RecA, and stabilizes the RecA filament (37). These observations suggest that regulation of filament dynamics is a fundamentally conserved theme in the governance of recombination, but that the means for establishing and maintaining the filament differ between eukaryotes and prokaryotes and seem to have evolved differently. Understanding the basis of these mechanisms is then central to revealing a precise picture of how recombination is regulated in eukaryotes versus prokaryotes.
Comparison of Brh2 with RecO reveals some intriguing similarities. While there is no relationship evident by sequence alignment, the architecture of the DNA-binding domain of each includes the OB fold (5, 38). Both proteins have been demonstrated to facilitate annealing of complementary ssDNAs complexed with their cognate single-strand DNA-binding protein, RPA or ssDNA-binding protein (SSB), respectively (11, 39), and in light of the findings presented in this current study, it appears that Brh2, like RecO protein (40), can promote D-loop formation in an ATP-independent manner. These DNA strand-pairing activities are also exhibited by Rad52 (18, 41, 42), which serves as the preeminent Rad51 mediator in yeast (43), and by Hop2 (44), which mediates Rad51 and Dmc1 filament formation in meiosis of yeast and higher organisms. Hop2 in heterodimeric complex with Mnd1 (45) stimulates Rad51 and Dmc1 homologous pairing reactions (46), promoting filament-directed capture of the duplex through the DNA-binding action of Hop2 (47, 48). This activity appears to be similar to the protein-mediated interaction between Rad51 filaments and Brh2-plasmid complexes that we observed in the present study. Analogy with Brh2 extends to the cryptic prophage-encoded RecT of Escherichia coli and bacteriophage lambda β protein, which are distantly related to Rad52 (49), exhibit similar homologous pairing capabilities (50, 51), and augment RecA-mediated reactions in vivo under some circumstances (52–55).
When highly overexpressed, RecO can partially compensate for the lack of RecA in promoting survival of E. coli after UV irradiation (40). However, compensation in activity appears to be the exception rather than the rule among the proteins in the mediator class. Certainly in the case of U. maydis there is no evidence to suggest redundancy in function between Brh2 and Rad51 despite the common ability of these proteins to promote D-loop formation. Expression of Brh2 from a strong promoter does not substitute for Rad51 in restoring DNA repair proficiency (56). Thus, the ability of Brh2 and the collection of genetically diverse proteins cited above to promote D-loop formation should not be taken as a touchstone meaning that they play a biological role equivalent to Rad51 (or RecA). Rather, it suggests that they have an undiscovered potential for roles beyond those previously considered. One possibility of a role for Brh2 might be as a trigger to initiate homologous pairing within the nuclear environment in which the conformation of DNA is largely double-stranded, a condition that appears to be extremely adverse for Rad51 activity. That Brh2 can promote D-loop formation in reactions without artificial ordering of the addition of components or with the preponderance of the DNA present in duplex form suggests an ability to initiate pairing reactions that is lacking in Rad51.
Materials and Methods
Reagents.
Brh2 in complex with Dss1 and Rad51 proteins were purified after overexpression in E. coli as described (11). Brh2F294A T296A protein with the indicated residues altered by site-directed mutagenesis (6) was prepared in the same manner. Oligonucleotides were synthesized and gel purified by Integrated DNA Technologies. Oligonucleotide sequences are based on plasmid pBluescript II SK+ DNA residues 2–101 or pUC19 DNA residues 142–221. Plasmid DNAs were purified without an alkaline denaturation step by a slight modification of a classic procedure (57). Briefly, cells converted to spheroplasts by digestion with lysozyme were lysed with sarkosyl and spun in an ultracentrifuge to produce a clear, nonviscous supernatant. This supernatant was extracted with phenol and treated with RNase, and the DNA was purified by chromatography using Qiagen proprietary resin. After elution with ethanolic saline (55% ethanol, 0.1 M NaCl) and concentration, the DNA was further purified by velocity sedimentation on a preparative scale in a neutral pH sucrose gradient. To denature covalently closed circular DNA irreversibly, samples were made alkaline by the addition of 0.5 M NaOH. After 15 min at room temperature an equivalent amount of HCl was added to reneutralize the sample. All DNA concentrations are expressed as moles of molecules rather than nucleotide, unless indicated otherwise. Oligonucleotides were 5′ end-labeled by using [γ-32P]ATP and T4 polynucleotide kinase.
D-Loop Reactions.
For standard D-loop reactions with Rad51, 6 nM 32P-labeled 100-mer was preincubated with protein at 37°C in a 15-μl reaction in a buffer containing 25 mM Tris·HCl (pH 7.5), 20 mM KCl, 1 mM DTT, 4 mM CaCl2, and 2 mM ATP. After 15 min reactions were started by the addition of 18 nM plasmid DNA, and incubation continued for an additional 30 min. D-loop reaction conditions with Brh2 were the same except that preincubation with ss100-mer was not performed unless otherwise indicated. Reactions were stopped by the addition of SDS and proteinase K to 1.2% and 1.7 mg/ml, respectively, and an additional incubation for 30 min. EDTA (20 mM) and loading dye were added, and DNA components were resolved by electrophoresis on 1% agarose gels. Dried gels were exposed to phosphor storage screens (Molecular Dynamics) and processed with a Typhoon 9400 PhosphorImager (Amersham Biosciences). Relative amounts of substrate and product bands were determined with ImagequaNT software (Molecular Dynamics). Percentage of D-loops indicates the fraction of radiolabeled oligonucleotide associated with the slow migrating D-loop band.
DNA Binding.
32P-labeled 100-mer at 6 nM (or 3.6 nM unlabeled plasmid DNA) was incubated with either Rad51 or Brh2 for 10 min at 37°C in buffer containing 25 mM Hepes, 1 mM DTT (with 4 mM CaCl2 and 2 mM ATP in the case of Rad51). Glutaraldehyde was then added to a final concentration of 0.2%, incubation continued for 10 min to cross-link protein–DNA complexes, and Tris·HCl (pH 8) was added to a final concentration of 120 mM to quench the cross-linking reaction. Products were resolved by electrophoresis on 1% agarose gels as above and visualized by phosporimaging or staining with ethidium bromide as required.
ACKNOWLEDGMENTS.
We thank Dr. Lorraine Symington of Columbia University for critical comments. This work was supported by National Institutes of Health Grants GM42482 and GM79859.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 3.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 5.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, et al. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol Cell Biol. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 8.Kojic M, Zhou Q, Lisby M, Holloman WK. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol Cell Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 11.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 14.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 15.Radding CM, Beattie KL, Holloman WK, Wiegand RC. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977;116:825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- 16.Vinograd J, Lebowitz J, Radloff R, Watson R, Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci USA. 1965;53:1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 18.Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Homologous pairing promoted by the human Rad52 protein. J Biol Chem. 2001;276:35201–35208. doi: 10.1074/jbc.M104938200. [DOI] [PubMed] [Google Scholar]
- 19.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 20.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51–DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst) 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 23.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 24.Galkin VE, et al. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivji MK, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J Mol Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Folta-Stogniew E, O'Malley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Gupta RC, Folta-Stogniew E, O'Malley S, Takahashi M, Radding CM. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- 29.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 30.Cox MM. The bacterial RecA protein as a motor protein. Annu Rev Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 31.Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata T, et al. Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: A possible advantage of DNA over RNA as genomic material. Proc Natl Acad Sci USA. 2001;98:8425–8432. doi: 10.1073/pnas.111005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lusetti SL, Drees JC, Stohl EA, Seifert HS, Cox MM. The DinI and RecX proteins are competing modulators of RecA function. J Biol Chem. 2004;279:55073–55079. doi: 10.1074/jbc.M410371200. [DOI] [PubMed] [Google Scholar]
- 34.Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 35.Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD, Cox MM. The DinI protein stabilizes RecA protein filaments. J Biol Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- 36.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 37.Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 2001;20:7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmins J, Leiros I, McSweeney S. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 2007;26:3260–3271. doi: 10.1038/sj.emboj.7601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc Natl Acad Sci USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luisi-DeLuca C. Homologous pairing of single-stranded DNA, superhelical double-stranded DNA catalyzed by RecO protein from Escherichia coli. J Bacteriol. 1995;177:566–572. doi: 10.1128/jb.177.3.566-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi B, Rybalchenko N, Golub EI, Radding CM. Human and yeast Rad52 proteins promote DNA strand exchange. Proc Natl Acad Sci USA. 2004;101:9568–9572. doi: 10.1073/pnas.0403205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung P, Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 44.Petukhova GV, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 45.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YK, et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc Natl Acad Sci USA. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory actioon of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta ERF, RAD52. BMC Genomics. 2002;3:8–18. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 51.Rybalchenko N, Golub EI, Bi B, Radding CM. Strand invasion promoted by recombination protein beta of coliphage lambda. Proc Natl Acad Sci USA. 2004;101:17056–17060. doi: 10.1073/pnas.0408046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 53.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poteete AR, Fenton AC. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages lambda and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clyman J, Belfort M. Trans and cis requirements for intron mobility in a prokaryotic system. Genes Dev. 1992;6:1269–1279. doi: 10.1101/gad.6.7.1269. [DOI] [PubMed] [Google Scholar]
- 56.Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cunningham RP, DasGupta C, Shibata T, Radding CM. Homologous pairing in genetic recombination: recA protein makes joint molecules of gapped circular DNA, closed circular DNA. Cell. 1980;20:223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]