Abstract
Coinfection of a host by multiple parasite species has important epidemiological and clinical implications. However, the direction and magnitude of effects vary considerably among systems, and, until now, there has been no general framework within which to explain this variation. Community ecology has great potential for application to such problems in biomedicine. Here, metaanalysis of data from 54 experiments on laboratory mice reveals that basic ecological rules govern the outcome of coinfection across a broad spectrum of parasite taxa. Specifically, resource-based (“bottom-up”) and predator-based (“top-down”) control mechanisms combined to determine microparasite population size in helminth-coinfected hosts. Coinfection imposed bottom-up control (resulting in decreased microparasite density) when a helminth that causes anemia was paired with a microparasite species that requires host red blood cells. At the same time, coinfection impaired top-down control of microparasites by the immune system: the greater the helminth-induced suppression of the inflammatory cytokine interferon (IFN)-γ, the greater the increase in microparasite density. These results suggest that microparasite population growth will be most explosive when underlying helminths do not impose resource limitations but do strongly modulate IFN-γ responses. Surprisingly simple rules and an ecological framework within which to analyze biomedical data thus emerge from analysis of this dataset. Through such an interdisciplinary lens, predicting the outcome of coinfection may become tractable.
Keywords: cytokine, disease ecology, metaanalysis, predation, resource limitation
Host–parasite interactions are rarely one-on-one. Instead, each host tends to harbor a community of parasite species, and these multispecies coinfections shape disease epidemiology [e.g., by altering the infectiousness (1–3) or availability (4) of hosts] as well as disease severity. Helminths (parasitic worms) and microparasites (viruses, bacteria, fungi, or protozoa), for example, widely cooccur and may interact either to the detriment or the benefit of the host (5, 6). Coinfection thereby raises the question of whether anthelmintic drugs would alter the severity of diseases like malaria (7, 8), and presents both challenges and opportunities for proposed “integrated control” programs that target multiple infections at once (5).
An ability to predict the outcome of coinfection (or at least to understand the context dependency of outcomes) would facilitate design of integrated control programs and could also aid understanding of the ecology and evolution of both parasites and hosts (9–11). However, the complexity of within-host interactions (12–16) can make it difficult to predict how one infection will affect the course of others. Disentangling such complexity is a core activity of community ecologists, who in recent years have successfully identified trophic rules that determine, for example, the abundance of organisms in marine (17) as well as tropical (18, 19) and temperate (20) terrestrial ecosystems. These trophic rules include “bottom-up” control of population size via resource limitation and “top-down” control via predation (21). Only by considering regulation by both resources and consumers have ecologists been able to explain the abundance of organisms across such diverse communities (17–20). One major finding is that the relative importance of top-down and bottom-up control mechanisms varies among species and ecosystems; a charismatic example of this is provided by African savannah systems, in which zebra populations are primarily controlled by predators, whereas wildebeest are more strongly regulated by the quality and quantity of plant matter available for them to eat (18). I applied these principles on a different scale in the present study, to investigate whether defined within-host ecological interactions, namely, competition for resources (bottom-up population control) and/or predation by the immune system (top-down control) (22), could explain changes in microparasite population size caused by underlying helminth coinfection. I essentially undertook analysis of the community ecology of the coinfected laboratory mouse.
Biomedical research provides evidence that helminths have the potential to alter both bottom-up and top-down control of microparasites. Helminths might impose resource limitation if, for example, they reduce the availability of surface area for microparasite attachment (23) or of the cell type essential for microparasite replication (24). Here, I focused on the extent to which helminth species affect the availability of host red blood cells (RBCs) as an example of the potential for bottom-up control: helminths that induce anemia might limit population growth of microparasites that depend on RBCs. Helminths are also expected to alter the efficacy of immune predation on microparasites. Ideally, the immune system would multitask with ease when concurrent challenges are anatomically separated or antigenically distinct (25). Indeed, membrane-bound molecules enable fine control of immune signaling, allowing deft multitasking by dendritic cells, for example (26). However, signaling molecules such as cytokines that are released into the environment are considered to be promiscuous (rather than antigen-specific) in their effects (27), and even gut-restricted helminths have systemic immunological effects via cytokines (28). The discovery of mutual inhibition (29) between the cytokine responses induced by helminths versus microparasites (30) therefore provided a fresh understanding of helminth–microparasite coinfection (13). I focused on the signature cytokines of helminth-induced and microparasite-induced immune responses [interleukin (IL)-4 and interferon (IFN)-γ, respectively (30)], to assess changes in the efficacy of top-down control of microparasites during helminth coinfection.
I conducted statistical metaanalysis of data from 54 experiments that compared microparasite densities in laboratory mice with and without helminth coinfection. I calculated log response ratios (31) and assessed whether the effect of coinfection on microparasite population size could be predicted by the potential for RBC limitation and/or the extent of cytokine alterations induced by coinfection. I also assessed whether changes in microparasite density were explained by host genotype, host sex, helminth or microparasite taxon, inoculating dose, or interval between infections. I found evidence for the bottom-up and top-down mechanisms only as significant predictors of peak microparasite densities during coinfection. The resulting framework helps to reconcile apparently contradictory laboratory findings and suggests a template for future work in the within-host ecology of infectious disease.
Results
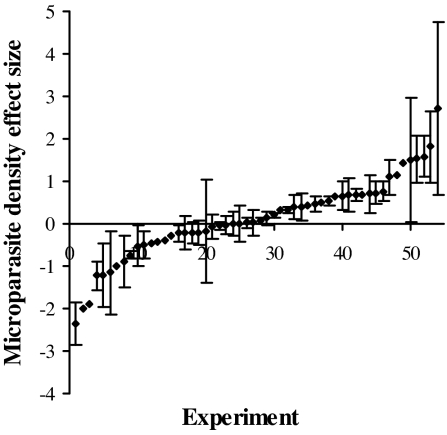
The metaanalysis first confirmed that effects of helminth coinfection vary significantly among species pairs, with 27% of coinfections resulting in no change, 23% resulting in reduced, and 50% in increased microparasite density (Fig. 1). In other words, the results quantitatively confirmed what qualitative biomedical reviews of coinfection biology (12–16) suggest, that there is no one general outcome observed when helminths and microparasites interact within a host. I next addressed the power of bottom-up and top-down ecological control mechanisms to explain differential outcomes and/or identify contexts in which to expect large growth of microparasite populations in helminth-coinfected hosts.
Fig. 1.
Effect sizes for changes in microparasite density induced by helminth coinfection, based on data from 54 experiments. For the 40 experiments for which data on variance were available, the 95% C.I. is depicted. Helminth coinfection resulted in significantly reduced microparasite density in 23% of experiments, increased density in 50%, and no effect (C.I. overlapping zero) in 27%.
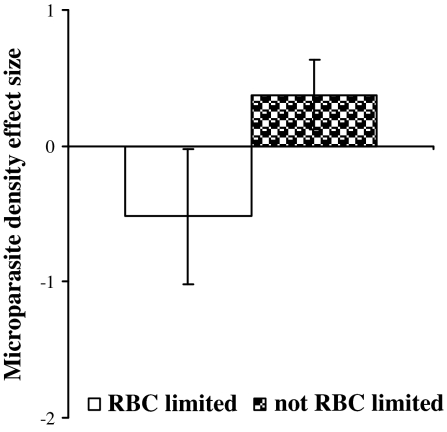
The analysis of bottom-up control revealed a general rule about how microparasites are affected by helminth coinfection. For species pairs without RBC limitation, microparasite density tended to be increased by helminth coinfection, whereas RBC limitation was associated with reduced peak microparasite density (Fig. 2). In other words, helminths that induce anemia reduced replication of microparasite species that require RBCs. In the absence of this effect, helminth coinfection led to significantly increased microparasite density. This conclusion was supported by weighted parametric analysis of the 40 experiments that provided variance estimates (Q1,38 = 5.90; P = 0.015) as well as unweighted nonparametric analysis of all 54 experiments (Q1,52 = 8.87; bootstrap P = 0.001).
Fig. 2.
Mean effect sizes for changes in microparasite density due to helminth coinfection, according to whether the helminth limits resource availability (here, host RBCs) for the microparasite. Data from 54 coinfection experiments were metaanalyzed to assess whether changes in peak microparasite density depended on helminth-induced RBC limitation (means ± bootstrapped 95% C.I. shown). Each pair of parasite species was identified a priori as possessing potential or no potential for RBC limitation (see Methods). RBC-limiting coinfection tended to decrease microparasite density, whereas microparasite density was increased when helminth coinfection did not impose RBC limitation for the microparasite.
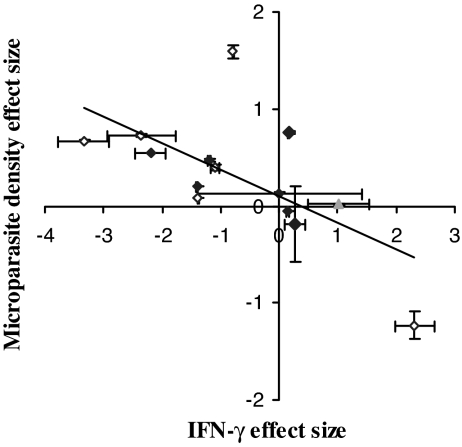
The analysis also revealed a rule about top-down control of microparasites during helminth coinfection, based on the 14 experiments for which data on microparasite densities and time-matched cytokines were available. Essentially, freedom from top-down control was associated with increased size of microparasite populations: the greater the helminth-induced reduction in microparasite-specific IFN-γ, the greater the increase in microparasite density (Fig. 3) (slope = −0.18 ± 0.05; Q1,12 = 13.54; P = 0.00023). Thus, helminth-induced changes in IFN-γ production by host cells significantly predicted the direction and magnitude of impact on microparasite density, across parasite taxa. However, changes in IFN-γ were not related to IL-4 production [see supporting information (SI) Text and SI Fig. 5].
Fig. 3.
Helminth-induced alterations in microparasite-specific immunity as a predictor of microparasite density effect size. IFN-γ is a potent immunological molecule involved in defense against microparasites. Metaanalysis of data from 14 experiments revealed a negative relationship between microparasite density and IFN-γ effect sizes induced by helminth coinfection (point estimate ± variance shown). The greater the helminth-induced reduction in microparasite-specific production of IFN-γ, the greater the increase in microparasite density. Host genotype [analyzed with A/J (gray triangle) omitted: tested for effects of C57BL/6 (filled diamonds) versus BALB/c (open diamonds) genotype] did not explain the pattern, and the pattern remained significant even when the data point at the bottom right was excluded from analysis.
Host genotypes (here, inbred strains within a species) did not differ in the efficiency with which they killed microparasites, nor did host sex, parasite taxa, dose, nor interval between infections influence control of microparasites during coinfection (see SI Text). Thus, of all predictor variables tested, only the bottom-up and top-down control factors of RBC limitation and IFN-γ production, respectively, appeared general enough to be predictive of outcome.
Discussion
Here, I show that resource limitation predictably decreased and suppression of inflammatory immune responses predictably increased microparasite population size in coinfected hosts. In other words, helminth-induced changes in RBC availability and IFN-γ production were predictive of whether microparasite density was decreased or increased by coinfection. Bottom-up and top-down ecological “rules” therefore detectably govern these microparasite populations (Fig. 4). In some senses, this is unsurprising; microparasites are organisms, after all, and there is no reason to suspect that they are exempt from ecological processes that shape the abundance of organisms in macroecosystems (e.g., refs. 17–20). Furthermore, IFN-γ is central to the clearance of virtually all microparasites, despite differences in the details of which cell types or anatomical location might be involved (27, 30). However, generalizations are rare in coinfection biology, and qualitative reviews of immunity to coinfection (12–16) never consider resource limitation when attempting to reconcile apparently contradictory findings of immunological studies. Indeed, in biomedicine, scientists tend to study resources or immune responses, but not both. There is thus a divide even among subdisciplines of biomedicine that is bridged here. The major contribution of this metaanalysis is the use of interdisciplinary thinking to reveal generalizations and to encourage holistic investigation of the within-host ecology of coinfection.
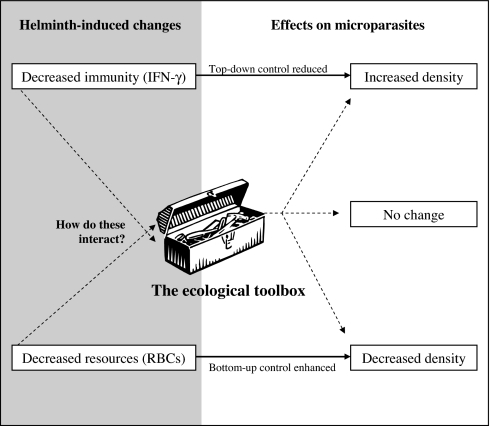
Fig. 4.
Conceptual diagram of a unified bottom-up/top-down ecological framework within which to analyze helminth–microparasite coinfection. Helminth-induced changes in immunity or resource availability (in gray region) have effects on microparasite density (in white region) that correspond to top-down and bottom-up ecological processes. For example, decreased production of the cytokine IFN-γ reduced top-down control, as evidenced by the increased density of microparasites (see Fig. 3). At the same time, decreased RBC density enhanced bottom-up control, as evidenced by the decreased density of microparasites requiring that cell type (see Fig. 2). The solid arrows correspond to these scenarios. However, how helminth-induced changes in immunity and resource availability might interact to shape microparasite populations is unknown, as represented by dashed lines. For example, in cases in which immunity fails to explain observed microparasite densities, might resource limitation be operating? In biomedicine, the two lines of inquiry are virtually never pursued together, whereas the science of ecology provides a conceptual and statistical/mathematical framework in which to understand interactions between bottom-up and top-down processes (represented by the ecological toolbox). Ecology may thus be able to explain and predict a wider range of coinfection outcomes than biomedical subdisciplines have yet managed, including cases in which helminth coinfection does not alter microparasite density or does so in apparently counterintuitive ways.
Strikingly, ecology provides a rigorous conceptual and statistical framework within which to analyze the relative strengths of top-down versus bottom-up control, as well as interactions or other nonlinearities in the mechanisms driving dynamics in within-host communities (Fig. 4). On the macroecosystem scale, for example, the strength of top-down control of Swedish foxes (20) and Panamanian caterpillars (19) has been shown to be strongest in the absence of bottom-up control: only when resources are not limiting do predators dominate prey dynamics. Parallel experimental methods (i.e., manipulating both resources and immunity) and statistical methods (i.e., explicitly testing for interactions) could reveal similar context dependencies in the strength of top-down control of microparasites during coinfection. Ecological methods can also identify which population control mechanism is most important for each species in an ecosystem. Zebras, for instance, appear to be primarily regulated by predation, whereas coherding wildebeest are regulated by the availability of food (18). Would analogous studies of parasite populations reveal that different, even closely related, parasite species are controlled within the host by distinct ecological mechanisms? Such information would be of both basic and applied scientific interest.
I found that the extent to which helminths reduced microparasite-specific production of IFN-γ was predictive of the direction and magnitude of effect on microparasite density (Fig. 3). However, I was unable to identify a general mechanism of IFN-γ suppression: neither antigen-specific nor polyclonal IL-4 was predictive of the observed changes induced by coinfection (see SI Text). Other anti-inflammatory cytokines such as the potent immunomodulator IL-10 (32) may also be involved in IFN-γ suppression. Indeed, differences among helminth species or inoculating doses in the propensity to induce immunomodulatory versus helminth-clearing cytokines (33) could differentially impact the strength of microparasite-specific IFN-γ responses and thus generate the x axis variability evident in Fig. 3. Too few coinfection studies to date have measured IL-10 to permit inclusion of that factor in this metaanalysis. In any case, there may be too many intermediate or system-specific steps between helminth-induced immune responses and their bystander effects on microparasites to be detected in such an analysis. What is remarkable here is that a detectable general rule about top-down control of microparasites during helminth coinfection emerged, despite the dynamic complexities of immune responses (34). It is important to highlight such cases of functional simplicity in complex systems, particularly in this era of high-throughput data collection in biomedicine (27).
At the same time, I found that helminths that caused anemia imposed resource limitation on RBC-dependent microparasites and thereby reduced their population size compared with what was observed during microparasite infection alone (Fig. 2). Given this RBC-based definition, bloodborne protozoa in phylum Apicomplexa (the parasites that cause babesiosis and malaria) dominated the resource-limited category. Still, neither kingdom nor phylum-level taxonomic distinctions among microparasites accounted for the pattern in Fig. 2, and when Apicomplexans were paired with helminths that did not induce anemia, their densities were increased, on average. Thus, the coarse-grained categories of resource limitation used here provided evidence of power to predict the direction of effects of helminth coinfection even within Apicomplexa. The results also support the hypothesis that the effect of resource limitation ought to be strongest for species that are averse to infection of the very young RBCs, or reticulocytes, that the host produces in homeostatic response to anemia (24) (see SI Text). It would be desirable to assess more general, and ideally quantitative, metrics of resource limitation in future studies: for example, competition for space among parasites (23) or for nutritional resources among host, helminths, and microparasites (35). Such data are not yet widely available in the published literature.
In summary, this analysis lends credence to the idea that the outcome of coinfection may be predictable from ecological first principles. The fact that the density of microparasites without resource limitation was positively rather than neutrally affected by helminth coinfection (Fig. 2) suggests that release of top-down control might have consequences only in the absence of resource limitation. This is exactly the sort of interaction that ecology is well poised to frame and address, as outlined above and in Fig. 4. These results also enable a priori prediction of which species combinations will result in explosive microparasite replication. The most explosive might involve a helminth that powerfully reduces IFN-γ responses but does not impose resource limitation for the microparasite species with which it is paired. Such predictions could be tested. Based on the RBC limitation framework, for example, a species pair that showed evidence of bottom-up but not top-down control could be introduced into animals with blood transfusion. Would alleviation of bottom-up control allow helminth-induced changes in IFN-γ to dominate dynamics? To enable formal assessment of the relative or context-dependent importance of each mechanism (see SI Text), it appears essential for immunological measurements in future studies to be routinely complemented by measurements of the resources essential to microparasite replication. Interestingly, both the bottom-up and top-down factors analyzed here appear to be direct host defense mechanisms: suppression of erythropoeisis and destruction of uninfected RBCs are part of the normal physiological response against malaria (36).
Unearthing generalities such as these ecological “rules” governing microparasite density during coinfection may be particularly useful in settings where not only pairwise but multivalent interactions among parasite species are common (5, 6). Community ecology has revealed the power of pairwise species interactions to illuminate community structure in a multispecies world, even if imperfectly. Similarly, understanding pairwise interactions in animal models of coinfection is a good starting point to render real-world complexity tractable. For example, once interactions are better understood in the laboratory, it will be more feasible to extend the findings to coinfected hosts in nature, including their nutritional challenges (35), seasonality in immunocompetence, or extrinsic factors that alter intensity of exposure to parasites (6, 37). Future metaanalyses may use data on laboratory mice to identify contexts in which helminths predictably alter the severity of microparasitic disease (refs. 5 and 6; also see the SI Text), the feasibility of drug treatment (38), or the efficacy of vaccination (2, 15). Of ecological and evolutionary interest, metaanalysis of coinfection might also reveal determinants of the species composition of parasite communities (11) and/or the probable consequences of selective regimens imposed by coinfection (9). Such analyses would complement ongoing, detailed biomedical investigations of system specificities and would arguably maximize the value of animal research. Viewing coinfection through the lens of community ecology may even reveal that coinfection is not as hopelessly complex as it first appears.
Methods
The data for this metaanalysis came from 54 experiments on helminth–microparasite coinfection in laboratory mice (see SI Tables 1 and 2). The range of infections included 9 helminth species and 28 microparasite species (6 viral, 8 bacterial, and 14 protozoal). Published studies were located via PubMed and ISI Web of Science searches on combinations of the following terms or truncated variants thereof: coinfection, concomitant infection, concurrent infection, polyparasitism, mixed infection, helminth, nematode, trematode, cestode, virus, bacteria, fungus, and protozoa. Electronic searches were followed by bibliographic searches of all retrieved articles.
Experiments were selected for inclusion in the analysis if all of the following were true: (i) the hosts were laboratory mice, whether inbred or outbred; (ii) the coinfection comprised a helminth–microparasite pairing; (iii) the helminth infection was given before or at the same time as, but not after, microparasitic infection; and (iv) microparasite density data as well as pathology or cytokine data, where available, were reported for both the control (i.e., microparasite alone) and experimental (i.e., coinfected) groups. However, experiments that were insufficiently quantitative (e.g., which pooled samples from individual mice) were excluded.
Summary statistics were gathered from tables, text, and figures of selected studies. To facilitate extraction of data from figures, scanned or pdf snapshot images were imported into Data Thief Digitizing Freeware for digital measurement. As for any metaanalysis, the data were constrained by the experimental protocols historically adopted, in this case, over 37 years of parasitological research. Infectious doses and the interval of time between infections thus varied among experiments. The dose of Schistosoma mansoni, for example, varied between 20 and 200 cercariae; still, most studies used 20–70 cercariae per mouse, and statistical analysis found no effect of dose on subsequent microparasite density. The interval of time between helminth and microparasite infection was remarkably consistent for a given helminth species and tended to follow the onset of chronicity, e.g., S. mansoni was present for 7–8 weeks before microparasite infection in nearly all cases. The net result was that the interval between infections was confounded with helminth species, but neither factor was a statistically significant predictor of microparasite density. Finally, to minimize the impact of differential infection kinetics among experiments, peak microparasite densities, rather than values observed on a given day, were analyzed. Cytokine data (IL-4 and IFN-γ concentrations measured after antigen-specific or polyclonal stimulation of lymphoid cells in vitro) were taken from the time point at which microparasite density peaked. This synchrony maximized the size of the dataset.
Data were standardized for metaanalytic comparison through calculation of log response ratios (31): lnRR = ln(xco + 1) − ln(xsolo + 1), where xco is mean microparasite density, pathology, or cytokine production in coinfected mice, and xsolo is the same metric in mice with microparasite infection alone. Thus, for example, effect sizes significantly greater than 0 correspond to increased microparasite density, disease severity, or cytokine production during coinfection. The response ratio was chosen because it represents proportionate change due to coinfection and is thus comparable across diverse data ranges; also, its statistical properties have been characterized (31).
Most statistical analysis was conducted by using MetaWin 2 statistical software (39) according to standard procedures (31, 40, 41). After effect sizes were calculated, it was essential to consider the “file-drawer” problem (42) that null results are less likely to be published than others. Data collated for this study were tested for publication bias, first via a test for correlation between effect sizes and sample sizes (39). For microparasite density and cytokine data, these were uncorrelated (Spearman's ρ = −0.22–0.13; P = 0.3–0.9). The second test for bias was Rosenthal's fail-safe number, an estimate of the number of nonsignificant, unpublished, or missing studies that would be required to change the conclusion. All fail-safe numbers exceeded the 5N + 10 rule (42): for example, the fail-safe for the microparasite density data was 590, more than double the 5N + 10 value of 275. The pathology data, however, gave evidence of publication bias. Although the fail-safe number (606) suggested no bias, the effect size–sample size correlation was significant (Spearman's ρ = 0.56; P = 0.013). This suggests that coinfection experiments resulting in increased disease severity were more likely to be published than others. Given this bias, the pathology data were excluded from further metaanalysis. All analyzed data conformed to normality assumptions.
Identification of the potential for RBC resource limitation was as follows. For a helminth to be considered RBC-limiting for the microparasite with which it was paired: (i) the helminth species must be known to induce anemia (e.g., by bloodfeeding or causing hemorrhage); and (ii) the microparasite species had to rely on RBCs for replication. When both (i) and (ii) were true, then the microparasite in the pair was considered RBC-limited (see SI Tables 1 and 2). If only one or neither was true, the pair was not considered RBC-limited. The RBC usage characteristics of helminth and microparasite species were identified via consultation of online parasitology and microbiology atlases, plus primary literature searches. The primary literature was particularly important for determining whether helminths cause anemia. Whenever possible, evidence was taken from the coinfection studies themselves. Helminths in genera Fasciola (43), Heligmosomoides (44), Nippostrongylus (45), Schistosoma (46), and Trichinella (47) were considered to alter RBC availability, whereas helminths in genera Echinostoma (48), Litomosoides (49), and Taenia (50) were not. In equivocal cases [e.g., Trichinella, which alters RBC availability by suppressing erythropoiesis rather than destroying mature RBCs (47)], statistical analysis was used to confirm that conclusions were robust to categorical reassignment.
Metaanalytic mixed models with fixed effects of categorical group or regression variable and random effects of studies (41) were applied to effect size data. For the subset of studies that provided data on within-study variability, P values were obtained by weighted parametric analysis of variance or regression. Separate, unweighted analyses included all effect size estimates, even from studies for which variance data were unavailable; P values for those analyses were obtained nonparametrically, via 999 bootstrap iterations (40, 41). Categorical models assessed whether host strain, host sex, parasite taxon, or the potential for RBC limitation were predictive of microparasite density during coinfection. Continuous models assessed the relationship between IL-4 and IFN-γ production as well as whether dose, time interval between infections, or cytokines were predictive of microparasite density. Weighted two-way generalized linear models (in SAS Systems 8.02) were then used to assess the relative importance of bottom-up and top-down factors as predictors of microparasite density during coinfection. Effect size data were weighted for two-way analysis by the inverse of each study's effect size variance (41).
Supplementary Material
ACKNOWLEDGMENTS.
I thank Q. Bickle (London School of Hygiene and Tropical Medicine, London), S. Borrow (The Edward Jenner Institute for Vaccine Research, Compton, UK), M. Edwards (Novartis Horsham Research Centre, Horsham, UK), N. Kumar (Johns Hopkins Bloomberg School of Public Health, Baltimore), A. Njunda (University of Buea, Buea, Cameroon), G. Noland (Johns Hopkins Bloomberg School of Public Health), M. Stevenson (Centre for the Study of Host Resistance, McGill University, Montreal, Canada), and Z. Su (Centre for the Study of Host Resistance) for sharing preprints and/or raw data; the librarians at the Universities of Edinburgh and Missouri at Columbia for assistance; A. Read, J. Allen, J. de Roode, G. Long, and T. Little for their commentary on draft manuscripts; and the Leverhulme Trust, the School of Biological Sciences, and the Biotechnology and Biological Sciences Research Council for fellowship support (Grant BB/D01977X/1).
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707221105/DC1.
References
- 1.Abu-Raddad LJ, Patnaik P, Kublin JG. Science. 2006;314:1603–1606. doi: 10.1126/science.1132338. [DOI] [PubMed] [Google Scholar]
- 2.Lello J, Boag B, Fenton A, Stevenson IR, Hudson PJ. Nature. 2004;428:840–844. doi: 10.1038/nature02490. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohani P, Green CJ, Mantilla-Beniers NB, Grenfell BT. Nature. 2003;422:885–888. doi: 10.1038/nature01542. [DOI] [PubMed] [Google Scholar]
- 5.Hotez PJ, Molyneux DH, Fenwick A, Ottesen E, Ehrlich Sachs S, Sachs JD. PLoS Med. 2006;3:e102. doi: 10.1371/journal.pmed.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mwangi TW, Bethony JM, Brooker S. Ann Trop Med Parasitol. 2006;100:551–570. doi: 10.1179/136485906X118468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Druilhe P, Tall A, Sokhna C. Trends Parasitol. 2005;21:359–362. doi: 10.1016/j.pt.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Nacher M. Trends Parasitol. 2006;22:350–351. doi: 10.1016/j.pt.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Read AF, Taylor LH. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- 10.Graham AL. Q Rev Biol. 2002;77:409–433. doi: 10.1086/344414. [DOI] [PubMed] [Google Scholar]
- 11.Pedersen AB, Fenton A. Trends Ecol Evol. 2007;22:133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Christensen NO, Nansen P, Fagbemi BO, Monrad J. Parasitol Res. 1987;73:387–410. doi: 10.1007/BF00538196. [DOI] [PubMed] [Google Scholar]
- 13.Cox FEG. Parasitology. 2001;122:S23–S38. doi: 10.1017/s003118200001698x. [DOI] [PubMed] [Google Scholar]
- 14.Hartgers FC, Yazdanbakhsh M. Parasite Immunol. 2006;28:497–506. doi: 10.1111/j.1365-3024.2006.00901.x. [DOI] [PubMed] [Google Scholar]
- 15.Kamal SM, El Sayed Khalifa K. Parasite Immunol. 2006;28:483–496. doi: 10.1111/j.1365-3024.2006.00909.x. [DOI] [PubMed] [Google Scholar]
- 16.Page KR, Scott AL, Manabe YC. Cell Microbiol. 2006;8:185–196. doi: 10.1111/j.1462-5822.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 17.Burkepile DE, Hay ME. Ecology. 2006;87:3128–3139. doi: 10.1890/0012-9658(2006)87[3128:hvncom]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Grange S, Duncan P. Ecography. 2006;29:899–907. [Google Scholar]
- 19.Richards LA, Coley PD. Oikos. 2007;116:31–40. [Google Scholar]
- 20.Elmhagen B, Rushton SP. Ecol Lett. 2007;10:197–206. doi: 10.1111/j.1461-0248.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- 21.Cappuccino N, Price PW, editors. Population Dynamics: New Approaches and Synthesis. San Diego: Academic; 1995. [Google Scholar]
- 22.Haydon DT, Matthews L, Timms R, Colegrave N. Proc R Soc Lond B. 2003;270:289–298. doi: 10.1098/rspb.2002.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts-Thomson IC, Grove DI, Stevens DP, Warren KS. Gut. 1976;17:953–958. doi: 10.1136/gut.17.12.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lwin M, Last C, Targett GAT, Doenhoff MJ. Ann Trop Med Parasitol. 1982;76:265–273. doi: 10.1080/00034983.1982.11687541. [DOI] [PubMed] [Google Scholar]
- 25.Ismail N, Bretscher PA. J Immunol. 1999;163:4842–4850. [PubMed] [Google Scholar]
- 26.Cervi L, MacDonald AS, Kane C, Dzierszinski F, Pearce EJ. J Immunol. 2004;172:2016–2020. doi: 10.4049/jimmunol.172.4.2016. [DOI] [PubMed] [Google Scholar]
- 27.Germain RN. Science. 2001;293:240–245. doi: 10.1126/science.1062946. [DOI] [PubMed] [Google Scholar]
- 28.Mohrs K, Harris DP, Lund FE, Mohrs M. J Immunol. 2005;175:5306–5313. doi: 10.4049/jimmunol.175.8.5306. [DOI] [PubMed] [Google Scholar]
- 29.Mosmann TR, Cherwinski HM, Bond MW, Giedlin MA, Coffman RL. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 30.Abbas AK, Murphy KM, Sher A. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 31.Hedges LV, Gurevitch J, Curtis PS. Ecology. 1999;80:1150–1156. [Google Scholar]
- 32.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Ann Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 33.Maizels RM, Balic A, Gomez-Escobar N, Nair M, Taylor MD, Allen JE. Immunol Rev. 2004;201:89–116. doi: 10.1111/j.0105-2896.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- 34.Frank SA. J Theor Biol. 2002;219:281–290. doi: 10.1006/jtbi.2002.3122. [DOI] [PubMed] [Google Scholar]
- 35.Koski KG, Scott ME. Annu Rev Nutr. 2001;21:297–321. doi: 10.1146/annurev.nutr.21.1.297. [DOI] [PubMed] [Google Scholar]
- 36.Evans KJ, Hansen DS, van Rooijen N, Buckingham LA, Schofield L. Blood. 2006;107:1192–1199. doi: 10.1182/blood-2005-08-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Booth M, Vennervald BJ, Kenty L, Butterworth AE, Kariuki HC, Kadzo H, Ireri E, Amaganga C, Kimani G, Mwatha JK, et al. BMC Infect Dis. 2004;4:13. doi: 10.1186/1471-2334-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu-Hua X, Utzinger J, Chollet J, Tanner M. Int J Parasitol. 2006;36:957–964. doi: 10.1016/j.ijpara.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg MS, Adams DC, Gurevitch J. MetaWin: Statistical Software for Meta-Analysis. Sunderland, MA: Sinauer Associates; 2000. Version 2.1.4. [Google Scholar]
- 40.Adams DC, Gurevitch J, Rosenberg MS. Ecology. 1997;78:1277–1283. [Google Scholar]
- 41.Gurevitch J, Hedges LV. Ecology. 1999;80:1142–1149. [Google Scholar]
- 42.Rosenthal R. Psych Bull. 1979;86:638–641. [Google Scholar]
- 43.Fagbemi BO, Christensen NO, Nansen P. Vet Parasitol. 1985;17:101–110. doi: 10.1016/0304-4017(85)90095-0. [DOI] [PubMed] [Google Scholar]
- 44.Baker NF. Exp Parasitol. 1955;4:526–541. doi: 10.1016/0014-4894(55)90041-2. [DOI] [PubMed] [Google Scholar]
- 45.McNeil KS, Knox DP, Proudfoot L. Parasite Immunol. 2002;24:15–22. doi: 10.1046/j.0141-9838.2001.00428.x. [DOI] [PubMed] [Google Scholar]
- 46.Lewinsohn R. Trans R Soc Trop Med Hyg. 1975;69:51–56. doi: 10.1016/0035-9203(75)90010-3. [DOI] [PubMed] [Google Scholar]
- 47.Ngwenya BZ. Parasite Immunol. 1982;4:197–207. doi: 10.1111/j.1365-3024.1982.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 48.Noland GS, Graczyk T, Fried B, Fitzgerald EJ, Kumar N. J Parasitol. 2005;91:944–948. doi: 10.1645/GE-456R.1. [DOI] [PubMed] [Google Scholar]
- 49.Graham AL, Lamb TJ, Read AF, Allen JE. J Infect Dis. 2005;191:410–421. doi: 10.1086/426871. [DOI] [PubMed] [Google Scholar]
- 50.Ansari A, Williams JF. J Parasitol. 1976;62:728–736. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.