Abstract
The process by which some eukaryotic organelles, for example the endomembrane system, evolved without endosymbiotic input remains poorly understood. This problem largely arises because many major cellular systems predate the last common eukaryotic ancestor (LCEA) and thus do not provide examples of organellogenesis in progress. A model is emerging whereby gene duplication and divergence of multiple “specificity-” or “identity-” encoding proteins for the various endomembranous organelles produced the diversity of nonendosymbiotically derived cellular compartments present in modern eukaryotes. To address this possibility, we analyzed three molecular components of the endocytic membrane-trafficking machinery. Phylogenetic analyses of the endocytic syntaxins, Rab 5, and the β-adaptins each reveal a pattern of ancestral, undifferentiated endocytic homologues in the LCEA. Subsequently, these undifferentiated progenitors independently duplicated in widely divergent lineages, convergently producing components with similar endocytic roles, e.g., β1 and β2-adaptin. In contrast, β3, β4, and all other adaptin complex subunits, as well as paralogues of the syntaxins and Rabs specific for the other membrane-trafficking organelles, all evolved before the LCEA. Thus, the process giving rise to the differentiated organelles of the endocytic system appears to have been interrupted by the major speciation event that produced the extant eukaryotic lineages. These results suggest that although many endocytic components evolved before the LCEA, other major features evolved independently and convergently after diversification into the primary eukaryotic supergroups. This finding provides an example of a basic cellular system that was simpler in the LCEA than in many extant eukaryotes and yields insight into nonendosymbiotic organelle evolution.
Keywords: adaptin, Rab, SNARE, trafficking, autogenous
Eukaryotic cells are characterized by internal membrane-bound compartments, which include the nucleus, mitochondria, plastids, peroxisomes, and organelles of the membrane-trafficking system. By contrast, prokaryotes (eubacteria and archaebacteria) generally lack such internal membranous organelles. Based on molecular (1), and paleontological evidence (2), prokaryotes most likely preceded the eukaryotes, making eukaryogenesis one of the most important developments in cellular history.
Two mechanisms have been proposed for the origin of novel organelles, i.e., organellogenesis. Mitochondria and plastids clearly derive from anciently captured α-proteobacteria and cyanobacteria, respectively, demonstrating that endosymbiosis played a pivotal role in organellar evolution (3, 4). This process also generated even more-complex membrane topologies in many algae by secondary enslavement (5). Mechanistic aspects of endosymbiosis, including genome reduction via transfer of genes to the host nucleus and organellar import and targeting of those gene products, are emerging from studies of organelles such as degenerate mitochondria (6) and nucleomorphs (5) that are at different stages in their transition from free-living organism to cellular compartment. However, some organelles appear to have evolved autogenously, from preexisting components in the protoeukaryote, without a significant contribution from endosymbiosis (7, 8). Understanding the process of autogenous organelle evolution is hampered by the lack of available intermediate forms because most basic eukaryotic cellular features appear established before the last common eukaryotic ancestor (LCEA) (9, 10). Incidents of proposed primitive simplicity in some eukaryotic lineages, with respect to mitochondria, peroxisomes, introns, and the Golgi complex, all now appear to be better explained via secondary loss (11–14), removing any potential transitional forms.
Organelles thought to have autogenous origins include those of the membrane-trafficking system (ref. 15 and references therein). The system encompasses the endoplasmic reticulum (ER), the Golgi complex, the plasma membrane, and a variety of endocytic compartments. Movement between compartments is accomplished by packaging of cargo into vesicular or tubular membranous carriers and delivery by fusion of the carrier membrane with the target organelle. The protein factors required by each transport pathway include SNAREs, coat proteins, and Rabs (16). SNAREs are coiled-coil trans-membrane proteins that function as components of the fusion machinery and also as identifiers in providing membrane specificity (17, 18). Rabs, members of the Ras small GTPase superfamily, regulate membrane fusion and coordinate specificity among the multiple transport pathways and between the factors responsible for membrane fusion (19). Coat proteins participate in formation of carriers through membrane deformation, cargo selection, and other interactions (16).
Many factors involved in membrane trafficking are the products of gene duplications, with each member of the protein family performing similar functional roles but at distinct cellular locations (16). Comparative genomic and phylogenetic analysis of Rabs (20), SNAREs (21, 22), vesicle coats (23, 24), and several other trafficking components (25), sampling a broad diversity of eukaryotes, resolved these protein families into clades that correspond to organelle-specific paralogues. Importantly, each paralogue clade encompasses the full diversity of the taxa sampled, in many cases from five of the six eukaryotic supergroups (26), strongly implying that the major organellar paralogue families of Rabs (20), SNAREs (22), and vesicle coats (27) were established before the divergence of the existing eukaryotic groups, i.e., before the LCEA.
Incorporation of phylogenetic data has allowed for a molecular model of autogenous organellogenesis to emerge explaining how a single ancestral endomembrane, formed by invagination of the plasma membrane, differentiated into a variety of chemically and topologically distinct membranes (8, 15, 28). In this model, an undifferentiated endomembrane organelle, possessing identity-encoding proteins, would have evolved into two or more distinct organelles via duplications of the genes encoding the identity proteins [supporting information (SI) Fig. 6]. As the organelles replicate at cell division, and the protein duplicates diverge in sequence and function over many generations, novel organelle identity is acquired (SI Fig. 6). Various authors have proposed vesicle coats (8), SNAREs (22), Qa-SNAREs or syntaxins (21), small GTPases (29), and Sec1/Munc18 (SM) proteins (30) as the evolutionarily critical identity-encoding protein. However, because organellar identity is likely to depend on multiple factors, and each of these protein families share the same basic evolutionary pattern of paralogous expansion before the LCEA, it is probable that they all contributed to the origins and evolution of novel endomembrane compartments.
If organellogenesis requires more than one protein family, and if the duplications giving rise to the organellar specific paralogues were not concurrent, then there would be periods when some protein families of an emerging organelle have duplicated although others have not. If, at that point, the protoeukaryotic lineage underwent a radiative speciation event, i.e., the eukaryotic “big bang” (31), then those proteins that had already duplicated would resolve into organelle-specific clades encompassing all taxa, whereas those proteins associated with the same organelles or pathway-specific complexes that had not yet duplicated, would resolve into lineage-specific clades (SI Fig. 6 A and C) or else would retain undifferentiated versions active at multiple organelles.
Given that the basic complement of eukaryotic membrane-trafficking organelles and major protein families were already established in the LCEA (10, 13, 20, 22, 27, 32), we focus here on a more comprehensive and critical phylogenetic analysis of three protein families of the endocytic system. Essentially, the endocytic system is composed of two interconnected pathways: (i) recycling machinery returning receptors and plasma membrane components to the cell surface and (ii) degradative machinery transporting molecules to the digestive organelle, i.e., the lysosome or vacuole. We performed phylogenetic analysis of endocytic syntaxins, Rab 5, and the large subunits of adaptin (AP) complexes from representatives of nearly all major lineages of eukaryotes for which genomes have been sequenced. In stark contrast to the evolutionary pattern observed for the other AP subunits and for the syntaxins and Rab paralogues specifically associated with other membrane-trafficking organelles, we have identified three endocytic proteins that have undergone gene duplications, coupled with convergent specialization in organellar location, substantially after the LCEA.
Results
Evolution of the Anterograde Endocytic Syntaxins (SynE).
In mammals, the SynE homologues, syntaxin 13 and syntaxin 7, are localized at the early endosome or late endosome/lysosome, respectively (33). In Saccharomyces cerevisiae, the syntaxin Pep12p acts at the prevacuolar compartment (PVC), whereas Vam3p acts at the vacuole (34). In Arabidopsis thaliana, AtPep12 (Syp21) acts at the PVC, whereas AtVam3 (Syp22) is localized at both the PVC and vacuole (35). Previous phylogenetic analyses showed that early and late endosomal syntaxins cluster in a single clade (21, 36). This topology distinguished the SynE subfamily from other endocytically associated syntaxins, such as Tlg2/Syntaxin 16, involved in post-Golgi transport, and marks SynE as one of five major syntaxin subfamilies that evolved before the LCEA.
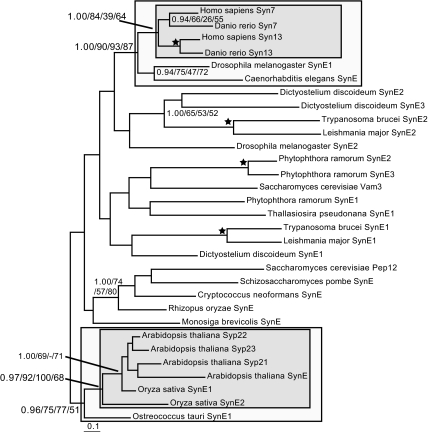
We sampled representative genomes from the five available eukaryotic supergroups (26) to obtain clear SynE homologues. Initial analysis to identify and remove long-branch sequences (SI Fig. 7) yielded a dataset that was then analyzed by four separate phylogenetic methods (Fig. 1). Surprisingly, SynE paralogues failed to resolve into organelle-specific clades but rather grouped into lineage-specific clades. For example, all flowering plant SynE homologues, including both Syp21 and 22, grouped together with the single green algal sequence as the immediate outgroup. In animals, syntaxins 13 and 7 are clearly the product of a duplication that took place after the proterostome/deuterostome divergence, i.e., entirely independently of the flowering plant duplications. The rest of the tree was poorly resolved. Nonetheless, at least two independent duplications of SynE, subsequent to the diversification of the major eukaryotic lineages but relatively early in each group, must have occurred.
Fig. 1.
Phylogenetic analysis of SynE genes with long-branch taxa removed, showing lineage-specific duplications in vertebrates and flowering plants. The inner box illustrates the timing of the duplication in those clades, whereas the outer box shows the well resolved preduplicates supporting that duplication. Nodes supporting clades of note are bolded. In this and subsequent phylogeny figures, node support values are given in the order of Bayesian posterior probabilities, PhyML bootstrap percentages, ML distance-corrected bootstrap percentages, and RaxML bootstrap percentages. Values are given for all nodes supported by >0.80 PP and >50% bootstrap support in two of three other methods. A star denotes a node supported by >0.95 posterior probability and >95% bootstrap support in two of three other methods. The − illustrates that the clade of interest was not reconstructed by the relevant method.
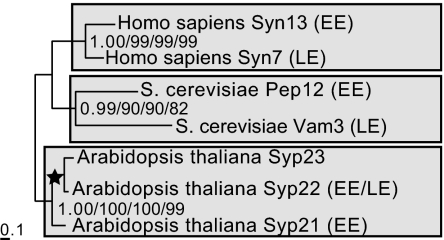
Because most SynEs have not been functionally characterized, their cellular locations remain uncertain. To explicitly test whether all SynE family members with known early endosomal or lysosomal/vacuolar locations resolve by organismal lineage or rather by location, we performed phylogenetic analysis on seven functionally characterized SynE homologues from three model organisms (Fig. 2). With very strong support, these SynE homologues resolve into lineage-specific clades, each containing syntaxins associated with both the early and late endocytic pathways. Thus, the current configuration of multiple endocytic syntaxins providing functionality for the degradative and recycling pathways is the result of gene duplications that occurred subsequent to the LCEA independently in animals, plants, and fungi. These syntaxin paralogues are derived from a common ancestral gene and have convergently evolved to localize and function at separate endocytic stages (SI Fig. 8A).
Fig. 2.
Phylogeny of SynE homologues that have been functionally characterized as associated with the early or late endocytic pathways. This clearly resolves the homologues into lineage-specific and not organellar clades. EE, associated with early endosomes or PVC; LE, associated with lysosome/vacuole.
Evolution of Early Endosomal Rabs (Rab 5).
The Rab families involved in recycling (Rabs 4, 5, 11) and degradative pathways (Rab 7) form organelle- or pathway-specific clades that evolved before the LCEA (20), with clear phylogenetic separation of Rab 7 and Rab 5 (37). Rab 5 mediates fusion of vesicles to early endosomes (38, 39). Trypanosome Rab 5A and Rab 5B localize to separate endosomal compartments and associate with distinct cargos (39). Rab 5 paralogues in yeast also appear to possess distinct functions, although some redundancy likely exists (40), and similarly nonredundant functionality has been described for mammalian Rab 5 isoforms (41). Previous phylogenetic analyses of Rab 5 paralogues from these three taxa suggested that the last common ancestor of this gene in trypanosomes, yeast, and humans are derived from a single Rab5 gene that was then duplicated in these lineages (42). However, the analysis did not encompass diverse eukaryotic taxa or use methods accounting for rate variation, an important factor that can cause artifacts in phylogenetic reconstruction (31).
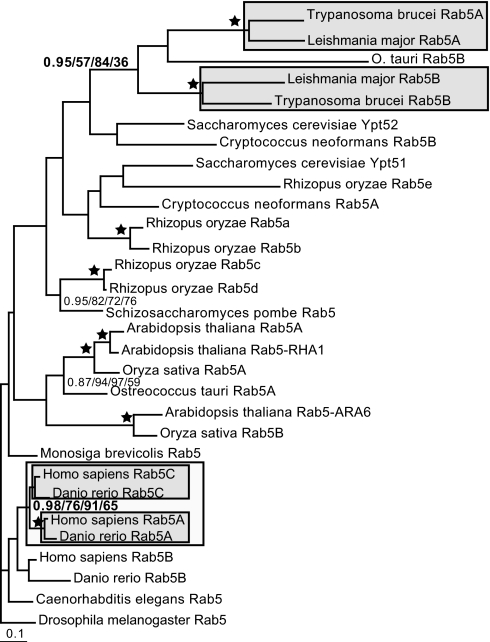
Sampling genomes from diverse taxa allowed us to build a more comprehensive Rab 5 dataset, which we subjected to phylogenetic analysis to identify and remove long-branch taxa (SI Fig. 9). Analysis of the resulting dataset shows that Rab 5 paralogues in vertebrates and in kinetoplastids (Fig. 3 and SI Fig. 10) are likely the result of independent lineage-specific duplications. Unlike the SynE proteins, the Rabs corresponding to the recycling (Rab 5) and degradative (Rab 7) pathways must have already been distinct genes very early in eukaryotic evolution and before the LCEA (20, 37). However, later lineage-specific functional expansion in the endocytic system appears to have continued for Rab 5 (SI Fig. 8B).
Fig. 3.
Phylogenetic analysis of Rab 5 sequences. Shown is the phylogenetic analysis of Rab 5 homologues with long-branch taxa removed. The independent duplication of Rab 5 homologues in vertebrates (node value in bold) with the duplicate clades marked by the inner box and the node supporting their monophyly marked by the outer box is shown. Similar duplications are likely to have occurred in the kinetoplastids (SI Fig 10); the placement of the O. tauri Rab 5B homologue is likely a result of long-branch attraction. Similar gene duplications may well have occurred in the fungi and Viridiplantae. However, in the latter two lineages, although the pattern is suggestive, it is not possible to state with certainty because of the lack of phylogenetic resolution.
Evolution of the β- and γ-Subunits of AP Complexes.
The AP complexes serve as cargo selectors for vesicles entering the endocytic system (43). There are four separate complexes, each composed of four subunits (small, medium, and the large β- and γ-subunits). These complexes derive from duplications of genes ancestrally related to the F-COP subcomplex of coatomer (44). The two large subunits are homologous, the product of an even more-ancient gene duplication (23). AP1, AP3, and AP4 function at the trans-Golgi network (TGN), and AP2 functions at the cell surface. Overall, all four complexes appear to have been present very early, and likely in the LCEA (23, 24, 45, 46, 47).
In mammals and yeast, separate β-subunits interact with the distinct AP1 and AP2 complexes (43, 47). Previous analyses of the β-subunits suggested that the evolutionary history of the β-subunits AP1 and AP2 may not be as simple as for the other AP complex components (23, 47). Whereas the large γ, the medium (υ), and the small (σ) subunits of AP1, AP2, AP3, and AP4 complexes resolve into discrete clades, the β-subunits form clear βAP3 and βAP4 clades but did not resolve separate β1 and β2 clades (23, 47). However, previous analyses did not include the full diversity of eukaryotes, and used methods susceptible to artifact (31). We therefore analyzed the phylogeny of the β-subunits of all four AP complexes with taxon sampling expanded to represent eukaryotic diversity. We also undertook phylogeny of the large γ-subunit to confirm that the other subunits had indeed diverged into four distinct complexes before the LCEA.
A broadly sampled dataset of the large γ-subunit was analyzed, allowing us to identify and remove long-branch sequences (SI Fig. 11). In the resulting dataset, the four AP complexes each formed separate clades encompassing the diversity of eukaryotes sampled, with support of 1.00/100%/100%/98%, 1.00/100%/100%/100%, 1.00/100%/100%/100%, and 0.99/79%/92%/87% for γ, α, δ, and ε clades, respectively (SI Fig. 12; support values given in same order as in figures). This finding robustly confirms that the duplications giving rise to AP complexes preceded the LCEA. Similar results were recently shown for the medium AP subunit (24, 46).
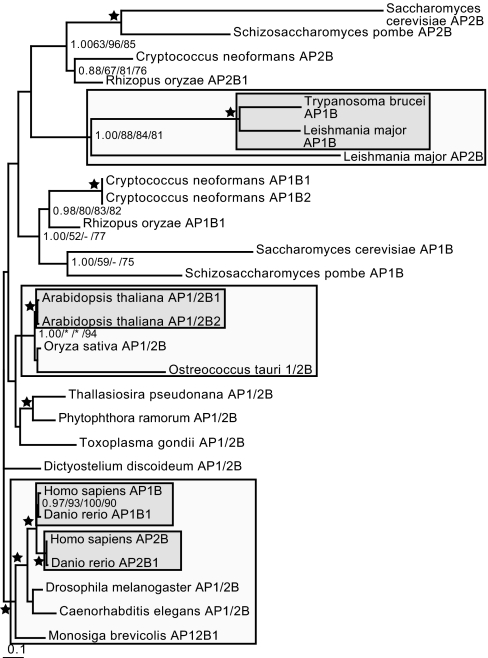
Upon phylogenetic analysis to remove long-branch taxa (SI Fig. 13), analysis of the β-subunits (SI Fig. 14) showed strongly resolved support for a joint β1/2 clade (1.00/93%/87%/94%) and separate clades of β3 (1.00/100%/100%/100%) and β4 (1.00/100%/100%/100%). Many taxa do not, in fact, possess separate β1 and β2 homologues (47) (SI Table 1). Phylogenetic analysis of the β1/2 sequences alone did not resolve clades corresponding to AP1 and AP2, instead showing lineage-specific duplications (Fig. 4). Within flowering plants, a duplication specific for A. thaliana was found, but Oryza sativa and Ostreococcus tauri each possess a single β1/2-subunit. In the kinetoplastids, a duplication into AP1 and AP2 before the divergence of Trypanosoma and Leishmania likely occurred. Even more clearly, in animals, the β1 and β2 paralogues duplicated after the proterostome/deuterostome divergence, because all other metazoa and sister lineages have a single paralogue; specifically the β1/2-subunits of the choanoflagellate Monosiga brevicolis and the proterostome invertebrates Caenorhabditis elegans and Drosophila melanogaster.
Fig. 4.
Phylogenetic analysis of the β1/2 clade. The lineage-specific duplicates are robustly resolved in Arabidopsis, in the vertebrates, and in the kinetoplastids (nodes shown in bold). The asterisks in the support values for the Viridiplantae denotes the fact that although the full clade of streptophytes and green algae is not reconstructed by PhyML and ML distance methods, a clade of the single O. sativa as an outgroup to the two A. thaliana duplicates is supported by 92% and 100%, respectively. This supports the conclusion of an independent duplication giving rise to β1- and β2-subunits in this lineage. Here, the inner box denotes the duplicate clades; the outer box illustrates the node supporting monophyly of the duplicates.
These data are most consistent with the duplications giving rise to all subunits of all AP complexes having occurred before the LCEA, with one exception. The β-subunit of AP1 and AP2 duplicated independently in multiple eukaryotic lineages, after their divergence from the common ancestor (SI Fig. 8C). Thus, it seems that in the LCEA, AP1 and AP2 shared a common β-subunit despite being fully differentiated with respect to all of the remaining subunits in each complex.
Discussion
The ancestral eukaryote appears to have been a remarkably complex cell, with mitochondria, peroxisomes, introns, and nearly all eukaryotic hallmarks, including an endomembrane system (10–12, 14). In particular, all major vesicle coats and the organelles associated with those coats (10, 13, 32), as well as aspects of a sophisticated endocytic system, were already established before the LCEA (27). Our new analysis of three endocytic components, SynE, Rab 5, and β-adaptin, by contrast, reveals features that differentiated much later.
Instead of respective paralogues clustering according to intracellular location, as observed for most paralogues functioning in membrane transport pathways originating before the LCEA, SynE, Rab 5, and β1/2 adaptin cluster by organismal lineage. It is not possible to reconstruct the exact number of endocytic homologues present in the LCEA, due in part to the lack of resolution in the deepest nodes of the phylogenies in Figs. 1 and 3 and in part to the formal possibility that other endocytic genes may have been present in this ancestor that have been subsequently lost in multiple lineages. However, the phylogenetic pattern observed is most consistent with there being fewer paralogues of these endocytic components in the ancestral eukaryote than there are in many eukaryotes today. Indeed, it is most simply explained by the presence of single members of the SynE, Rab 5, and β1/2 adaptin families in the LCEA that subsequently underwent several independent lineage-specific duplications (SI Fig. 8).
The products of the duplicate genes encoding SynE, Rab 5, and β1/2 adaptin subunit differ in their localizations and, for β-adaptin, inclusion into separate protein complexes. Thus, the evidence suggests that these genes also diverged similarly in function in each lineage, indicative of convergent evolution. Because these examples involve basic cellular machinery, they differ from gene duplications involving specialized tissue-specific machinery (e.g., syntaxins 1, 2, 3, and 4 and their plant equivalents, syp 111, 121, 131, and relatives) (48, 49) and also from cases such as SSO1 and SSO2 in yeast, which encode plasma-membrane syntaxins with no discernable difference in function (50).
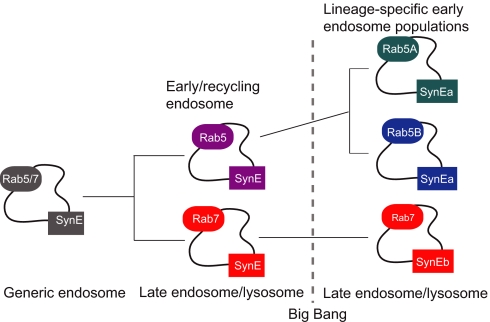
The endocytic system of the LCEA consisted, most parsimoniously, of an ancestral SynE homologue and at least one endocytic Rab (Fig. 5). Although the order in which the Rab subfamilies evolved from a single prototypic Rab protein is unknown, it is clear that Rab 5 and Rab 7, which today demarcate the early endosomes and lysosomes/vacuoles (19), were already present and distinct from one another (37). Thus, the LCEA differentiated recycling and degradative endocytic functions (Fig. 5). However, because the LCEA may have possessed only a single SynE homologue, this implies that the Rab 5 and Rab 7 compartments/subdomains were serviced by an undifferentiated ancestral SynE protein. The duplications giving rise to the endosomal and lysosomal/vacuolar SynE paralogues therefore occurred independently, with convergence of function in at least vertebrates, streptophytes, and ascomycetes. The ability of the A. thaliana AtVAM3 and AtPEP12 to complement vam3 and pep12 yeast mutants, respectively, are examples of experimentally demonstrated functional convergence (51, 52). Thus, in this respect, early endosomes and lysosomes/vacuoles were likely less distinct than they appear in some modern systems.
Fig. 5.
Evolution of endocytic organelles with rabs and syntaxins. The phylogenetic data here are most consistent with the endocytic system having evolved by the process of autogenous organelle evolution illustrated in SI Fig. 6 but being interrupted by the eukaryotic big bang. A single undifferentiated endocytic compartment would have initially been serviced by both undifferentiated endocytic rabs and syntaxins. The rabs duplicated and diverged before the eukaryotic big bang, however the endocytic syntaxins did not. Post-LCEA SynE and Rab 5 continued the trajectory of gene duplication and functional divergence, yielding increased specificity of function.
The duplications producing multiple Rab 5 paralogues also occurred independently in different lineages, implying continued evolution and specialization of the early endosomal system in many lineages subsequent to the LCEA (Fig. 5). That the β-adaptin duplications giving rise to distinct AP1 and AP2 subunits had not yet occurred in the LCEA suggests that the process of organellar differentiation begun by the other AP subunits was incomplete before the divergence of the major eukaryotic supergroups (SI Fig. 8). Consistent with this idea of evolutionary plasticity in the eukaryotic endocytic system are recent studies of dynamin that suggest that its role in endocytosis likely evolved convergently in animals and ciliates (24). The loss of key endocytic components in eukaryotic lineages and the novel endocytic adaptors in opisthokonts, such as GGAs, and epsins also suggest evolutionary lability in the endocytic machinery (45).
Understanding the process by which eukaryotic organelles arose is a fundamental aim of evolutionary biology. If a cellular system evolving by paralagous organellar expansion (8, 28) was interrupted by a speciation event, a phylogenetic pattern could result whereby some of the organelle-specific machinery would resolve into clades by cellular function and some by lineage (SI Fig. 6). This pattern is precisely what is observed for the endocytic factors analyzed here. One important step to understanding the origins of eukaryotic cell organelles is to reconstruct the LCEA by comparative analysis. To achieve this, one must differentiate between truly ancestral eukaryotic characters and convergently evolved features restricted to well studied model organisms. Here, we show that certain gene duplications during the evolution of the endocytic system, previously considered ancestral for all eukaryotes, actually occurred much later. It will be important to assess in more detail when the various pieces of the endocytic machinery evolved relative to the LCEA, emphasising a need for experimental work in multiple and evolutionarily disparate organisms to determine which details of endocytosis are common to all eukaryotic cells.
Methods
Sequence Sampling Strategy.
To ensure representation from the major eukaryotic supergroups, to allow taxonomic comparability between analyses of the various proteins, to avoid issues of paralogy, and to maintain taxon cardinality amenable to rigorous computational analysis, an a priori sampling strategy was used. Amino acid sequences of Rab 5, SynE, and the large AP subunits were retrieved from representatives of the five eukaryotic supergroups with completed genome sequences. Homology was assessed by BLAST (53) and reciprocal BLAST, whereby candidates retrieved the initial query or named homologues thereof with at least five orders of magnitude better than the next named homologue of a different subfamily. Otherwise, the putative candidate was excluded from analyses to avoid paralogy issues. Sequences were retrieved from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov), the Joint Genome Institute [(JGI) www.jgi.doe.gov/], or from the URLs given in brackets. Accession numbers for proteins used for the analyses can be found in SI Table 2.
The taxa included in preliminary phylogenetic analyses were A. thaliana (NCBI), C. elegans (NCBI), Chlamydomonas reinhardtii (JGI), Cryptococcus neoformans (NCBI), Cyanidioschyzon merolae (http://merolae.biol.s.u-tokyo.ac.jp), Danio rerio (NCBI), Dictyostelium discoideum (NCBI), D. melanogaster (NCBI), Entamoeba histolytica (NCBI), Giardia intestinalis (NCBI), Homo sapiens (NCBI), Leishmania major (www.genedb.org/), M. brevicolis (JGI), Naegleria gruberi (JGI), O. sativa (NCBI), O. tauri (JGI), Phytophthora ramorum (JGI), Plasmodium falciparum (NCBI), Rhizopus oryzae (JGI), S. cerevisiae (NCBI), Schizosaccharomyces pombe (NCBI), Thallasiosira pseudonana (JGI), and Trypanosoma brucei (NCBI). To maintain comparability between analyses, C. merolae, N. gruberi, T. thermophila, and C. reinhardtii, although sampled, were excluded, because in each case at least two of the four possible genes examined could not be confidently identified by the above criteria. Trichomonas vaginalis was also excluded because of massive expansion of its endocytic machinery (54).
Alignment and Phylogeny.
Amino acid sequences were aligned by using T_Coffee v.1.37 (55) and were manually adjusted. Only unambiguously aligned regions of homology were retained. Details of the datasets, including size and models of sequence evolution, are in SI Table 3. Alignments are available on request. For each protein, an initial phylogenetic analysis was performed to identify long-branch sequences, which were likely to contribute artifacts to subsequent analyses. These sequences were then removed, and a new mask was created, allowing for more alignment positions to be analyzed and for more-intense phylogenetic analysis involving Bayesian, two methods of protein maximum-likelihood (ML), and ML-corrected distance (MLD) methodologies. All datasets contained representatives from the five eukaryotic supergroups with sequenced genomes. Further subalignments were created and analyzed to test specific clade robustness (SI Fig. 10) and to test the resolution of functionally characterized homologues into either organelle- or lineage-specific groups (Fig. 2).
The model of sequence evolution for each dataset was determined by using Prot-Test version 1.3 (56) and incorporated corrections for rate variation and invariable sites where relevant. Trees were built by using MrBayes v.3.1.2 (57) for Bayesian analysis to determine optimal tree topology and posterior probability (PP) values for the nodes, with 1,000,000 Markov Chain Monte Carlo generations and the burn-in value determined graphically by removing trees before the plateau. ML bootstrap values were obtained by PhyML v.2.4.4 (58) and by RaxML (59) from 100 pseudoreplicate datasets. MLD bootstrap values were generated by using Fitch from the PHYLIP v.3.6a3 package (60) based on distance matrices calculated by Tree-Puzzle (61) and puzzleboot (http://hades.biochem.dal.ca/Rogerlab/Software/software.html) from 100 pseudoreplicate datasets. Apart from the datasets for SI Figs. 10 and 11, which were not analyzed by MLD for computational reasons, all datasets were analyzed by all four methods. Nodes with >0.95 posterior probability and >80% bootstrap support were considered robust, although, on tree figures, all nodes with support values >0.80 posterior probability and 50% bootstrap are shown.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Thomas Cavalier-Smith (University of Oxford, Oxford) for extensive critical and constructive comments on the manuscript and the many genome projects that made their data publicly available; and W. Ford Doolittle for comments on and support of early work on this project. This work was funded by a Wellcome Trust program grant (to M.C.F.) and a Wellcome Trust/Canadian Institutes of Health Research (CIHR) Joint Traveling Research Fellowship (to J.B.D.), as well as funding from the CIHR (to P.P.P.). We wish to acknowledge the use of the CamGrid computational resource on which these analyses were performed.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707318105/DC1.
References
- 1.Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, et al. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butterfield NJ. Paleobiology. 2000;26:386–404. [Google Scholar]
- 3.Gray MW, Doolittle WF. Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. New Haven, CT: Yale Univ Press; 1970. [Google Scholar]
- 5.Cavalier-Smith T. Curr Opin Microbiol. 2002;5:612–619. doi: 10.1016/s1369-5274(02)00373-9. [DOI] [PubMed] [Google Scholar]
- 6.van der Giezen M, Tovar J, Clark CG. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. Nature. 1975;256:463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 9.Dacks JB, Doolittle WF. Cell. 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 10.Roger AJ. Am Nat. 1999;154:S146–S163. doi: 10.1086/303290. [DOI] [PubMed] [Google Scholar]
- 11.Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. Mol Biol Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 12.Vanacova S, Yan W, Carlton JM, Johnson PJ. Proc Natl Acad Sci USA. 2005;102:4430–4435. doi: 10.1073/pnas.0407500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dacks JB, Davis LA, Sjogren AM, Andersson JO, Roger AJ, Doolittle WF. Proc Biol Sci. 2003;270(Suppl 2):S168–S171. doi: 10.1098/rsbl.2003.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Embley TM, Martin W. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 15.de Duve C. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 16.Bonifacino JS, Glick BS. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen YA, Scheller RH. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 18.Rothman JE, Wieland FT. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 19.Seabra MC, Wasmeier C. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Pereira-Leal JB, Seabra MC. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 21.Dacks JB, Doolittle WF. J Cell Sci. 2002;115:1635–1642. doi: 10.1242/jcs.115.8.1635. [DOI] [PubMed] [Google Scholar]
- 22.Yoshizawa AC, Kawashima S, Okuda S, Fujita M, Itoh M, Moriya Y, Hattori M, Kanehisa M. Traffic. 2006;7:1104–1118. doi: 10.1111/j.1600-0854.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 23.Schledzewski K, Brinkmann H, Mendel RR. J Mol Evol. 1999;48:770–778. doi: 10.1007/pl00006521. [DOI] [PubMed] [Google Scholar]
- 24.Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koumandou VL, Dacks JB, Coulson RM, Field MC. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 27.Dacks JB, Field MC. In: Organelles, Genomes and Eukaryote Phylogeny: An Evolutionary Synthesis in the Age of Genomics. Hirt RP, Horner DS, editors. London: CRC; 2004. pp. 309–334. [Google Scholar]
- 28.Dacks JB, Field MC. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- 29.Jekely G. Bioessays. 2003;25:1129–1138. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- 30.Arac D, Dulubova I, Pei J, Huryeva I, Grishin NV, Rizo J. J Mol Biol. 2005;346:589–601. doi: 10.1016/j.jmb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H. Proc R Soc London Ser B. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JJ, Leedale GF, Bradbury P, editors. The Illustrated Guide to Protozoa. Lawrence, Kansas: Society of Protozoologists; 2002. [Google Scholar]
- 33.Collins RF, Schreiber AD, Grinstein S, Trimble WS. J Immunol. 2002;169:3250–3256. doi: 10.4049/jimmunol.169.6.3250. [DOI] [PubMed] [Google Scholar]
- 34.Pelham HR. Curr Opin Cell Biol. 2002;14:454–462. doi: 10.1016/s0955-0674(02)00352-6. [DOI] [PubMed] [Google Scholar]
- 35.Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV. Mol Biol Cell. 2003;14:361–369. doi: 10.1091/mbc.E02-08-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dacks JB, Doolittle WF. Mol Biochem Parasitol. 2004;136:123–136. doi: 10.1016/j.molbiopara.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Langford TD, Silberman JD, Weiland ME, Svard SG, McCaffery JM, Sogin ML, Gillin FD. Exp Parasitol. 2002;101:13–24. doi: 10.1016/s0014-4894(02)00037-1. [DOI] [PubMed] [Google Scholar]
- 38.Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. J Cell Biol. 2000;151:539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pal A, Hall BS, Nesbeth DN, Field HI, Field MC. J Biol Chem. 2002;277:9529–9539. doi: 10.1074/jbc.M110055200. [DOI] [PubMed] [Google Scholar]
- 40.Singer-Kruger B, Stenmark H, Dusterhoft A, Philippsen P, Yoo JS, Gallwitz D, Zerial M. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez-Dominguez C, Stahl PD. J Biol Chem. 1999;274:11459–11462. doi: 10.1074/jbc.274.17.11459. [DOI] [PubMed] [Google Scholar]
- 42.Field H, Farjah M, Pal A, Gull K, Field MC. J Biol Chem. 1998;273:32102–32110. doi: 10.1074/jbc.273.48.32102. [DOI] [PubMed] [Google Scholar]
- 43.Robinson MS. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Duden R, Griffiths G, Frank R, Argos P, Kreis TE. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- 45.Field MC, Gabernet-Castello C, Dacks JB. In: Evolution of the Eukaryotic Endomembrane System and Cytoskeleton. Jekely G, editor. Vol 1. Austin, TX: Landes Bioscience Publishing; 2006. pp. 84–96. [Google Scholar]
- 46.Singh B, Gupta RS. Biochem J. 2004;378:519–528. doi: 10.1042/BJ20031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boehm M, Bonifacino JS. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leyman B, Geelen D, Quintero FJ, Blatt MR. Science. 1999;283:537–540. doi: 10.1126/science.283.5401.537. [DOI] [PubMed] [Google Scholar]
- 49.Low SH, Miura M, Roche PA, Valdez AC, Mostov KE, Weimbs T. Mol Biol Cell. 2000;11:3045–3060. doi: 10.1091/mbc.11.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aalto MK, Ronne H, Keranen S. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bassham DC, Gal S, da Silva Conceicao A, Raikhel NV. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato MH, Nakamura N, Ohsumi Y, Kouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Wada Y. J Biol Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- 53.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Notredame C, Higgins DG, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 56.Abascal F, Zardoya R, Posada D. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 57.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 58.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 59.Stamatakis A. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 60.Felsenstein J. Cladistics. 1993;5:164–166. [Google Scholar]
- 61.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.