Abstract
Reliable predictive rules that relate protein sequence to structure would facilitate postgenome predictive biology and the engineering and de novo design of peptides and proteins. Through a combination of experiment and analysis of the protein data bank (PDB), we have deciphered and rationalized new rules for helix–helix interfaces of a common protein-folding and association motif, the antiparallel dimeric coiled coil. These interfaces are defined by a specific pattern of interactions among largely hydrophobic side chains often referred to as knobs-into-holes (KIH) packing: a knob from one helix inserts into a hole formed by four residues on the partner. Previous work has focused on lateral interactions within the KIH motif, for example, between an a position on one helix and a d′ position on the other in an antiparallel coiled coil. We show that vertical interactions within the KIH motif, such as a′-a–a′, are energetically important as well. The experimental and database analyses concur regarding preferred vertical combinations, which can be rationalized as leading to favorable side-chain interactions that we call constellations. The findings presented here highlight an unanticipated level of complexity in coiled-coil interactions, and our analysis of a few specific constellations illustrates a general, multipronged approach to addressing this complexity.
Keywords: backbone thioester exchange, knobs-into-holes packing, peptides, protein design, protein folding
The α-helical coiled coil is a ubiquitous oligomerization domain found in a wide variety of proteins (1, 2). Coiled-coil interactions direct and stabilize the multimerization of transcription factors, motor proteins, cytoskeleton components, extracellular matrix components, and membrane-fusion machinery (3, 4). It has been estimated that 3–5% of translated protein sequences encode coiled-coil domains (5); indeed, we find that, of the current 46,968 entries in the protein databank (PDB), 2,738 (5.8%) contain coiled-coil interactions of some type (O.D.T. and D.N.W., unpublished data). Thus, deciphering rules that link the sequences of coiled coils to their assembly would have an impact on many aspects of postgenome predictive biology (6) and aid our ability to engineer or even design protein–protein interactions (1, 7, 8).
The effects of sequence variation on coiled-coil stability have been widely studied (9–16), but substantial gaps in our understanding remain. For example, the vast majority of prior efforts have focused on parallel coiled-coil homo dimers, but other coiled-coil states are seen in Nature. Oligomers ranging from trimer through dodecamer have been observed, as have structures with different types (hetero-oligomers) or arrangements (parallel vs. antiparallel) of helices (3, 13, 17). In particular, antiparallel coiled-coil dimers have received little attention despite increasing recognition of their structural and functional importance both as stand-alone motifs and as components of globular protein architectures. In the PDB, for example, we find 1,273 coiled-coil structures with ≤70% sequence identity; of these, 800 (63%) are antiparallel dimeric coiled coils (O.D.T. and D.N.W., unpublished data). Among this subset, 661 (83%) are intramolecular and the remainder are intermolecular helix associations (13, 18). Even among the widely studied parallel coiled coils, only a limited set of factors has been shown to influence stability (1–4), and it seems likely that important determinants of stability and selectivity remain to be identified.
Here, we show that largely overlooked patterns of side-chain packing at the interhelical interface contribute to antiparallel coiled-coil dimerization selectivity. We use a unique experimental approach to evaluate the energetic significance of certain side-chain juxtapositions that have not been examined. We show how seemingly subtle side-chain variations can lead to surprising levels of dimerization preference. Furthermore, we demonstrate that findings from the model system reflect preferences among coiled-coil proteins of known three-dimensional structure.
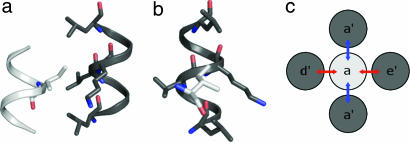
As is true for all coiled coils, the formation of antiparallel coiled-coil dimers is driven by the burial of hydrophobic side chains arranged along one side of each participating α-helix. The helices typically display a “heptad repeat” of amino acid residues, with the heptad positions designated abcdefg. Hydrophobic side chains at a and d align to create the dimerization interface. Two or more such interfaces come together through the interdigitation of side chains, which Crick described as “knobs-into-holes packing” (KIH) (19). As illustrated in Fig. 1, each buried a or d side chain (a “knob”) has a local packing environment (a “hole”) that is defined by four side chains from the partner. We divide the four interhelical side-chain–side-chain contacts into two types (Fig. 1): lateral interactions, for which a line between the two side chains is approximately perpendicular to the helix axis; and vertical interactions, for which a line between the two side chains is approximately parallel to the helix axis.
Fig. 1.
Knobs-into-hole packing at antiparallel coiled-coil interfaces. (a and b) Orthogonal views of a knob (light gray) into hole (dark gray) interaction observed in an experimentally determined protein structure (residues 411 and 58–65 of PDB ID code 2ic6). (c) Diagram illustration of b with the heptad register assignment superimposed. Lateral interactions of the knob residue are indicated with red arrows, and vertical interactions with blue arrows.
Most prior analyses of a or d side-chain interactions at the interface have focused on lateral interactions with a′ or d′ partners (the prime indicates that the residue resides in a different helix). Extensive work by Vinson et al., for example, has shown that the contribution of a given buried a side chain to parallel coiled-coil dimer stability depends on the identity of the lateral hydrophobic packing partner (a-a′ interaction) (9, 12). We have recently shown that lateral partners (a-d′) influence antiparallel coiled-coil stability (20). To our knowledge, however, the contribution of vertical side-chain contacts to coiled-coil stability and pairing selectivity has not been analyzed in detail for either parallel or antiparallel coiled coils (21). Indeed, given the amount of work necessary for previous studies of lateral pairing, the prospect of extending such an effort to examine vertical contacts might seem operationally daunting (9–16).
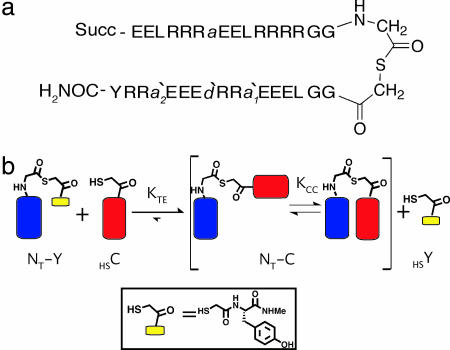
We have recently developed an experimental design that facilitates the rapid study of sequence–stability relationships among antiparallel coiled coils (20, 22–24). Our model system (NT-C; Fig. 2) consists of two short α-helix-prone segments connected with a flexible linker that allows, but does not enforce, intramolecular coiled-coil association. The linking segment contains a central thioester bond to allow thiol-thioester exchange. When NT-C is combined with a small thiol-containing molecule in aqueous solution at neutral pH, thiol-thioester exchange occurs rapidly. The equilibrium constant (KTE) provides insight on the noncovalent attraction between the two α-helical segments (Fig. 2b) (20, 22). This system is well suited to exploring how the interplay between mutations within each helix influences antiparallel coiled-coil stability. As illustrated in Fig. 3, our original study focused on a lateral (a-d′) pair at the interface: an a position on the N-terminal helix, and a d′ position on the C-terminal helix (20). Synthesis of 10 short peptides (5 alternative residues at each guest site) enabled us to obtain ΔGTE values (from KTE) for 25 coiled-coil variants.
Fig. 2.
Thioester model system. (a) Design and sequence of NT-C; Succ = N-terminal succinyl group. Residues a/d′/a′ correspond to mutations sites. (b) Thioester exchange process for NT-C. The thioester-thiol pair on the left comprises N- (blue) and C-terminal (red) segments, whereas the pair on the right contains the full-length coiled coil and a small thiol.
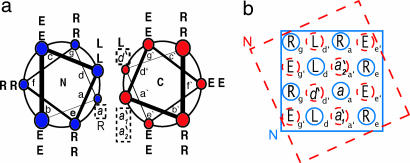
Fig. 3.
Coiled-coil interactions in thioester model system. (a) Helical-wheel diagram showing the helical regions of NT-C. (b) Partial helical net for NT-C. In each diagram, the N-terminal segment is shown in blue and the C-terminal segment is shown in red. Note that the numbering of the mutation sites is different from that in ref. 20.
Here, we build on this experimental design to show that: (i) certain combinations of residues in the vertical a′-a–a′ constellations are favored over others; (ii) vertical a′-a–a′ constellations contribute significantly to stability and specificity in antiparallel coiled-coil dimers; (iii) our findings are robust and not system-dependent; (iv) the preferred combinations are mirrored in coiled coils of known three-dimensional structure; and (v) there is an energetically significant interplay among lateral and vertical interactions.
Vertical Contacts Affect Antiparallel Coiled-Coil Specificity and Stability.
The vertical contact residues for the guest site at a in our model system are designated a1′ and a2′; Figs. 2 and 3). In our original study a1′ = a2′ = Leu (20). To evaluate the importance of vertical contacts, we changed a1′ and a2′ to Ile for the 25 a–d′ variants considered previously (Leu, Ile, Val, Ala, or Asn at a or d′). As shown in Table 1, switching between a1′ = a2′ = Leu and a1′ = a2′ = Ile causes a significant change in stability for a number of lateral a–d′ pairings. For example, the most stable lateral pairing changes from Ile-Leu when a1′ = a2′ = Leu to Leu-Leu when a1′ = a2′ = Ile. Interpretation of these differences, however, is complicated by the fact that Leu → Ile mutations at a1′ and a2′ lead to multiple changes in lateral and vertical contacts at the antiparallel coiled-coil interface.
Table 1.
Thermodynamic data obtained from thioester exchange of NT-C mutants
| ΔGCC | d′ | |||||
|---|---|---|---|---|---|---|
| Leu | Ile | Val | Asn | Ala | ||
| a′ = a′ = Leu | ||||||
| a Leu | −1.4 | −1.3 | −0.9 | 0.2 | −0.4 | |
| Ile | −1.7 | −1.0 | −0.8 | −0.1 | −0.9 | |
| Val | −1.4 | −0.8 | −0.6 | 0.0 | −0.9 | |
| Asn | 0.3 | 0.2 | 0.4 | 0.5 | 0.8 | |
| Ala | −0.7 | −0.7 | −0.5 | 0.4 | 0.0 | |
| a′ = a′ = Ile | ||||||
| a Leu | −2.1 | −1.9 | −1.5 | 0.0 | −0.7 | |
| Ile | −1.6 | −1.4 | −0.7 | −0.1 | −0.5 | |
| Val | −1.6 | −1.2 | −0.8 | 0.2 | −0.3 | |
| Asn | 0.2 | 0.1 | 0.3 | 0.3 | 0.3 | |
| Ala | −0.7 | −0.6 | −0.4 | 0.4 | 0.0 | |
Values are reported in kilocalories per mole. Uncertainty ≈ ±0.1 kcal/mol.
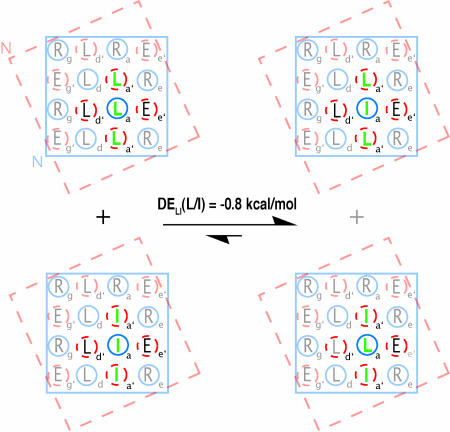
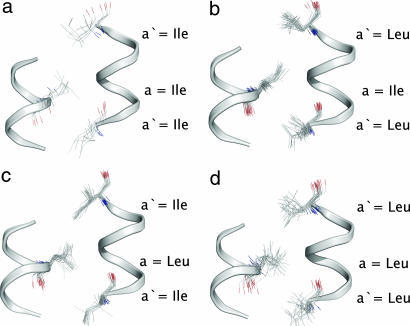
To isolate the energetic contribution to coiled-coil pairing preference of vertical contacts within the a2′-a-a2′ triad, we can consider hypothetical equilibria such as the one illustrated in Fig. 4. These partial helix net diagrams represent the original NT-C design and three mutants thereof. In each of these thioesters d′ = Leu, whereas positions a, a1′, and a2′ vary between Leu and Ile, with a1′ = a2′. The figure explicitly indicates the entire hole into which each a, a1′, or a2′ knob inserts on intramolecular coiled-coil formation. The only difference between the pair of coiled coils on each side of the equilibrium arrows is the identity of the vertical contacts within each a2′-a-aa2′ triad (Leu-Leu-Leu and Ile-Ile-Ile “homo-triads” on the left, Leu-Ile-Leu and Ile-Leu-Ile “hetero-triads” on the right).
Fig. 4.
Partial helical-net diagrams for NT-C and three mutants used to calculate the discrimination energy (DE). The reported DE value was derived from the thermodynamic information in Table 1.
ΔGTE values for each of the four coiled coils in Fig. 4 indicate ΔΔG = −0.8 kcal/mol for this equilibrium. Below, we use the term “discrimination energy” (DE) to describe this type of thermodynamic comparison, because DE tells us about the extent to which one pattern of interhelical pairing is favored relative to another. The result of the equilibrium in Fig. 4 is designated DELI because we are comparing the impact of the vertical environments provided by a1′ = a2′ = Leu vs. a1′ = a2′ = Ile. More specifically, we can designate this result as DELI(L/I) to indicate that the left side has a = Leu in the Leu-a-Leu vertical triad, and a = Ile in the Ile-a-Ile vertical triad. The deduced DELI(L/I) value indicates that the sum of vertical contacts in the Leu-Ile-Leu plus Ile-Leu-Ile vertical hetero-triads is 0.8 kcal/mol more favorable than the sum of vertical contacts in the Leu-Leu-Leu plus Ile-Ile-Ile vertical homo-triads.
Testing the Ile/Leu Triads in an Independent System.
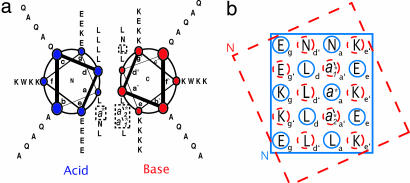
The information gleaned from our thioester-based model system will not be useful unless it is broadly applicable to antiparallel coiled-coil dimers. We turned our attention to a different system to test generality. Very few heterodimeric antiparallel coiled coils have been subjected to careful biophysical scrutiny (18); among the best-characterized examples is a design generated by Oakley et al. (25) This system consists of negatively charged (“acid”) and positively charged (“base”) partners. The hydrophobic core comprises primarily leucine residues, with Asn at a single a position on each component to control relative helix orientation. The Oakley system is illustrated in Fig. 5, which identifies the sites we used for substitutions. The vertical triad on which we focus comprises the residues designated a*, a1′*, and a2′* in Fig. 5; in the original sequence, all three of these residues are Leu. This system should be sufficiently different from our thioester system to represent a valid test for generality because the Oakley helices are twice as long as the helical segments in NT-C, and the residues at the b, c, and e–g positions vary significantly between the two systems. A key similarity between the two model systems is a leucine-rich hydrophobic interface, which enables the comparison of specific interfacial mutations in these different antiparallel coiled-coil contexts.
Fig. 5.
Oakley model system. (a) Helical-wheel diagram showing the helical regions of Oakley's heterodimeric antiparallel coiled coil (25). (b) Partial helical net for the peptides shown in a. In each diagram, the “acid” peptide is shown in blue and the “base” peptide is shown in red. The boxes highlight interactions discussed in the text.
In the Oakley heterodimer, the side chain from residue a*, an a position on the acid peptide, fits into a hole on the base peptide defined by one lysine and three leucine residues (which include a1′* and a2′*). We prepared the a* = Ile mutant in addition to the original acid peptide. Two base peptides were prepared, the original sequence (a1′* = a2′* = Leu) and a double mutant (a1′* = a2′* = Ile), so that we could change the vertical contact residues for a*. We examined all four acid–base peptide pairings, that is, the coiled coils that contain all four possible vertical triads among Leu-Leu-Leu (the original Oakley design), Ile-Ile-Ile, Leu-Ile-Leu, and Ile-Leu-Ile. Analytical ultracentrifugation showed that each acid–base pair is dimeric [see supporting information (SI) Text].
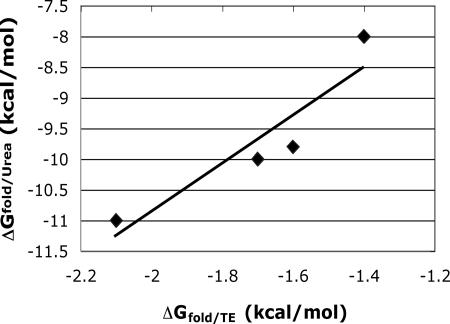
Urea-denaturation experiments revealed significant differences in the four coiled-coil stabilities (Table 2). There is a reasonable correlation between ΔGUrea for the four acid–base peptide pairs and ΔGTE for the analogous members of the thioester system (Fig. 6), which is noteworthy in light of the substantial differences between the two coiled-coil systems. (These differences include variation in the magnitude of ΔG for coiled-coil formation: −8 to −11 kcal/mol for the Oakley system vs. −1.4 to −2.1 for the thioester system.) The four ΔGUrea values can be used to calculate DELI(L/I) = −3.2 kcal/mol for the Oakley system. In a complementary study, we prepared derivatives of the two acid and two base peptides bearing N- or C-terminal cysteine residues, respectively, to allow direct determination of the discrimination energy under native conditions by disulfide exchange (26–28). This approach indicated DELI(L/I) = −2.6 kcal/mol, in good agreement with the value deduced from the unlinked species. Overall, the experiments based on the Oakley antiparallel coiled coil are consistent with the thioester experiments in revealing that vertical hetero-triads containing Leu and Ile are preferred relative to the homo-triads. The quantitative differences between the two systems suggest that the thioester approach underestimates vertical contact energetics, perhaps because in this case the a2′ position is near the “open” end of the helical hairpin and thus may be subject to fraying (29), which could reduce the measured free energies.
Table 2.
Comparison of thermodynamic data obtained for Oakley acid/base mutants and from thioester exchange of the corresponding NT-C mutants
| a/a1,2′ | NT-C |
Oakley heterodimeric coiled-coil |
|
|---|---|---|---|
| ΔGfold/TE, kcal/mol | ΔGfold/Urea, kcal/mol | Tm, °C | |
| L/L | −1.4 | −8.0 | 45 |
| I/I | −1.6 | −9.8 | 53 |
| I/L | −1.7 | −10.0 | 55 |
| L/I | −2.1 | −11.0 | 58 |
Fig. 6.
Correlation of ΔGfold/Urea for the Oakley acid–base mutations and ΔGfold/TE for the corresponding mutations in NT-C. The line corresponds to a linear regression fit (y = 3.9321x − 3.0308, r2 = 0.855).
Analysis of Antiparallel Coiled-Coil Structures from the PDB.
The energetic importance of vertical contacts in two antiparallel coiled-coil model systems led us to ask whether this pattern of side-chain interactions plays a role in natural coiled coils. We used the program SOCKET, which searches protein-structure coordinate files for KIH interactions characteristic of coiled-coil structures, and the SOCKET-derived database of structurally verified coiled coils, CC+ (13). Recently, we created an interface to search CC+ for various coiled-coil parameters, such as oligomer state and helix orientation, and also for more specific features, such as sequence motifs (O.D.T. and D.N.W., unpublished data). By using these resources, we searched the PDB for examples of antiparallel two-helix coiled-coil structures with 70% sequence identity or less. From this subset we culled examples of vertical residue triads (a′-a-a′) in which a was Leu or Ile and the residues at a′ were either both Ile or both Leu; these vertical triads allow comparison with our experimental studies. Defining the residues at each site in the a′-a–a′ vertical triads limited the number of coiled-coil substructures returned in our searches: there were 2,072 KIH interactions for all types of a′-a–a′ vertical triads in the subset of antiparallel two-helix coiled-coil structures, but only 162 that matched our constraints. Nonetheless, the number of structures was sufficient to perform reliable analyses and draw conclusions.
Table 3 gives the raw counts for each of the residues at the central a position in the different vertical backgrounds, a′ = a′ = Ile or a′ = a′ = Leu. Expected numbers for each combination were calculated based on the frequencies of these residues at the a sites of all KIH interactions in the coiled-coil database with sequences ≤70% identical. The ratio of the observed/expected numbers is effectively a propensity for a side chain to be in the specified environment. Gratifyingly, the hetero-triads have favorable propensities (>1) and the homo-triads are disfavored, which is consistent with the model system results. Furthermore, by analogy with the theoretical equilibrium shown in Fig. 4, the data of Table 3 can be used to calculate a theoretical discrimination energy DELI(L/I)′ = −0.81 kcal/mol. The similarity of this result to the experimental DE value is remarkable, although both values are subject to errors. Thus, the trend highlighted by the experimental model studies is precisely mirrored in the coiled-coil structures of the protein databank: the vertical hetero-triads Leu-Ile-Leu and Ile-Leu-Ile are preferred relative to the homo-triads Leu-Leu-Leu and Ile-Ile-Ile.
Table 3.
Numbers of specific a′-a-a′ combinations in natural antiparallel coiled-coil structures
| Amino acid at a |
a′ = a′ = Ile |
a′ = a′ = Leu |
||||
|---|---|---|---|---|---|---|
| Observed | Expected | Observed/expected | Observed | Expected | Observed/expected | |
| Ile | 6 | 9.4 | 0.6 | 26 | 16.5 | 1.6 |
| Leu | 19 | 14.2 | 1.3 | 21 | 24.7 | 0.9 |
| Sum (all amino acids) | 59 | 103 | ||||
Expected numbers were calculated by using the percentage of occurrence of Ile and Leu at all a sites of coiled-coil structures in the CC+ database with 70% sequence identity or lower. These values are 16% and 24%, respectively. Note: the theoretical DELI(L/I)′ value is equal to −RT(ln(26/6) − ln(21/19)); the ratios of the raw numbers for Ile at both sites or Leu at both sites are self-normalizing, though the same result is obtained by using the normalized values (−RT(ln1.6 − ln 0.9 + ln1.3 − ln 0.6)).
Preferred Side-Chain Constellations.
For each of the homotypic and heterotypic vertical triads containing Leu and Ile residues, the structures found in the CC+ database were superimposed (Fig. 7). We have used such superimpositions for rationalizing statistically favored interstrand side-chain–side-chain interactions in β-sheets (30). The overlaid structures reveal that the statistically favored heterotypic combinations, Leu-Ile-Leu and Ile-Leu-Ile, are the “tightest” structurally, as indicated by two types of analysis. First, we calculated the rmsd values measured across the side-chain heavy atoms for each of the four sets of superimposed structures. The rmsd values were smaller for the statistically favored vertical hetero-triads (0.96 Å for Leu-Ile-Leu and 0.77 Å for Ile-Leu-Ile) relative to the homo-triads (1.58 Å for Ile-Ile-Ile and 1.52 Å for Leu-Leu-Leu). This trend suggests that the favored vertical triads lead to preferred conformations of side chains, or what we term preferred side–chain constellations.
Fig. 7.
Superposition of structures culled from the PDB for vertical triads containing Ile and Leu in antiparallel two-helix coiled coils. Superpositions were performed by using the MMTSB toolset (41), and rendered in PYMOL (42). The PDB and associated chain and residue identifiers for these structures are given in SI Table 12. For each overlay, a representative backbone is shown to guide the eye (backbone rmsd = 0.33–0.86 Å for the four Ile-Leu combinations at a′aa′).
In a second mode of analysis, we examined the side-chain dihedral angles for all four vertical triads culled from the CC+ database (see SI Table 12). The results indicate that the hetero-triads sample fewer of the possible side-chain conformations than do the homo-triads. This trend supports the notion that the conformational space sampled is tighter for the favored hetero-triads than for the homo-triads. Indeed, for the hetero-triads, preferred side-chain conformations were apparent. For instance, the dominant constellation for Leu-Ile-Leu had Ile in the g −t conformation and both Leu in t g+; for Ile-Leu-Ile, the dominant constellation had the side-chain conformation pattern g −t, t g+, g −t. Each side-chain conformation in these dominant constellations is fully compatible with the α-helix. All Ile display the g −t conformation, which is the favored rotamer (81%) for Ile in α-helices (31), and all Leu display t g+, the second-most populated conformer (30%) for Leu in α-helices. Further inspection of the structures revealed that in the homo-triads only “glancing” contacts are made between individual methyl groups from different side chains. In contrast, in the favored hetero-triads more intimate contacts occur between side chains, with the terminal methyl groups of the knob making multiple contacts with the vertical partners to give “nested-packing” interactions (32).
Interplay Among Vertical and Lateral Interactions.
The analyses described above show that the data obtained by using our thioester-based antiparallel coiled-coil model system provide insight that is relevant to side-chain–side-chain packing preferences in natural coiled coils. This correlation encouraged further analysis of the data in Table 1. These ΔGTE data allow the calculation of 50 independent DELI(X/Y) values, for X = Leu, Ile, Val, Asn, or Ala and Y = Leu, Ile, Val, Asn, or Ala (see SI Tables 7–11). Each of these DELI(X/Y) values reflects an energetic comparison of two different antiparallel coiled-coil pairings: Leu-X-Leu plus Ile-Y-Ile vs. Leu-Y-Leu plus Ile-X-Ile. Because DELI(X/Y) is meaningless if X = Y, and because DELI(X/Y) = −DELI(Y/X), 10 independent DELI(X/Y) values result when X and Y each vary among five residues. The number of DELI(X/Y) values that can be calculated from our ΔGTE data expands by a factor of five because we have examined five different residues (Leu, Ile, Val, Asn, or Ala) as the lateral partner (d′) for the central residue in the a′-a-a′ vertical triads under scrutiny. Comparing these 5 sets of 10 DELI(X/Y) values should indicate whether there is an energetically significant interplay between the lateral a-d′ and the vertical a′-a-a′ interactions experienced by a given a residue when it occupies its hole on the partner helix.
As a prelude to exploring the interplay between lateral and vertical interactions, we used an absolute DELI value of ≥0.4 kcal/mol (twice the estimated experimental uncertainty) as the criterion to identify those cases in which there is a significant preference for one pair of dimers relative to the alternative pair. By this criterion, 21 of the DELI values generated from our ΔGTE data (42%) are significant. This proportion indicates that the impact of vertical interactions within antiparallel coiled coils is not limited to the Leu-Ile combinations examined above, but rather that many vertical triads can exert a substantial influence on antiparallel coiled-coil dimerization selectivity.
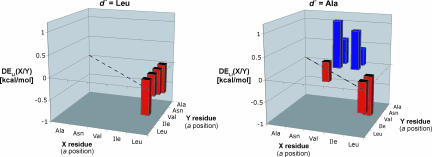
Comparison among the five sets of 10 DELI(X/Y) values indicates that there is an energetically significant interplay between the lateral partner d′ and the vertical partners a′ in their interactions with an a residue from the partner helix (a knob fitting into an a′, d′, a′, e′ hole). We illustrate this point in Fig. 8 by comparing the significant DELI values for d′ = Leu and for d′ = Ala. (Analogous results for d′ = Ile, Val, and Asn may be found in SI Fig. 18.) This comparison shows that the identity of d′ can determine whether DELI is significant for an X/Y pair as the knob residues (a). To our knowledge, all published efforts to account for or predict sequence–stability or sequence–selectivity relationships among coiled coils focus on pairwise interactions between side chains that are brought into contact on coiled-coil formation. The distinctions among our d′ = Leu and d′ = Ala datasets, however, show that the energetic impact of having both vertical neighbors (a′) Ile or both Leu on the knob residue at a is influenced by the identity of the d′ residue. Thus, treating vertical contacts in a simple pairwise or even triad fashion is not adequate for complete rationalization of coiled-coil stability or pairing preference. Similarly, treating lateral interactions in a simple pairwise fashion is not adequate, because of interplay involving vertical contacts (see SI Fig. 19).
Fig. 8.
Significant discrimination energies DELI(X/Y) for d′ = Leu and d′ = Ala (see Fig. 4). Values were determined to be significant if DE ≥0.4 kcal/mol (two times the expected error). Positive values (blue) indicate a preference for X paired with leucine vertical contacts, whereas negative values (red) indicate a preference for X paired with isoleucine vertical contacts. Note that only one side of the diagonal (dotted line) is shown because DELI(X/Y) = −DELI(Y/X). These graphs are intended to show qualitatively that the residue at d′ can strongly influence vertical pairing preferences of the residue at a′. DE data are available in SI Tables 7–11.
Conclusions.
We have uncovered a substantial thermodynamic influence exerted by previously unexplored patterns of intermolecular side-chain contacts at antiparallel coiled-coil interfaces. Our results provide the first quantitative analysis of vertical contact effects on coiled-coil pairing preference. Data derived from model systems indicate that vertical packing interactions involving buried side chains at a positions of the coiled-coil heptad repeat can have a significant effect on antiparallel dimerization selectivity. This trend is found in natural antiparallel coiled-coil dimers of known structure, with certain a′-a-a′ hetero-triads, Ile-Leu-Ile and Leu-Ile-Leu, favored relative to the corresponding homo-triads. Analyses of the favored structural motifs reveal preferred constellations in which the side chains adopt favorable dihedral angles and form intimate contacts.
Based on these results, it seems likely that vertical interactions represent an important but largely unrecognized component of the “code” that determines coiled-coil interaction preferences in complex biological environments. Past studies of coiled-coil pairing preferences, mostly involving parallel dimers, have identified relatively few mechanisms by which selectivity can be achieved. This limited set of rules has allowed considerable progress in the areas of coiled-coil prediction and design (1, 3), but our findings suggest that additional advances will be possible based on a more sophisticated treatment. Previously elucidated factors include lateral pairing of nonpolar side chains (9–16), which is driven by stereochemical complementarity, and lateral pairing of polar side chains (9–16, 33), which is driven by hydrogen bonding (at least for Asn-Asn pairs). In addition, pairing preferences are influenced by the electrostatic interactions (attractive or repulsive) between ionized side chains (34–39). Our findings indicate that vertical interactions must be considered as well if we are to understand the origins of dimerization selectivity among natural coiled-coil sequences and expand current abilities to predict and design such interactions. Here, we have explicitly addressed interactions associated with a′-a-a′ vertical triads in antiparallel dimers, but the approaches we have used can readily be extended to d′-d-d′-type vertical interactions at antiparallel coiled-coil interfaces, to d′-a-d′ and a′-d-a′ vertical interactions at parallel coiled-coil interfaces, and to other more-complex combinations of side chains.
In addition to highlighting the role of vertical contacts at coiled-coil interfaces, our results show that simple pairwise analysis of side-chain–side-chain interactions at such interfaces does not fully capture the factors that control stability or partner preference. High-fidelity interactions are common and necessary in biology, and proteins must find their partners in the cellular milieu efficiently and without engaging in promiscuous associations. Coiled-coil motifs represent a natural mechanism for guiding and cementing protein–protein interactions. Because 3–5% of the proteome is estimated to encode such motifs (5), however, selectivity in coiled-coil pairing constitutes an extraordinary feat of molecular recognition and discrimination. Our results provide an expanded framework for exploring partner selectivity among natural coiled-coil sequences and for designing new sequences that display high-fidelity pairing with a specified partner.
Methods
Backbone Thioester Exchange Assays.
Assays were typically initiated by mixing ≈0.1 mM each peptide in 50 mM buffer (phosphate, pH 7) and by allowing equilibration for 90 min. Details are provided in SI Text.
Analysis of PDB Structures.
The November 1, 2007, release of the PDB (40) was searched for helix–helix interactions with knobs-into-holes packing by using the program SOCKET (14) with a packing cutoff of 7.0 Å. More sophisticated searches were used to create a subset comprising two-helix, antiparallel coiled-coil motifs with 70% sequence identity at the most, and with α-helices longer than 11 aa. Full details of CC+ and our analysis interface will be presented elsewhere. Amino acid frequencies from this subset of interactions were normalized as described in the text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Josh Price for assistance with the CD data workup and Beth Bromley for valuable discussions on mutant cycles. T his work was supported National Institutes of Health Grant GM-61238, and by a Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council (United Kingdom) studentship (to O.D.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709068105/DC1.
References
- 1.Woolfson DN. Adv Protein Chem. 2005;70:79–116. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 2.Mason JM, Arndt KM. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 3.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 4.Gruber M, Lupas AN. Trends Biochem Sci. 2003;28:679–685. doi: 10.1016/j.tibs.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Wolf E, Kim PS, Berger B. Prot Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong JH, Keating AE, Singh M. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill RB, Raleigh DP, Lombardi A, DeGrado WF. Acc Chem Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 9.Acharya A, Rishi V, Vinson C. Biochemistry. 2006;45:11324–11332. doi: 10.1021/bi060822u. [DOI] [PubMed] [Google Scholar]
- 10.Mason JM, Schmitz MA, Muller K, Arndt KM. Proc Natl Acad Sci USA. 2006;103:8989–8994. doi: 10.1073/pnas.0509880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havranek JJ, Harbury PB. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 12.Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C. Biochemistry. 2002;41:14122–14131. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- 13.Walshaw J, Woolfson DN. J Mol Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 14.Keating AE, Malashkevich VN, Tidor B, Kim PS. Proc Natl Acad Sci USA. 2001;98:14825–14830. doi: 10.1073/pnas.261563398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripet B, Wagschal K, Lavigne P, Mant CT, Hodges RS. J Mol Biol. 2000;300:377–402. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- 16.Wagschal K, Tripet B, Lavigne P, Mant C, Hodges RS. Protein Sci. 1999;8:2312–2329. doi: 10.1110/ps.8.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walshaw J, Woolfson DN. J Struct Biol. 2003;144:349–361. doi: 10.1016/j.jsb.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Oakley MG, Hollenbeck JJ. Curr Opin Struct Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 19.Crick FHS. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 20.Hadley EB, Gellman SH. J Am Chem Soc. 2006;128:16444–16445. doi: 10.1021/ja067178r. [DOI] [PubMed] [Google Scholar]
- 21.Kwok SC, Hodges RS. J Biol Chem. 2004;279:21576–21588. doi: 10.1074/jbc.M401074200. [DOI] [PubMed] [Google Scholar]
- 22.Woll MG, Gellman SH. J Am Chem Soc. 2004;126:11172–11174. doi: 10.1021/ja046891i. [DOI] [PubMed] [Google Scholar]
- 23.Woll MG, Hadley EB, Mecozzi S, Gellman SH. J Am Chem Soc. 2006;128:15932–15933. doi: 10.1021/ja0634573. [DOI] [PubMed] [Google Scholar]
- 24.Hadley EB, Witek AM, Freire F, Peoples AJ, Gellman SH. Angew Chem Int Ed. 2007;46:7056–7059. doi: 10.1002/anie.200702449. [DOI] [PubMed] [Google Scholar]
- 25.McClain DL, Woods HL, Oakley MG. J Am Chem Soc. 2001;123:3151–3152. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]
- 26.Lumb KJ, Kim PS. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 27.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 28.Bilgicer B, Xing X, Kumar K. J Am Chem Soc. 2001;123:11815–11816. doi: 10.1021/ja016767o. [DOI] [PubMed] [Google Scholar]
- 29.Rohl CA, Baldwin RL. Biochemistry. 1994;33:7760–7767. doi: 10.1021/bi00191a003. [DOI] [PubMed] [Google Scholar]
- 30.Hutchinson EG, Sessions RB, Thornton JM, Woolfson DN. Protein Sci. 1998;7:2287–2300. doi: 10.1002/pro.5560071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovell SC, Word JM, Richardson JS, Richardson DC. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 32.Wouters MA, Curmi PMG. Proteins. 1995;22:119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez L, Woolfson DN, Alber T. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 34.Krylov D, Mikhailenko I, Vinson C. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grigoryan G, et al. PloS Comp Biol. 2006;2:551–563. [Google Scholar]
- 36.Campbell KM, Lumb KJ. Biochemistry. 2002;41:7169–7175. doi: 10.1021/bi025559l. [DOI] [PubMed] [Google Scholar]
- 37.Burkhard P, Ivaninskii S, Lustig A. J Mol Biol. 2002;318:901–910. doi: 10.1016/S0022-2836(02)00114-6. [DOI] [PubMed] [Google Scholar]
- 38.McClain DL, Gurnon DG, Oakley MG. J Mol Biol. 2002;324:257–270. doi: 10.1016/s0022-2836(02)01072-0. [DOI] [PubMed] [Google Scholar]
- 39.Ryan SJ, Kennan AJ. J Am Chem Soc. 2007;129:10255–10260. doi: 10.1021/ja073717w. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein FC, et al. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 41.Feig, et al. MMTSB Tool Set. La Jolla, CA: MMTSB NIH Research Resource, The Scripps Research Institute; 2001. [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.