Abstract
Heat-shock protein 90α (Hsp90α) is a member of the molecular chaperone family involved in protein folding and assembly. The role of Hsp90α in the developmental process, however, remains unclear. Here we report that zebrafish contains two Hsp90α genes, Hsp90α1, and Hsp90α2. Hsp90α1 is specifically expressed in developing somites and skeletal muscles of zebrafish embryos. We have demonstrated that Hsp90α1 is essential for myofibril organization in skeletal muscles of zebrafish embryos. Knockdown of Hsp90α1 resulted in paralyzed zebrafish embryos with poorly organized myofibrils in skeletal muscles. In contrast, knockdown of Hsp90α2 had no effect on muscle contraction and myofibril organization. The filament defects could be rescued in a cell autonomous manner by an ectopic expression of Hsp90α1. Biochemical analyses revealed that knockdown of Hsp90α1 resulted in significant myosin degradation and up-regulation of unc-45b gene expression. These results indicate that Hsp90α1 plays an important role in muscle development, likely through facilitating myosin folding and assembly into organized myofibril filaments.
Keywords: myofibrillogenesis, unc45, myosin chaperone, Hsp90
The myofibril of skeletal muscles, consisting of thick and thin filaments, is one of the most organized macromolecular structures in muscle cells. The thick and thin filaments are primarily made of myosin and actin, respectively. The myosin and actin filaments are organized to form highly repetitive structures called sarcomeres, the basic contractile unit in muscles. The precise folding and assembly of filament proteins into organized sarcomeres are critical for muscle development and contraction. Over 20 skeletal muscle diseases are caused by mutations or defective assembly of filament proteins (1, 2). An understanding of the mechanisms controlling the folding and assembly of filament proteins is the key to understanding many skeletal muscle diseases.
Genetic studies in Caenorhabditis elegans and biochemical analyses in vitro indicate that chaperone-mediated myosin folding is an integral part of myofibril assembly during muscle development (3). Mutations of C. elegans UNC-45, a myosin chaperone, result in paralyzed animals with severe myofibril disorganization in body wall muscles (4, 5). UNC-45 associates with heat-shock protein 90 (Hsp90) and acts as a cochaperone in myosin folding and assembly in C. elegans (6). Hsp90, consisting of Hsp90α and Hsp90β, is a member of the molecular chaperone family that plays an important role in cell proliferation, differentiation, stress management, and cancer (7–10). Hsp90α and Hsp90β are ubiquitously expressed in all eukaryotic cells. They represent some of the most abundant proteins in the eukaryotic cell. Expression of Hsp90α is inducible in response to temperature upshift or stress, whereas Hsp90β expression is more constitutive.
Hsp90α and Hsp90β function as molecular chaperones involved in the correct folding of newly synthesized proteins, preventing misfolding and aggregation of unfolded proteins, and assisting the correct assembly and localization of intracellular and secreted proteins (11). Knockout studies in mice have demonstrated that Hsp90β is required for the development of the placental labyrinth (12). The Hsp90β null mutant mouse embryos failed to form a fetal placental labyrinth and died around embryonic day 9.0/9.5. The in vivo function of Hsp90α in vertebrate development, however, has yet to be determined.
Recent studies in zebrafish demonstrated that Hsp90α is expressed strongly in developing somites and skeletal muscles during development (13). In vitro studies in myocytes indicated that Hsp90 forms a complex with newly synthesized myosin proteins and is involved in myosin folding and assembly (14). Inhibition of Hsp90 function by geldanamycin blocks myofibril assembly and triggers accumulation of myosin folding intermediates in C2C12 myocytes (14). Pharmacologic inhibition of Hsp90 function disrupted somite development in zebrafish (15). To determine whether Hsp90α plays an important role in myofibril assembly during muscle development, we used the knockdown approach to inhibit Hsp90α expression in zebrafish embryos. Knockdown studies in zebrafish have been successfully used to analyze gene functions involved in myofibril assembly that are difficult to study in mice via the knockout approach due to early embryonic lethality (16–18).
We report here that Hsp90α1, one of the two Hsp90α isoforms in zebrafish, plays a key role in myofibril assembly during muscle development. Knockdown of Hsp90α1 expression resulted in paralyzed zebrafish embryos without skeletal muscle contraction. Assembly of thick and thin filaments, as well as the M- and Z-lines, was disrupted in skeletal muscles. The myofibril defect could be specifically rescued in a cell autonomous manner by a transient expression of a Hsp90α1 transgene. Biochemical analysis revealed that the filament defect was probably caused by myosin degradation, because myosin heavy chain (MHC) proteins were barely detectable by Western blot in Hsp90α1 knockdown embryos. Together, these data suggest that Hsp90α1 plays an essential role in muscle development.
Results
Zebrafish Contains Two Hsp90α Genes, Hsp90α1 and Hsp90α2.
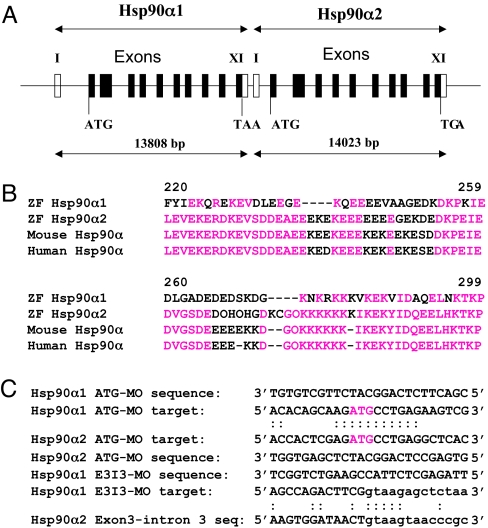
Hsp90α is ubiquitously expressed in all eukaryotic cells and its expression is inducible in response to temperature upshift or stress. However, Sass and colleagues have demonstrated that Hsp90α is specifically expressed in developing somite and skeletal muscles of zebrafish embryos (13). To better characterize Hsp90α expression, we analyzed the Hsp90α gene structure and expression in zebrafish. Genome sequence analysis revealed that zebrafish contains two highly related Hsp90α genes, Hsp90α1 and Hsp90α2. These two genes are closely linked on the same region of chromosome 20, separated by only 698 base pairs from the polyadenylation sequence of Hsp90α1 to the transcription start site of Hsp90α2 (Fig. 1A). Hsp90α1 encodes a protein of 725 aa, whereas Hsp90α2 encodes a protein of 734 aa [supporting information (SI) Fig. 6]. Sequence comparison revealed that Hsp90α1 and Hsp90α2 share 88% identity at the mRNA level and 91% identity at the protein level. The major difference between Hsp90α1 and Hsp90α2 lies between positions 237–277 that contain many charged glutamic acid and lysine amino acid residues (Fig. 1B). Sequence alignment revealed that Hsp90α2 is more closely related to human and mouse Hsp90α (Fig. 1B) and thus likely represents the ortholog of Hsp90α in zebrafish. Hsp90α1 represents a more diverse homolog of Hsp90α in zebrafish.
Fig. 1.
Gene structure and sequence comparison of zebrafish Hsp90α1 and Hsp90α2. (A) Zebrafish Hsp90α1 and Hsp90α2 share similar gene structures with 11 exons and 10 introns. Open boxes represent the 5′- and 3′-UTR regions. (B) Sequence comparison showing the region with more diverged sequences in Hsp90α1 and Hsp90α2 proteins. Sequence alignment indicates that zebrafish Hsp90α2 is more closely related to mouse and human Hsp90α. (C) MO antisense oligos and their target sequence in Hsp90α1 and Hsp90α2. The ATG start codon is identified in red. The intron sequences are shown in lowercase letters.
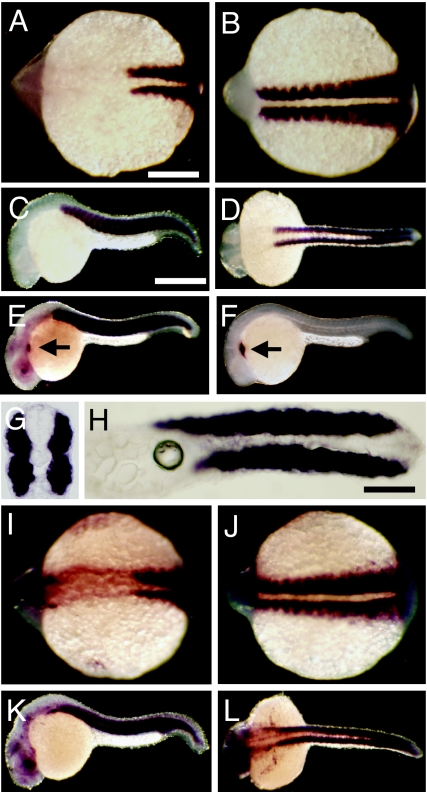
The temporal and spatial patterns of Hsp90α1 and Hsp90α2 expression were determined by whole mount in situ hybridization in zebrafish embryos. Hsp90α1 and Hsp90α2 exhibited overlapping patterns of expression in developing somites, skeletal and cardiac muscles (Fig. 2).
Fig. 2.
Temporal and spatial expression of Hsp90α1 and Hsp90α2 in zebrafish embryos. (A–D) Muscle-specific expression of Hsp90α1 in somites and skeletal muscles at 13 (A and B) and 24 hpf (C and D). (A and B) Dorsal view of anterior (A) and posterior (B) regions of a 13-hpf embryo. (C and D) Side (C) and dorsal (D) view of a 24-hpf embryo. The enzymatic reaction was carried out for 3 h during the whole-mount in situ hybridization. (E and F) Longer in situ staining (12 h) shows the cardiac expression of Hsp90α1 (arrow in E) in comparison with the cardiac muscle specific expression of myosin light chain (F). A weak staining of Hsp90α1 was also found in the brain and eye regions (E). (G and H) Cross (G) and horizontal (H) sections showing the muscle-specific expression of Hsp90α1 in zebrafish embryos at 24 hpf. (I–L) Expression of Hsp90α2 in somites, skeletal muscles and other tissues at 13 (I and J) and 24 hpf (K and L). In addition to muscle expression, Hsp90α2 expression was also detected in the head and eye regions (I). The enzymatic reaction was carried out for12 h during the whole-mount in situ hybridization. (Scale bars: A, 250 μm; C, 150 μm; and H, 50 μm.)
Knockdown of Hsp90α1 Expression Resulted in Paralyzed Zebrafish Embryos.
To determine whether Hsp90α1 plays a role in muscle development, we knocked down Hsp90α1 expression in zebrafish embryos. The Hsp90α1 translational blocker, ATG-MO (MO, morpholino), was specifically targeted to the sequence flanking the ATG start codon of the Hsp90α1 transcripts (Fig. 1C). The Hsp90α1 ATG-MO was injected into zebrafish embryos. The injected embryos were examined morphologically for 10 days after injection. Although the injected embryos appeared morphologically normal (SI Fig. 7 A and B), one striking phenotype was noted in all Hsp90α1 ATG-MO injected embryos (n = 289). Hsp90α1 knockdown embryos were paralyzed, unable to swim, and failed to show any signs of skeletal muscle contraction in response to physical stimulation by touch (SI Movie 1). In contrast, cardiac muscle contraction appeared normal (SI Movie 1). Injection of a higher dose (10 ng) of Hsp90α1 ATG-MO resulted in edema in most of the injected embryos (94%, n = 65; SI Fig. 7 C and D); however, cardiac muscle contraction appeared normal.
Sequence comparison revealed that Hsp90α2 shares 11 contiguous bases pairings (of 25) with the Hsp90α1 ATG-MO (Fig. 1C). This raises the question of whether the muscle phenotype was due to the specific knockdown of Hsp90α1 or both Hsp90α1 and Hsp90α2. To address the question of specificity, a Hsp90α1-specific splicing MO (E3I3-MO) was injected into zebrafish embryos. Hsp90α1 E3I3-MO could not knock down Hsp90α2, because only five contiguous base pairings were found between the E3I3-MO and the sequence at the exon 3 and intron 3 junction of Hsp90α2 (Fig. 1C). Injection of the Hsp90α1 E3I3-MO produced the same muscle phenotype as the Hsp90α1 ATG-MO in zebrafish embryos. To further confirm the specificity, we made a Hsp90α2-specific ATG-MO in the identical region as the Hsp90α1 ATG-MO. Injection of the Hsp90α2 ATG-MO had no effect on the muscle contraction of zebrafish embryos. Moreover, coinjection of Hsp90α1 and Hsp90α2 ATG-MOs showed no additional muscle phenotype compared with injection of Hsp90α1 ATG-MO alone. Together, these results indicate that the paralyzed muscle defects were indeed caused by the specific knockdown of Hsp90α1.
Knockdown of Hsp90α1 Expression Disrupted Myofibril Organization in Skeletal Muscles of Zebrafish Embryos.
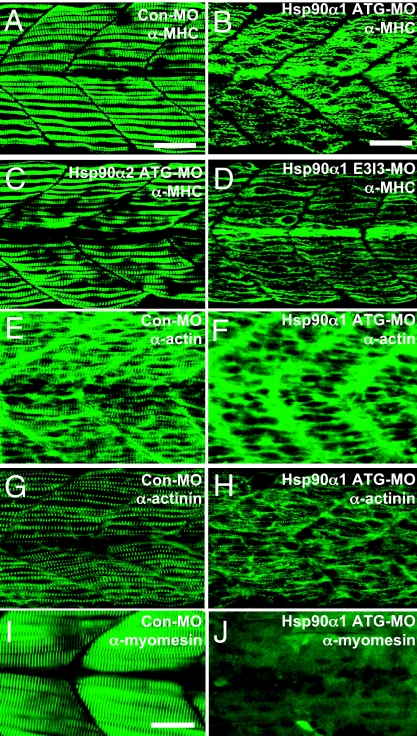
To determine which step of muscle development was affected, Hsp90α1 knockdown embryos were analyzed for myoblast specification and differentiation using several molecular markers. Expression of an early myogenic specification marker (MyoD) and a differentiation marker (MHC) appeared normal although at reduced levels for MHC in Hsp90α1 knockdown embryos (SI Fig. 7), indicating that Hsp90α1 is not required for myoblast specification and early differentiation. To determine whether blocking Hsp90α1 expression might disrupt myofibril organization during myofiber maturation, the Hsp90α1 knockdown embryos were examined by immunostaining with an anti-MHC antibody. Knockdown of Hsp90α1 expression severely disrupted the sarcomere formation and thick filament organization in slow muscles. Very few sarcomeres could be detected in Hsp90α1 knockdown myofibers (Fig. 3B). Identical phenotypes were obtained with the injection of the Hsp90α1 splicing E3I3-MO (Fig. 3D). In contrast, injection with the control MO (Con-MO) or Hsp90α2 specific ATG-MO had no effect on the sarcomere formation and myofibril organization (Fig. 3 A and C), further confirming that the skeletal muscle defects were specific to Hsp90α1 knockdown.
Fig. 3.
Knockdown of Hsp90α1 expression resulted in myofibril disorganization in skeletal muscles of zebrafish embryos. (A and B) Anti-MHC antibody (F59) staining shows the organization of thick filaments in trunk slow muscles of control-MO (A) or Hsp90α1-ATG-MO (B) injected embryos at 24 hpf. (C and D) Anti-MHC antibody (F59) staining shows the organization of thick filaments in trunk slow muscles of Hsp90α2 ATG-MO (C) or Hsp90α1-E3I3-MO (D) injected embryos at 24 hpf. (E and F) Anti-actin antibody staining shows the organization of thin filaments in control-MO (E) or Hsp90α1-ATG-MO (F) injected embryos at 24 hpf. (G and H) Anti-α-actinin antibody staining shows the organization of the Z-line in control-MO (G) or Hsp90α1-ATG-MO (H) injected embryos at 24 hpf. (I and J) Anti-myomesin antibody staining shows the organization of the M-line in control-MO (I) or Hsp90α1-ATG-MO (J) injected embryos at 72 hpf. (Scale bars: A and B, 25 μm and I, 15 μm.)
To test whether the thin filament was also affected by Hsp90α1 knockdown, the Hsp90α1 knockdown embryos were stained with anti-α-actin antibody. Compared with the control-MO injected embryos (Fig. 3E), Hsp90α1 knockdown embryos showed few or no thin filaments (Fig. 3F). To test whether other sarcomeric structures, such as the M and Z lines, were also affected in the Hsp90α1 knockdown of slow muscles, we analyzed the localization of myomesin and α-actinin, the respective M- and Z-line-specific proteins, by antibody staining. Both M- and Z-line organization was significantly disrupted (Fig. 3 H and J). To determine whether fast muscles located in the medial region of the myotone were affected by Hsp90α1 knockdown, the Hsp90α1 morphant embryos were analyzed by histological staining. The results showed that fast muscles contained highly disorganized myofibrils (SI Fig. 8B). Immunostaining with the anti-myomesin antibody confirmed that the M-line was disorganized in fast muscles of Hsp90α1 knockdown embryos (SI Fig. 8D). Together, these data indicate that Hsp90α1 is required for myofibril organization in both slow and fast muscles during muscle development in zebrafish embryos.
Rescue of Myofibril Defects by Ectopic Expression of Hsp90α1.
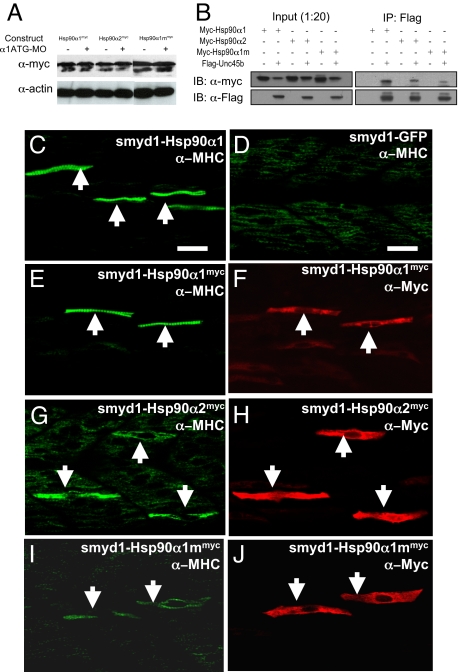
To confirm the specificity of phenotype, we performed a rescue experiment by a transient expression of Hsp90α1 in the knockdown zebrafish embryos. Muscle-specific expression was directed using a muscle-specific smyd1 promoter (19). The Hsp90α1 ATG-MO was unable to inhibit expression of the smyd1-Hsp90α1 transgene because the 5′-UTR sequence targeted by the ATG-MO was replaced with a 5′-UTR sequence from the β-globin gene. Myofibril organization was analyzed in zebrafish embryos coinjected with the Hsp90α1 ATG-MO and the smyd1-Hsp90α1 construct. Over 90% (n = 47) of the coinjected embryos contained normal myofibers with clearly organized sarcomeres (Fig. 4C, Table 1). In contrast, coinjection of the Hsp90α1 ATG-MO with smyd1-GFP vector showed no rescue (Fig. 4D). The rescue by Hsp90α1 appeared to be mosaic, likely due to the mosaic pattern of gene expression from DNA injection.
Fig. 4.
Rescue of myofibril organization defect in Hsp90α1 knockdown embryos by ectopic expression of a Hsp90α1 transgene. (A) Western blot analysis shows the expression of myc-tagged Hsp90α1-, Hsp90α2-, or MEEVD-deleted Hsp90α1 (Hsp90α1m) in wild type or Hsp90α1 ATG-MO coinjected embryos. Expression of the myc-tagged proteins was analyzed by Western blot using anti-myc tag antibody (9E10). Each lane contains protein extract from 10 embryos. Actin was used as a loading control. (B) Coimmunoprecipitation shows binding of Hsp90α1, Hsp90α2, or MEEDV domain deleted Hsp90α1 to UNC-45b. (C and D) F59 antibody staining shows the mosaic rescue of myofibers in a Hsp90α1 knockdown embryo co-injected with the smyd1-Hsp90α1 construct (C) or smyd1-GFP vector control (D) at 24 hpf. The rescued myofibers are indicated by arrows. (E and F) Double immunostaining with anti-MHC (E) and anti-myc (F) antibodies shows the rescued myofibers expressing Hsp90α1myc. (G and H) Double staining with anti-MHC (G) and anti-myc (H) antibodies shows partial rescue in Hsp90α2myc expressing myofibers. (I and J) Double staining with anti-MHC (I) and anti-myc (J) antibodies shows the partially rescued myofibers expressing the MEEVD domain deleted Hsp90α1. (Scale bars: 30 μm.)
Table 1.
Rescue of myofibril assembly defect by transient expression of Hsp90α1
| No. of embryos | With rescued myofibers | Without rescued myofibers | Rescue, % | |
|---|---|---|---|---|
| Hsp90α1-ATG-MO (2.5 ng) + smyd1-gfp (0.1 ng) | 34 | 0 | 34 | 0 |
| Hsp90α1-ATG-MO (2.5 ng) + smyd1-Hsp90α1 (0.1 ng) | 47 | 42 | 5 | 90 |
| Hsp90α1-ATG-MO (2.5 ng) + smyd1-Hsp90α1myc (0.1 ng) | 37 | 33 | 4* | 89 |
| Hsp90α1-ATG-MO (2.5 ng) + smyd1-Hsp90α2myc (0.1 ng) | 26 | 5† | 21 | 19 |
| Hsp90α1-ATG-MO (2.5 ng) + smyd1-Hsp90α1mmyc (0.1 ng) | 53 | 27† | 26 | 51 |
hsp90α1-ATG-MO was coinjected with smyd1-gfp, smyd1-Hsp90α1, smyd1-Hsp90α1myc, smyd1-Hsp90α1mmyc, or smyd1-Hsp90α2myc DNA construct at the one- or two-cell stages. The injected embryos were scored by myofibril assembly and sarcomere formation using anti-myosin (F59) and anti-myc antibody staining. Coinjection with smyd1-Hsp90α1 or smyd1-Hsp90α1myc rescued the myofibril defect in Hsp90α1 knockdown embryos.
*Embryos that showed no rescue had no myc-tagged Hsp90α1 expression in slow muscles.
†Partially rescued myofibers.
To confirm that the rescued myofibers indeed expressed the Hsp90α1 transgene, a gene construct (smyd1-Hsp90α1myc) expressing a myc-tagged Hsp90α1 was coinjected with the Hsp90α1 ATG-MO into zebrafish embryos. Double immunostaining analysis using the anti-MHC (F59), and the anti-myc antibodies showed that only those myofibers expressing myc-tagged Hsp90α1 were rescued (Fig. 4 E and F), suggesting that Hsp90α1 functions in a cell autonomous manner. To test whether the myofibril defect could be rescued by ectopic expression of Hsp90α2, a similar rescue experiment was performed using a smyd1-hsp90α2myc DNA construct. Double immunostaining revealed that Hsp90α2 could partially rescue the myofibril defects resulting from Hsp90α1 knockdown (Table 1). Myofibers expressing myc-tagged Hsp90α2 showed increased myosin expression (Fig. 4 G and H). However, no organized sarcomeres could be observed in these myofibers (Fig. 4G). Together, these data indicate that Hsp90α1 plays a critical role in myosin expression and assembly.
Deletion of MEEVD Domain Reduced Hsp90α1 Activity.
The highly conserved C-terminal MEEVD domain in Hsp90α has been implicated in binding with the tetratricopeptide repeat (TPR) in cochaperones (20–24). Mutation of the MEEVD domain in Hsp90α reduced binding to co-chaperones (6, 20). We therefore tested whether deletion of the C-terminal MEEVD domain could affect the activity of Hsp90α1 in myofibril organization. A DNA construct expressing a MEEVD domain deleted Hsp90α1 (Hsp90α1mmyc) was generated and used in the rescue assay. Expression of the MEEVD truncated Hsp90α1 could not be blocked by the Hsp90α1 ATG-MO (Fig. 4A). Double staining with anti-MHC and -myc antibodies revealed that MEEVD deletion significantly reduced the activity of Hsp90α1 in the rescue experiment. Myofibers expressing the truncated Hsp90α1 exhibited a partially rescued phenotype (Fig. 4 I and J). Coimmunoprecipitation analysis demonstrated that deletion of the MEEVD domain significantly reduced Hsp90α1 binding to UNC-45B (Fig. 4B). Together, these data indicate that that the MEEVD domain is likely involved in Hsp90α1 function in myofibril assembly.
Knockdown of Hsp90α1 Resulted in Increased Myosin Degradation in Zebrafish Embryos.
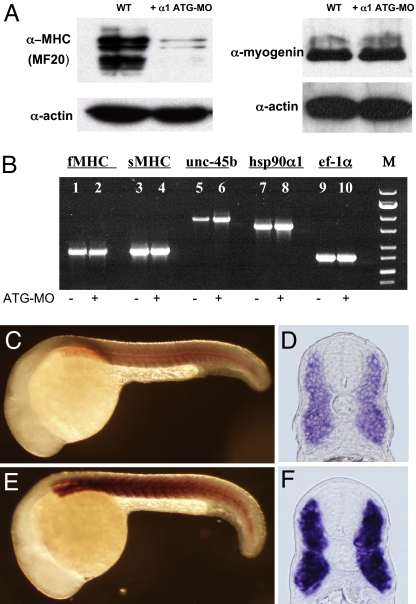
In vitro studies have suggested that Hsp90α associates with myosin proteins, and is required for myosin folding and assembly in C2C12 myoblast cells (14). To test whether knockdown of Hsp90α1 might disrupt myosin folding and result in myosin degradation, we analyzed the levels of myosin protein in the Hsp90α1 knockdown embryos by Western blot using the anti-MHC antibody (MF20). MHC protein levels were significantly reduced in Hsp90α1 knockdown embryos compared with wild type control (Fig. 5A). In contrast, expression of actin, the key thin filament protein, appeared normal (Fig. 5A).
Fig. 5.
Knockdown of Hsp90α1 results in MHC degradation and up-regulation of unc-45b expression. (A) Western blot analysis shows the low levels of MHC proteins in Hsp90α1 knockdown embryos. In contrast, expression levels of actin and myogenin were not affected. (B) RT-PCR analysis of fast-muscle MHC (fMHC), slow-muscle MHC (sMHC), unc-45b, Hsp90α1, and ef-1α expression in control (lanes 1, 3, 5, 7, and 9) or Hsp90α1 ATG-MO injected (lanes 2, 4, 6, 8, and 10) embryos. (C–F) Side views (C and E) or cross-sections (D and F) show unc-45b expression in control (C and D) or Hsp90α1 ATG-MO (E and F) injected embryos at 24 hpf.
The lower levels of MHC proteins could be due to myosin protein degradation or decreased MHC gene transcription and translation. To clarify this question, we analyzed the expression of MHC mRNAs in Hsp90α1 knockdown embryos by RT-PCR and whole-mount in situ hybridization. The results showed that knockdown of Hsp90α1 expression did not alter MHC gene expression in either slow or fast muscles (SI Fig. 9). This was further confirmed by real-time RT-PCR (SI Table 2). Together, these data suggest that the low levels of MHC in Hsp90α1 knockdown embryos were likely caused by protein degradation or poor translation.
Knockdown of Hsp90α1 Increases unc-45b Expression in Zebrafish Embryos.
It has been shown that myosin chaperone UNC-45 acts as a Hsp90α cochaperone in myosin assembly in C. elegans (6). A zebrafish unc-45 ortholog, unc-45b, is specifically expressed in developing somites and skeletal muscles of zebrafish embryos (25). Knockdown or mutation of unc-45b resulted in paralyzed zebrafish embryos with muscle defects in thick filaments (17, 26), a phenotype very similar to that of Hsp90α1 knockdown.
Highly regulated unc-45 expression is critical for myosin folding and assembly (27–29). To test whether knockdown of Hsp90α1 might alter unc-45b expression, we analyzed unc-45b expression in Hsp90α1 knockdown embryos by RT-PCR, whole-mount in situ hybridization, and real-time PCR. The results showed that unc-45b expression was significantly increased in Hsp90α1 knockdown embryos (Fig. 5 B–F). Analysis by real-time PCR revealed a 3- to 4-fold increase of unc-45b mRNA levels in Hsp90α1 knockdown embryos (SI Table 2). These data indicate a close interaction between Hsp90α1 and unc-45b in the control of myofibril assembly.
Discussion
Organized myofibril assembly is essential for muscle contraction. In this study, we have characterized the expression and function of Hsp90α in zebrafish embryos. Our studies have demonstrated that zebrafish contains two Hsp90α genes with similar patterns of expression. Hsp90α1 plays a key role in myofibril assembly. Knockdown of Hsp90α1 expression resulted in MHC degradation, which led to paralyzed fish embryos with disorganized myofibrils in skeletal muscles.
Zebrafish Contains Two Distinct Hsp90α Genes, Hsp90α1 and Hsp90α2.
We demonstrated that zebrafish Hsp90α1 and Hsp90α2 are closely linked on the same region of chromosome 20, separated by only 698 base pairs. Hsp90α1 is almost identical to the previously reported Hsp90α (AF068773 or NM_131328), with the exception of the 5′-UTR sequence and the 30-bp coding region for the first 10 amino acids at the N terminus. Ironically, the 5′-UTR sequence and the first 30-bp coding region in the previously reported zebrafish Hsp90α are identical to the corresponding sequence reported here for the Hsp90α2. To determine whether zebrafish embryos express such chimerical mRNA transcripts, we performed RT-PCR analysis using a 5′ primer from the Hsp90α2 5′-UTR region and a 3′ primer around the stop codon of Hsp90α1. No PCR product was generated from such PCR analysis (data not shown). In contrast, a PCR product of the predicted size was generated when the Hsp90α2 5′-UTR primer was used with its own 3′ primer (data not shown). Similarly, a PCR product was amplified from the Hsp90α1 transcripts when its own 5′ and 3′ primers were used (Fig. 5B). Together, these data suggest that the chimerical Hsp90α transcript previously reported (AF068773 or NM_131328) may not exist or at least is not expressed in early zebrafish embryos.
Distinct Functions of Hsp90α1 and Hsp90α2 in Muscle Development.
It appears that Hsp90α1 and Hsp90α2 may have different functions in muscle development. Knockdown of Hsp90α2 using its specific ATG-MO gave no detectable muscle phenotype. This is consistent with previous report that knockdown of Hsp90α2 alone in zebrafish embryos did not give rise to severe phenotypes unless Hsp90β was knocked down at the same time, producing heart and notochord defects (30). The lack of muscle phenotype from Hsp90α2 knockdown is consistent with its poor activity in our rescue assay, which shows that, unlike Hsp90α1, which consistently showed a strong rescue, ectopic expression of Hsp90α2 showed poor or no rescue in myofibril organization in Hsp90α1 knockdown embryos. Together, these data suggest that Hsp90α1 may play a more critical role in muscle development and its functions cannot be replaced by Hsp90α2 in zebrafish embryos.
Interestingly, we noted that knockdown of Hsp90α1 alone or in combination of Hsp90α2 had little effect on cardiac muscle contraction. Immunostaining with anti-myosin and -actin antibodies further confirmed that cardiac muscles were normal although the same embryo showed a clear skeletal muscle defect (SI Fig. 10). A high dose of Hsp90α1 ATG-MO injection caused edema in the injected embryos. However, edema is a very common phenotype associated with high doses of MO injection. The lack of a clear heart phenotype from the knockdown of Hsp90α1 and Hsp90α2 could be due to the incomplete knockdown of these genes. Alternatively, this could be caused by functional redundancy from Hsp90β, consistent with a previous report that double knockdown of Hsp90α2 and Hsp90β were required to produce a heart defect in zebrafish embryos (30).
Chaperones and Myosin Assembly.
We showed that knockdown of Hsp90α1 expression resulted in MHC degradation in zebrafish embryos. Our in vivo data are consistent with previous studies suggesting that Hsp90α1 may act as a myosin chaperone (14). In vitro studies have shown that nascent myosin filaments form a complex with Hsp90 and Hsp70 (13). Efficient folding and assembly of myosin are possible only when it is expressed in muscle cells, suggesting that unique components from muscle cells are required for myosin folding and assembly (31). These studies are also consistent with previous findings from fission yeast demonstrating that Swo1p (the fission yeast Hsp90 homolog) is important for myosin II assembly (32). Together, these studies may shed light into the mechanism(s) by which heat-shock proteins are involved in muscle development and atrophy. These studies may also have a biomedical significance because heat shock proteins have been implicated in myopathies in aging muscles. Down-regulation of heat-shock protein expression has been linked to muscle atrophy (33). Overexpression of Hsp70 in muscles of old mice significantly enhanced their recovery after damage (34).
It should be noted that, in addition to myosin folding, Hsp90 has been implicated in signal transduction in differentiating C2C12 myoblasts (35). Inhibition of Hsp90 function by geldamycin results in destabilization of Hsp90-dependent kinases ErbB2, Fyn, and Akt and decreased expression of Myogenin, a key transcription factor in myoblast differentiation (35). Our studies by Western blot analysis showed that expression of Myogenin protein was not affected in Hsp90α1 knockdown embryos (Fig. 5A). However, we could not rule out the possibility that there may be an effect on Myogenin protein phosphorylation.
Although our studies indicate that Hsp90α1 likely functions as a myosin chaperone, we cannot rule out the possible impact of knockdown Hsp90α1 on myosin translation. It has been reported that Heme-regulated eIF-2a Kinase (HRI) is a client protein of Hsp90α (36, 37). The phosphorylation of eIF-2α by HRI results in the inhibition of protein synthesis in vitro. If Hsp90α1 is obligatory for HRI kinase activity in muscle cells, we should expect that knockdown of Hsp90α may decrease HRI kinase activity, which will result in increased protein synthesis in fish embryos. This contradicts our finding that the knockdown of Hsp90α1 reduced the levels of myosin proteins. The effect of Hsp90α1 knockdown on protein synthesis may be more complicated in fish embryos than that observed in vitro, and the role of Hsp90α1 on myosin protein translation remains to be determined.
UNC-45 and Myosin Assembly.
UNC-45, a myosin-binding protein and Hsp90 cochaperone, is required for myosin assembly in body wall muscles in C. elegans (6). UNC-45 mutation causes myosin degradation in C. elegans (28). In vertebrates, two UNC-45 isoforms have been identified: a general cell ubiquitous isoform and a striated muscle-specific isoform (3, 38). Knockdown or mutation of the muscle-specific isoform unc-45b resulted in paralyzed zebrafish embryos with a loss of myosin filaments in trunk muscles (17, 26). The similar, if not identical, phenotypes from Hsp90α1 or unc-45b knockdown are consistent with a close interaction between these two proteins in myofibril assembly.
Association of Hsp90α with UNC-45 is mediated, in part, by interaction with the highly conserved C-terminal MEEVD domain (6, 39). Consistent with this notion, we showed that the deletion of the C-terminal MEEVD domain decreased Hsp90α1 binding to UNC-45b and its activity in myosin assembly. Myofibers overexpressing the MEEVD truncated Hsp90α1 showed a partially rescued phenotype (Fig. 4I). Interestingly, we also noted that Hsp90α2, which contains the MEEVD domain, showed little or no activity in myosin assembly, consistent with its weak binding to UNC-45b (Fig. 4 B and G). Together, these data indicate that association with UNC-45B is critical for the function in myosin assembly. Moreover, in addition to the MEEVD domain, other Hsp90α1-specific domains are likely involved in UNC-45b binding and myofibril assembly.
We demonstrated in this study that knockdown of Hsp90α1 significantly increased the levels of unc-45b mRNA transcripts in the morphant embryos. The molecular mechanism underlying this up-regulation of unc-45b expression is not clear. The up-regulation of unc-45b expression may be a compensatory response to the increased protein misfolding induced by Hsp90α1 knockdown. Interestingly, a reciprocal up-regulation of Hsp90α expression has also been found in unc-45b knockdown or mutant zebrafish embryos (17, 26). It has been shown that highly regulated unc-45 expression is critical for myosin folding and assembly in body wall muscles of C. elegans (27–29). Strikingly, overexpression of UNC-45 also leads to myosin degradation in body wall muscles of C. elegans, as in unc-45 mutants (28). This raises the question of whether increased unc-45b expression in zebrafish embryos could also contribute to myosin degradation in Hsp90α1 knockdown embryos. This remains to be analyzed in future studies through a direct overexpression of unc-45b in zebrafish embryos using a transgenic approach.
Materials and Methods
Synthesis of Morpholino Antisense Oligos.
Morpholino antisense oligos were synthesized by Gene Tools. The Hsp90α1 and Hsp90α2 translation blockers (ATG-MO) were targeted to sequence near the ATG start codon. The Hsp90α1 E3I3 splicing blocker was targeted to sequence at the exon 3 and intron 3 junction. The control MO was the standard control oligo purchased from Gene Tools.
Hsp90α1 ATG-MO: 5′-CGACTTCTCAGGCATCTTGCTGTGT-3′; Hsp90α2 ATG-MO: 5′-GTGAGCCTCAGGCATCTCGAGTGGT-3′; and Hsp90α1 E3I3-MO: 5′-TTAGAGCTCTTACCGAAGTCTGGCT-3′.
Microinjection in Zebrafish Embryos.
Morpholino antisense oligos were dissolved in 1× Danieau buffer to a final concentration of 0.1 mM, 0.25 mM, 0.5 mM or 1 mM. Approximately 1–2 nl (1, 2.5, or 5 ng) was injected into each zebrafish embryo at the one- or two-cell stages. For rescue assay, 2.5 ng of Hsp90α1 ATG-MO was coinjected with 0.1 ng of DNA construct into each zebrafish embryo.
Regular and Real-Time RT-PCR Analysis of Gene Expression.
Total RNA was extracted from 100 control or Hsp90α1 ATG-MO injected zebrafish embryos at 24 hours postfertilization (hpf). Standard PCR was carried out by using primers described in SI Text. Real-time PCR was carried out as described in SI Text.
Whole-Mount in Situ Hybridization and Immunostaining.
In situ hybridization was carried out as described (40). Immunostaining was carried out as described (16) with the following antibodies: anti-α-actinin (clone EA-53, #A7811, Sigma), anti-MHC for slow muscles (F59, Developmental Studies Hybridoma Bank), anti-MHC for fast muscles (A4.1025, DSHB), anti-MHC (MF-20, DSHB), anti-myomesin (mMaC myomesin B4, DSHB), and anti-actin (Ac1–20.4.2, Progen).
Construction of cmv-Hsp90α1, cmv-Hsp90α1myc, cmv-Hsp90α1mmyc, cmv-Hsp90α2myc, smyd1-Hsp90α1, smyd1-Hsp90α1myc, smyd1-Hsp90α1mmyc, and smyd1-Hsp90α2myc Plasmid Constructs.
Construction of the above expression plasmids is described in detail in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Rebecca Petre and John Stubblefield for editing, Oluwasina Folawiyo for assistance in whole-mount in situ hybridization, Shenyun Fang for assistance in coimmunoprecipitation, and J. Adam Frederick for video taping the zebrafish embryos. This research was supported by Grants IS-3703-05 and MB-8705-04 from Binational Agricultural Research and Development Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707330105/DC1.
References
- 1.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 2.Laing NG, Nowak KJ. BioEssays. 2005;27:809–822. doi: 10.1002/bies.20269. [DOI] [PubMed] [Google Scholar]
- 3.Hutagalung AH, Landsverk ML, Price MG, Epstein HF. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- 4.Epstein HF, Thomson JN. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- 5.Barral JM, Bauer CC, Ortiz I, Epstein HF. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 7.Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 8.Terasawa K, Minami M, Minami Y. J Biochem (Tokyo) 2005;137:443–447. doi: 10.1093/jb/mvi056. [DOI] [PubMed] [Google Scholar]
- 9.Voellmy R, Boellmann F. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 10.Rutherford S, Knapp JR, Csermely P. Adv Exp Med Biol. 2007;594:190–197. doi: 10.1007/978-0-387-39975-1_16. [DOI] [PubMed] [Google Scholar]
- 11.Pearl LH, Prodromou C. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 12.Voss AK, Thomas T, Gruss P. Development. 2000;127:1–11. doi: 10.1242/dev.127.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Sass JB, Martin CC, Krone PH. Int J Dev Biol. 1999;43:835–838. [PubMed] [Google Scholar]
- 14.Srikakulam R, Winkelmann DA. J Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 15.Lele Z, Hartson SD, Martin CC, Whitesell L, Matts RL, Krone PH. Dev Biol. 1999;210:56–70. doi: 10.1006/dbio.1999.9262. [DOI] [PubMed] [Google Scholar]
- 16.Tan X, Rotllant J, Li H, DeDeyne P, Du SJ. Proc Natl Acad Sci USA. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wohlgemuth SL, Crawford BD, Pilgrim DB. Dev Biol. 2007;303:483–492. doi: 10.1016/j.ydbio.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Hinits Y, Hughes SM. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du SJ, Rotllant J, Tan X. Dev Dyn. 2006;235:3306–3315. doi: 10.1002/dvdy.20984. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Sullivan WP, Toft DO, Smith DF. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young JC, Obermann WM, Hartl FU. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 22.Carrello A, Ingley E, Minchin RF, Tsai S, Ratajczak T. J Biol Chem. 1999;274:2682–2689. doi: 10.1074/jbc.274.5.2682. [DOI] [PubMed] [Google Scholar]
- 23.Russell LC, Whitt SR, Chen MS, Chinkers M. J Biol Chem. 1999;274:20060–20063. doi: 10.1074/jbc.274.29.20060. [DOI] [PubMed] [Google Scholar]
- 24.Ramsey AJ, Russell LC, Whitt SR, Chinkers M. J Biol Chem. 2000;275:17857–17862. doi: 10.1074/jbc.M001625200. [DOI] [PubMed] [Google Scholar]
- 25.Etheridge L, Diiorio P, Sagerstrom CG. Dev Dyn. 2002;224:457–460. doi: 10.1002/dvdy.10123. [DOI] [PubMed] [Google Scholar]
- 26.Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, Strahle U. Dev Biol. 2007;308:133–143. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Hoppe T, Cassata G, Barral JM, Springer W, Hutagalung AH, Epstein HF, Baumeister R. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Landsverk ML, Li S, Hutagalung AH, Najafov A, Hoppe T, Barral JM, Epstein HF. J Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 30.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow D, Srikakulam R, Chen Y, Winkelmann DA. J Biol Chem. 2002;277:36799–36807. doi: 10.1074/jbc.M204101200. [DOI] [PubMed] [Google Scholar]
- 32.Mishra M, D'souza VM, Chang KC, Huang Y, Balasubramanian MK. Eukaryot Cell. 2005;4:567–576. doi: 10.1128/EC.4.3.567-576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McArdle A, Broome CS, Kayani AC, Tully MD, Close GL, Vasilaki A, Jackson MJ. Exp Gerontol. 2006;41:497–500. doi: 10.1016/j.exger.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Broome CS, Kayani AC, Palomero J, Dillmann WH, Mestril R, Jackson MJ, McArdle FASEB J. 2006;20:1549–1551. doi: 10.1096/fj.05-4935fje. [DOI] [PubMed] [Google Scholar]
- 35.Yun BG, Matts RL. Exp Cell Res. 2005;307:212–223. doi: 10.1016/j.yexcr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Uma S, Hartson SD, Chen JJ, Matts RL. J Biol Chem. 1997;272:11648–11656. doi: 10.1074/jbc.272.17.11648. [DOI] [PubMed] [Google Scholar]
- 37.Matts RL, Hurst R. J Biol Chem. 1989;264:15542–15547. [PubMed] [Google Scholar]
- 38.Price MG, Landsverk ML, Barral JM, Epstein HF. J Cell Sci. 2002;115:4013–4023. doi: 10.1242/jcs.00108. [DOI] [PubMed] [Google Scholar]
- 39.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 40.Du SJ, Dienhart M. Dev Dyn. 2001;222:655–666. doi: 10.1002/dvdy.1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.