Abstract
Friedreich's ataxia (FRDA) is a neurodegenerative disorder arising from a deficit of the mitochondrial iron chaperone, frataxin. Evidence primarily from yeast and mammalian cells is consistent with the hypothesis that a toxic hydroxyl radical generated from hydrogen peroxide (H2O2) via iron-catalyzed Fenton chemistry at least partially underlies the pathology associated with this disease. However, no whole-organism studies have been presented that directly test this hypothesis. We recently developed a Drosophila model that recapitulates the principal hallmarks of FRDA [Anderson PR, Kirby K, Hilliker A, Phillips JP (2005) Hum Mol Genet 14:3397–3405]. Using the Drosophila FRDA model, we now report that ectopic expression of enzymes that scavenge H2O2 suppresses the deleterious phenotypes associated with frataxin deficiency. In contrast, genetic augmentation with enzymes that scavenge superoxide is without effect. Augmentation of endogenous catalase restores the activity of the reactive oxygen species (ROS)-sensitive mitochondrial enzyme, aconitase and enhances resistance to H2O2 exposure, both of which are diminished by frataxin deficiency. Collectively, these data argue that H2O2 is an important pathogenic substrate underlying the phenotypes arising from frataxin deficiency in Drosophila and that interventions that reduce this specific ROS can effectively ameliorate these phenotypes. The therapeutic implications of these findings are clear and we believe warrant immediate clinical investigation.
Keywords: phenotypic rescue, catalase, Fenton chemistry, RNA interference
Friedreich's ataxia (FRDA) is a neurodegenerative disorder that arises from a genetic deficit of the mitochondrial iron chaperone protein, frataxin (1–3). The progressive loss of coordination of limb movements, dysarthria, nystagmus, scoliosis, and diabetes characteristic of FRDA usually manifest before adolescence. The peripheral nervous system (PNS) and the heart are among the most severely affected tissues, and more than half of those afflicted eventually succumb to cardiac-related complications (4–6).
Yeast, Drosophila, mouse, and cell culture models of frataxin deficiency have revealed important roles for frataxin and its homologs in mitochondrial iron storage (7–15), regulation of intracellular iron trafficking (7, 8, 10, 16, 17), iron-sulfur (Fe-S) cluster (16–21) and heme biogenesis (14, 22), and reactivation of the labile Fe-S cluster of mitochondrial aconitase (mACON) (22, 23). A role for frataxin in preventing formation of deleterious reactive oxygen species (ROS) has been well established (12, 15, 24), invoking a paradigm of FRDA pathology in which ROS toxicity leads to mitochondrial dysfunction with subsequent cell death (for review, see refs. 25 and 26).
The loss of intracellular iron chaperone capacity imposed by frataxin deficiency could elicit a Fenton chemistry-based mechanism of ROS toxicity. Elevated ROS production, including generation of the highly reactive hydroxyl radical, by iron-catalyzed Fenton chemistry with endogenous H2O2 as the substrate would be consistent with the susceptibility to H2O2 challenge noted in yeast and cell culture models of frataxin deficiency (3, 27–29). Yet no whole-organism animal studies have been presented that investigate the role of H2O2 in mediating frataxin deficiency phenotypes.
We recently reported the development of a Drosophila model of FRDA (17) that takes advantage of Gal4/UAS transgene-based RNAi-methodology to impose down-regulation of the Drosophila fraxatin homolog (Dfh) (30). In this model, Dfh suppression recapitulates the principal biochemical hallmarks of FRDA, including diminished activity of Fe-S-containing enzymes, susceptibility to iron toxicity, loss of intracellular iron homeostasis, and early-onset adult mortality.
The hypothesis that H2O2 plays a critical role in FRDA pathogenesis predicts that interventions that diminish the availability of this potential reactant will reduce the severity of at least some aspects of the disease. We tested the validity of this hypothesis by using the Drosophila model to overexpress a set of H2O2-scavenging enzymes [peroxisomal and mitochondrial catalases (CATs) and a mitochondrial peroxiredoxin] in DFH-deficient flies. This report details the outcome of that investigation.
Results
The Drosophila FRDA model used in this work employs the C96-Gal4 driver to reduce endogenous DFH via RNAi and to coordinately augment native levels of antioxidant enzymes (17). As reported earlier, DFH deficiency imposed by expression of UAS-DfhIR transgenes controlled by the C96-Gal4 driver confers marked early-onset adult mortality that is reflected as a 40% reduction in media adult life span (17). The C96-Gal4 driver promotes robust expression in the PNS and oenocytes with low-level expression in many other tissues (A.J.H., unpublished data). Use of this driver favors, but is not exclusively restricted to, expression in one of the two principal focal tissues of FRDA. Unlike drivers that promote robust widespread expression, the more focused expression of UAS-DfhIR by C96-Gal4 permits vigorous development and viable but short-lived adults (17).
CAT Restores Life Span.
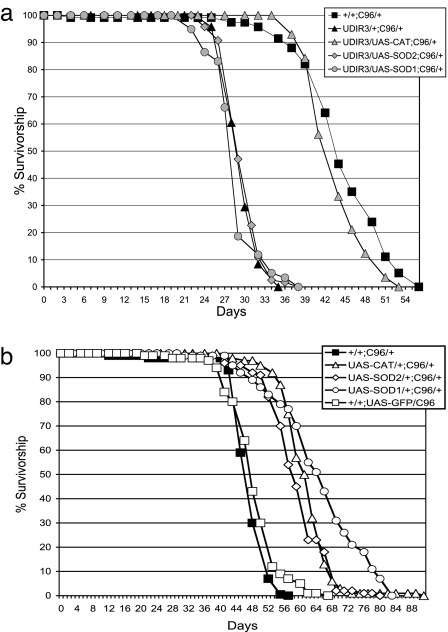
To investigate the role of H2O2 in the pathophysiology of DFH depletion, we first asked whether augmentation of CAT would suppress the early-onset mortality that characterizes flies deficient in DFH. In Drosophila, CAT localizes to the cytoplasm/peroxisomal compartment, and transgenic augmentation of CAT is known to enhance resistance to oxidative stress (31). Using the Gal4/UAS transgene expression system, we augmented flies deficient in DFH with Drosophila CAT. It should be noted that because CAT augmentation is confined essentially to the peripheral nervous system, its activity is not detectable above endogenous CAT activity in whole-fly extracts (data not shown). However, in previous studies (17), ubiquitous expression of the same UAS-Cat transgene used here elevated CAT activity by nearly 5-fold. As illustrated in Fig. 1a, the reduced life span of flies deficient in DFH is restored to control duration by expression of a UAS-CAT transgene. This result supports the proposed pathophysiological role of H2O2 and suggests that interventions that reduce this ROS have the potential to suppress the phenotypic outcomes of DFH deficiency.
Fig. 1.
CAT rescues life span in DFH-deficient Drosophila. (a) Augmentation of CAT, but not SOD1 or SOD2, restores life span in DFH-deficient Drosophila. (b) Expression of CAT, SOD1, and SOD2 in DFH-sufficient Drosophila. Survival of 125 males of each genotype on standard cornmeal food was followed at 25°C with enumeration and transfer of survivors to fresh bottles every 2–3 days. UDIR3 indicates UAS-DfhIR. C96 indicates C96 Gal4 driver. UAS-SOD1 indicates UAS-Cu,Zn-superoxide dismutase. UAS-SOD2 indicates UAS-Mn-superoxide dismutase. UAS-CAT indicates UAS-catalase. Note: Because CAT augmentation is confined essentially to the PNS, its activity is not detectable above background levels in whole-fly extracts (data not shown). However, in previous studies (17), ubiquitous expression of the same UAS-Cat transgene used here was shown to elevate CAT activity by nearly 5-fold.
Neither SOD1 nor SOD2 Restores Life Span.
We then asked whether the life span of DFH-deficient flies could be restored by attenuating the availability of a different ROS, namely superoxide. However, expression of the superoxide scavengers, SOD1 or SOD2, is without effect (Fig. 1a). This result is important for two reasons. First, it provides evidence against the possibility that the “rescue” observed with UAS-CAT (above) is a technical artifact arising from the reduced expression of UAS-DfhIR by competition for a limited pool of Gal4 (As described in a subsequent section, this point is reinforced through the use of a UAS/Gal4-independent CAT transgene). Second, and more importantly, it suggests that unlike H2O2, superoxide, whether in the cytoplasm (the domain of SOD1) or mitochondrial matrix (the domain of SOD2), is not directly involved in the life span-shortening effects of DFH deficiency.
As one of the controls for the above experiments, we examined the effects of overexpression of SOD1, SOD2, or CAT in DFH-sufficient flies by using the C96-Gal4 driver and were surprised to find that they all confer a modest but significant extension in life span compared with the control (Fig. 1b). However, when expressed in the context of DFH deficiency, only CAT restores the shortened life span phenotype. How DFH depletion suppresses the enhanced life span conferred by overexpression of SOD1 and SOD2 is not clear (see Discussion). The UAS-SOD1, UAS-SOD2, and UAS-CAT trangenes used in these studies all express their respective enzymatic activities at high levels in response to Gal4 induction (17).
Mitochondrial CAT Restores Life Span.
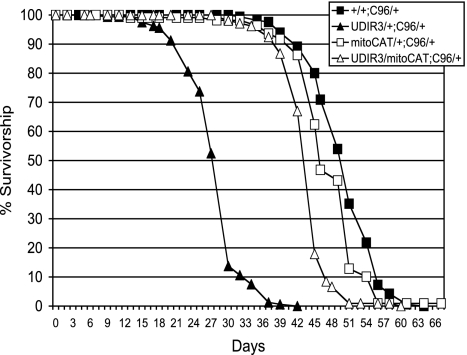
That overexpression of the H2O2-scavenging enzyme, CAT, restores the short life span of DFH-deficient Drosophila strongly implicates H2O2 as a critical element in the mechanism responsible for this early adult mortality phenotype. Although CAT provides robust restoration of life span in DFH-deficient flies, the disruption of iron metabolism arising from DFH deficiency should also occur in the mitochondrial matrix where DFH also resides (17, 32) and where mitochondrial superoxide emanating from the respiratory chain is reduced to H2O2 by SOD2 (33). In other words, all of the reactants for the generation of hydroxyl radical via Fenton chemistry should be present in the mitochondrial matrix of DFH-deficient flies, and enhanced scavenging of H2O2 in the mitochondrial matrix should equal or surpass that in the cytoplasm. However, unlike in mammals, CAT does not normally occur in the mitochondrial matrix of Drosophila. We therefore asked whether CAT ectopically expressed in the mitochondrial matrix would mitigate the life span-reducing effects of DFH deficiency. To determine the answer, we used a synthetic CAT transgene (mitoCAT) that targets CAT to the mitochondrial matrix via the 22-aa putative mitochondrial-targeting motif of the ornithine aminotransferase gene and that is expressed from the Drosophila Cat genomic promoter (34). This genomic promoter expresses in many adult tissues, including the adult PNS (35). As seen in Fig. 2, the mitoCAT transgene restores the life span of DFH-deficient flies to within 10% of controls. Thus, CAT expressed ectopically in mitochondria can restore the life span of flies deficient in DFH nearly as well as cytoplasmic/peroxisomal CAT. Moreover, the rescue of DFH deficiency by the mitoCAT transgene, with its native genomic promoter, reduces potential concerns about the potential confounding effects of multiple UAS transgenes. Overall, the mitoCAT data provide further support for the conclusion that H2O2 is an important substrate in the pathology of DFH deficiency and that enzymatic scavenging of this oxidant in the biological context of DFH insufficiency can suppress the development of some of the associated deleterious phenotypes.
Fig. 2.
mitoCAT rescues life span in DFH-deficient Drosophila. Note that mitoCAT does not extend life span in normal control adults. Survival of 125 males of each genotype on standard cornmeal food was followed at 25°C with enumeration and transfer of survivors to fresh bottles every 2–3 days. CAT activities in whole-fly extracts [normalized to +/+;C96/+ (100%)] were as follows: mitoCAT/+;C96/+ = 340% ± 20%; mitoCAT/UDIR3;C96/+ = 370% ± 20%. Note that CAT activity is not impaired in flies deficient in DFH. This finding is important because frataxin is required for heme synthesis, and CAT requires heme for activity. UDIR3 indicates UAS-DfhIR. C96 indicates C96-Gal4 driver. mitoCAT indicates mitochondrial catalase.
Mitochondrial Peroxiredoxin (mTPx) Restores Life Span.
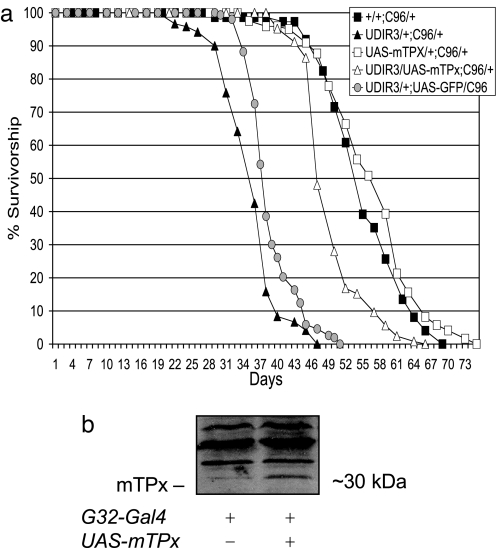
Among the reported biochemical consequences of DFH deficiency is the loss of iron homeostasis as reflected in the reduction in the level of the iron-binding protein, ferritin [FerHCH (17)]. Because CAT is a heme-containing protein, it is possible that the beneficial effects that accrue from the augmentation of CAT (or from the ectopic expression of mitoCAT) in DFH-deficient flies are not caused by the H2O2-scavenging activity of the enzyme but arise instead from the sequestering of potentially deleterious iron for the heme prosthetic groups required by the transgenic CAT. To examine this possibility, we overexpressed a H2O2-scavenging mTPx (36) in DFH-deficient flies. Because peroxiredoxins do not contain iron cofactors, restoration of life span by mTPx would argue against the possibility that supplemental CAT rescues by acting as a repository for reactive iron. Fig. 3 demonstrates that overexpression of UAS-mTPx restores the life span of the DfhIR flies to within 10% of controls. In support of the conclusion that H2O2 scavenging rather than iron sequestration underlies the observed rescue, we have also observed that chelation of dietary iron by bathophenanthrolinedisulfonic acid (37) does not improve life span of DFH-deficient flies (data not shown).
Fig. 3.
Mitochondrial peroxiredoxin (mTPx) restores life span in DFH-deficient Drosophila. (a) Survival of 125 males of each genotype at 25°C with enumeration and transfer of survivors to fresh bottles every 2–3 days. Note that expression of UAS-mTPx does not extend life span of normal control adults. (b) Mitochondrial extracts from control (genotype +/+; daG32/+) or adults (2–3 days old) expressing UAS-mTPx driven by the daG32-Gal4 driver were electrophoresed, transferred to nitrocellulose membranes, and probed for mTPx with anti-TPx IgG. Approximate molecular masses are indicated. G32-Gal4 indicates daG32 driver. C96 indicates C96 Gal4 driver. UDIR3 indicates UAS-DfhIR. UAS-mTPx indicates UAS-mitochondrial peroxiredoxin. Note that in b the daG32 driver was used to demonstrate functionality of the UAS-mTPx transgene because, unlike C96, it provides the level and breadth of mTPx expression required for immunoblot detection.
The issue raised earlier regarding possible confounding effects arising from the concurrent expression of multiple UAS transgenes can be seen in the data presented in Fig. 3a. As a control for this possibility, we expressed a control UAS transgene, UAS-GFP, along with UAS-DfhIR. In this experiment, the second transgene did exert a small positive impact on adult life span that likely arose not from the presence of GFP per se but from reduced expression of UAS-DfhIR through competition by UAS-GFP for Gal4. Although we believe that our interpretations of the effects of coexpression of UAS-CAT and UAS-mTPx with UAS-DfhIR are fundamentally correct, these data illustrate the potential for experimental artifacts with this system.
Adults Deficient in DFH Are Hypersensitive to H2O2.
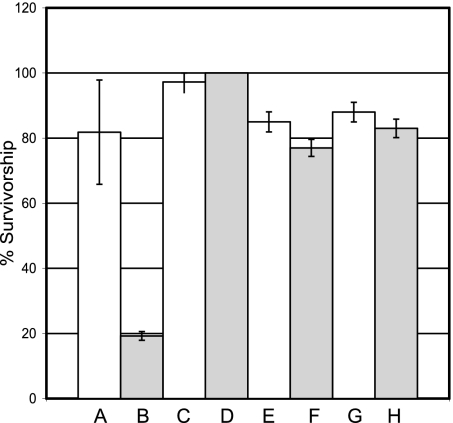
If endogenous H2O2 is an important factor in pathogenesis arising from DFH deficiency in Drosophila, we would predict that DFH-deficient flies would be hypersensitive to H2O2 exposure. Although susceptibility to H2O2 challenge typifies yeast and cell culture models of frataxin deficiency (3, 27–29), it has yet do be demonstrated in a whole-animal model of FRDA. Fig. 4 shows that DFH-deficient flies are indeed hypersensitive to H2O2 exposure. Moreover, this hypersensitivity can be alleviated by expression of the H2O2-scavenging enzymes CAT, mTPx, or mitoCAT.
Fig. 4.
DFH-deficient Drosophila are hypersensitive to H2O2 toxicity. Survival of males (7–9 days old) of each genotype on 1% sucrose containing 50 mM H2O2 was followed at 25°C with transfer of survivors to fresh vials every 12 h and final enumeration at 80 h. NB: +/+;C96/+ and UDIR3/+;C96/+ flies survive on 1% sucrose with no H2O2 beyond day 15 with little or no mortality. (A) +/+;C96/+, (B) UDIR3/+;C96/+, (C) mitoCAT/+;C96/+ (D) UDIR3/mitoCAT;C96/+, (E) +/+;UAS-mTPx/C96, (F) UDIR3/+;UAS-mTPx/C96, (G) UAS-CAT/+;C96/+, (H) UDIR3/UAS-CAT;C96/+. mitoCAT indicates mitochondrial catalase. UAS-mPTx indicates UAS-mitochondrial peroxiredoxin. UAS-CAT indicates UAS-catalase, C96 indicates the C96 Gal4 driver.
CAT Restores mACON Activity.
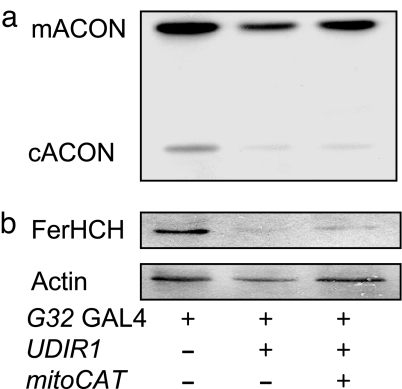
The aconitases are subject to reversible inactivation by ROS (38, 39). The vulnerability of the solvent-exposed Fe-S cluster of the these enzymes to ROS attack provides a sensitive indicator of changes in ROS flux in vivo (38, 39). To determine further whether ectopically expressed CAT actually reduces the ROS burden in DFH-deficient flies, we used the activities of mACON and cytoplasmic aconitase (cACON) to assess the level of ROS flux in DFH-deficient flies expressing mitoCAT. To allow a better visualization of the impact of mitoCAT on aconitase activity, we used flies carrying the daG32-Gal4 driver (Flybase: daG32 Gal4) because it provides more widespread expression of UAS transgenes than the C96-Gal4 driver, thereby exposing a greater proportion of the total aconitases in the fly to inactivation by the conditions imposed by DfhIR. In addition, the mitoCAT transgene was used to target the H2O2 scavenger to the mitochondrial matrix where mACON resides and, as above, to avoid possible Gal4 dilution artifacts that might occur in combination with the UAS-DfhIR transgene. As shown in Fig. 5a, the expression of mitoCAT restores mACON activity in DFH-deficient flies. In contrast, cACON (Fig. 5a) and the iron response protein/FerHCH couplet (Fig. 5b), both of which are constituents of the cytoplasmic compartment, remain unaffected.
Fig. 5.
mitoCAT restores the activity of mACON but not cACON in DFH-deficient Drosophila. (a) ACON activities in extracts of 2- to 3-day-old adult males were assayed after electrophoretic separation. (b) Total protein extracts of 2- to 3-day-old adult males were electrophoresed, transferred to nitrocellulose membranes, probed with anti-FerHCH antibodies, and then reprobed with anti-actin antibodies as a loading control. G32 GAL4 indicates daG32-Gal4 driver; mitoCAT indicates the mitoCat transgene; FerHCH indicates ferritin heavy-chain homolog. mACON activities normalized to the G32/+ control (100%) are: UDIR1/G32, 43 ± 2%; mitoCat/+;UDIR1/G32, 66 ± 2%. cACON activities normalized to the G32/+ control (100%) are: UDIR1/G32, 25 ± 6%; mitoCAT/+;UDIR1/G32, 28 ± 5%. ACON activities were compiled from densitometric scans of three separate experiments
Discussion
Deficiency of the mitochondrial iron chaperone, frataxin, is the primary molecular lesion in FRDA. Discovering how this deficiency leads to the clinical outcomes of the disease is urgent. Understanding the etiology of the disease has been aided by two salient findings, namely, that frataxin deficiency leads in turn to impaired intracellular iron homeostasis (7, 17, 40–42) and that oxidative stress plays a role as either a primary or secondary effector of FRDA pathology (for review, see refs. 25 and 26). At the outset of the present work, these and other findings had raised the possibility that at least part of the pathophysiology of FRDA arises from ROS (unspecified) emanating from impaired Fe-S-containing components of the respiratory chain. This work narrows the paradigm of ROS-mediated FRDA pathology with evidence from a whole-animal model that H2O2, specifically, is an important substrate in the pathology of frataxin deficiency, and perhaps most importantly, is an efficacious target for alleviating the deleterious effects of frataxin deficiency.
In an earlier study with our Drosophila FRDA model (17), we showed that widespread RNAi-mediated DFH deficiency with the ubiquitous Gal4 driver, daG32, caused delayed metamorphosis, impaired eclosion, and severe early mortality of the few surviving adults, and that overexpression of SOD1, SOD2, or CAT by the daG32-Gal4 driver did not improve the capacity of DFH-deficient larvae to develop into adults. In the present work, we have focused DFH deficiency on the adult PNS, which spares preadult effects, leaving reduced adult life span as the predominant phenotype. Using this system, we now report that overexpression of the H2O2-scavenging enzymes CAT, mitoCAT, or mTPx rescues the shortened life span brought about by DFH deficiency (Figs. 1–3). Each of these enzymes is known to be cytoprotective against H2O2 when overexpressed in either whole flies or in Drosophila cells in culture (31, 36, 43). Furthermore, although all of these enzymes reduce H2O2 to water, mTPx overexpression was not as effective as CAT or mitoCAT in rescuing the life span of DFH-deficient Drosophila. Differential levels of expression of the enzymes could underlie this difference, or the differential rescue could be caused by the reliance of mTPx on a secondary substrate, reduced thioredoxin, not required by either CAT or mitoCAT. Further to this point, we should add a cautionary note against comparing the relative “efficiencies” of rescue by the three enzymes. The domain of expression of these transgenes was very different: mitoCAT and mTPx were expressed ubiquitously, whereas cytoplasmic CAT was confined to the PNS, and we do not know what the relative expression of mitoCAT and mTPx actually was in the PNS.
Two ancillary observations merit comment. First, augmentation of SOD1, SOD2, or CAT provides a modest extension of life span in DFH-sufficient adults (Fig. 1b); however, of these three, only augmentation with CAT (and mitoCAT and mTPX) rescues the early mortality brought about by DFH deficiency. Second, the extended life span provided by augmentation with SOD1 or SOD2 (but not by CAT) in DFH-sufficient adults is suppressed in DFH-deficient adults (Fig. 1a). We propose that these apparently disparate observations share a common mechanistic origin. Resistance of DFH-deficient adults to rescue by augmentation with SOD1 or SOD2 argues that superoxide is not a component of the pathophysiological mechanism of DFH deficiency, which means that although DFH deficiency may suppress overall respiratory chain activity through the impaired synthesis and restoration of essential Fe-S clusters (17, 32), such suppressed respiratory chain activity is likely not accompanied by an increased flux of superoxide. If it were, augmentation with SOD2 and/or SOD1 would have restored life span of DFH-deficient adults. By this reasoning, the life span extension of DFH-sufficient adults by augmentation with SOD1 or SOD2 would appear to require a normally functioning respiratory chain. In this context, it might be informative in future experiments to examine the impact of SOD1 or SOD2 augmentation on cytoplasmic and mitochondrial aconitase activities in DFH-deficient flies. In keeping with our observations, it has been reported that increased expression of SOD1 and exposure to a small-molecule SOD2 mimetic does not improve murine FRDA cardiomyopathy (44).
It has recently been demonstrated that FRDA patient lymphoblasts exhibit increased generation of H2O2 (45). Although it is uncertain whether the deleterious phenotypes arising in the Drosophila model of FRDA are caused by increased levels of H2O2, increased susceptibility to H2O2-mediated damage, or a combination of these factors, it is clear that increasing H2O2-scavenging capacity improves pathology (as measured by life span). It is possible that the observed improvement in pathology is due at least in part to increased cellular energy production. The observed restoration of mACON activity in DfhIR flies expressing mitoCAT implies that the loss of mACON activity (and perhaps loss of mitochondrial respiratory complex activity) reported in ref. 17 could be at least partially caused by H2O2-mediated oxidative damage to iron cofactors. The exquisite sensitivity of the mACON Fe-S cluster to frataxin deficiency has been well established (17, 23, 32, 46). Thus, the increased activity of ACON by H2O2 scavenging may foreshadow discovery of protection of other iron cofactor enzyme activities important in electron transport and oxidative phosphorylation and improve the noted decrease in ATP production associated with FRDA (47).
Although H2O2 scavenging improved pathology, there were aspects of the phenotype not improved by this intervention. cACON activity levels and FerHCH levels were unaffected by H2O2-scavenging capacity, indicating that intracellular iron homeostasis was not impacted (Fig. 5b). These data underscore the complex nature of frataxin deficiency phenotypes and likely underlie the inability of increased mTPx and mitoCAT levels to correct for DFH deficiency completely (Figs. 2 and 3). Furthermore, these data underscore the need for investigation of combinatorial therapies addressing the many symptoms associated with FRDA.
FRDA manifests its most debilitating effects in the PNS and the heart. For this reason, we chose to use the C96-Gal4 driver as a central element of the Drosophila model of FRDA. Although C96, like most Gal4 drivers, does not exhibit strict tissue specificity, the results of its use in this work are nominally consistent with the notion that, as in mammals, the PNS of Drosophila is particularly susceptible to frataxin deficiency. To test further the equivalence of tissues sensitive to frataxin deficiency in mammals and Drosophila, we recently examined the consequences of RNAi-mediated Dfh deficiency by using Gal4 drivers that focus expression in several other tissues, including the motor neurons, skeletal muscle, and the heart. Of these tissues, only frataxin deficiency in the heart has thus far proved to be deleterious in the Drosophila model (P.R.A. and J.P.P., unpublished data). If confirmed, it will be of interest from a therapeutic perspective to determine whether the symptoms arising from cardiac-focused DFH deficiency can be also be mitigated by H2O2 scavenging.
Our results underscore the importance of discriminating between specific ROS involved in disease pathology. Several potential therapeutics for FRDA are in advanced clinical trials (48). Among the most promising are CoQ10 (ubiquinone) and the parabenzoquinone derivatives idebenone and MitoQ, which are designed to scavenge or reduce escape of electrons from complex I and complex II of the electron transport chain, which leads in turn to the formation of ROS. Our results support continued development of ROS-based therapeutics and suggest that a new focus on H2O2 may be clinically rewarding.
Materials and Methods
Drosophila Stocks and Culture Methods.
Drosophila stocks were maintained at 25°C on standard cornmeal agar medium unless otherwise stated. CO2 was used to anesthetize adult flies throughout, allowing at least 5 h of recovery at room temperature before the onset of experimentation to avoid potential latent effects of the anesthesia (49). The broadly expressing daG32-Gal4 driver (Flybase: daG32 Gal4) and the C96-Gal4 (50) driver (which promotes robust expression in the PNS on a background of widespread, low-level expression) were originally obtained from G. Boulianne (University of Toronto, Toronto, ON). UAS-Sod1, UAS-Cat, UAS-Sod2, and UAS-Dfh Inverted-Repeat UDIR1 flies were developed in this laboratory as described in ref. 17; UAS-mTPx was generated in the Orr laboratory using similar methods. Expression of UDIR3 leads to phenotypes as severe, or more severe, than those reported for UDIR1 (17). UAS-GFP flies were obtained from the Bloomington Stock Center. To avoid potential recessive effects, Gal4 drivers, UAS constructs, and the mitoCat transgene were used in hemizygous configuration for all experiments. Genetic backgrounds containing the C96-Gal4 driver, UDIR3, and either a second UAS transgene or mitoCAT were obtained by first generating UDIR3/SM5;C96/TM3 males that were then crossed to the appropriate UAS transgene or mitoCAT females. Controls for these genetic backgrounds were generated by substituting the second chromosome of the UDIR3 w1 progenitor stock for that containing UDIR3 in the above crossing scheme. The genetic backgrounds containing the daG32-Gal4 driver, UDIR1, and mitoCAT were obtained by first generating mitoCAT/SM5;daG32/TM3 males that were subsequently crossed to UDIR1 females. Controls for this genetic background were generated by substituting the second chromosome of the mitoCAT progenitor stock for that containing mitoCAT in the above crossing scheme. mitoCAT/SM5;daG32/TM3 and UDIR3/SM5;C96/TM3 stocks were expanded and briefly maintained by crossing males of these genotypes to Xa/SM5;TM3.
Western Immunoblotting.
Preparation of Drosophila samples for Western immunoblotting was performed as described in ref. 51. For this and all other procedures, protein levels were determined with the Bio-Rad protein assay. Samples were separated on 4% stacking, 15% separating SDS/polyacrylamide gels. The protein was transferred to Hybond-ECL nitrocellulose membrane (Amersham Pharmacia), blocked with 3% gelatin, and probed with appropriate primary antibody and secondary antibody conjugates. Rabbit polyclonal antibodies against the Drosophila ferritin heavy-chain homolog (a gift from F. Missirlis), or peroxiredioxin DPX-4783 (from W.C.O., described in ref. 36) were used in combination with goat anti-rabbit IgG horseradish peroxidase conjugate (Stressgen) and detected with ECL Western blotting detection reagents (Amersham Pharmacia). Mouse anti-actin monoclonal IgM antibodies (Developmental Studies Hybridoma Bank) were used in combination with anti-mouse IgM alkaline phosphatase conjugate and detected according to the manufacturer's instructions (Sigma). Prestained protein ladders (Invitrogen) were run in parallel with samples to estimate protein molecular weight.
Catalase Assay.
Catalase activity was determined spectrophotometrically by following H2O2 breakdown at 230 nm as described in ref. 52.
ACON Assay.
Mitochondrial and cytoplasmic ACON activities in whole-animal extracts were assayed jointly after electrophoretic separation (51).
Life Span Determinations.
Survival of 125–150 adult males on standard cornmeal medium was followed at 25°C with transfer of survivors to fresh bottles every 2–3 days. The life span data shown are representative of three to five independent replicates.
H2O2 Toxicity.
Survival of 7- to 9-day-old males in groups of 10 was assessed on 1% sucrose or 1% sucrose fortified with 50 mM H2O2 at 25°C. Sucrose solution (450 μl) was absorbed into eight layers of 1.5-cm2 squares of KimWipe placed in standard fly vials. Vials were changed every 48 h with enumeration between 80 and 90 h.
ACKNOWLEDGMENTS.
This work was supported by grants from the Friedreich's Ataxia Research Alliance and the Natural Sciences and Engineering Research Council of Canada (to J.P.P.), by a grant from the Canadian Institutes of Health Research (to J.P.P. and A.J.H.), and by a grant from the National Institutes of Health (to W.C.O.). P.R.A. is the recipient of a Doctoral Research Award from the Canadian Institutes of Health Research and a Postgraduate Fellowship Award from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, et al. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 2.Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M. Nat Genet. 1997;16:345–351. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 3.Tan G, Chen L, Lonnerdal B, Gellera C, Taroni FA, Cortopassi GA. Hum Mol Genet. 2001;10:2099–2107. doi: 10.1093/hmg/10.19.2099. [DOI] [PubMed] [Google Scholar]
- 4.Delatycki MB, Williamson R, Forrest SM. J Med Genet. 2000;37:1–8. doi: 10.1136/jmg.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel PI, Isaya G. Am J Hum Genet. 2001;69:15–24. doi: 10.1086/321283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alper G, Narayanan V. Pediatr Neurol. 2003;28:335–341. doi: 10.1016/s0887-8994(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 7.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 8.Radisky DC, Babcock MC, Kaplan M. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- 9.Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. Am J Hum Genet. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen OP, Kaplan J. FEBS Lett. 2001;509:131–134. doi: 10.1016/s0014-5793(01)03137-4. [DOI] [PubMed] [Google Scholar]
- 11.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 12.Park S, Gakh O, Mooney SM, Isaya G. J Biol Chem. 2002;277:38589–38595. doi: 10.1074/jbc.M206711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichol H, Gakh O, O'Neill HA, Pickering IJ, Isaya G, George GN. Biochemistry. 2003;42:5971–5976. doi: 10.1021/bi027021l. [DOI] [PubMed] [Google Scholar]
- 14.Park S, Gakh O, O'Neill HA, Mangravita A, Nichol H, Ferreira GC, Isaya G. J Biol Chem. 2003;278:31340–31351. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- 15.O'Neill HA, Gakh O, Park S, Cui J, Mooney SM, Sampson M, Ferreira GC, Isaya G. Biochemistry. 2005;44:537–545. doi: 10.1021/bi048459j. [DOI] [PubMed] [Google Scholar]
- 16.Chen OP, Hemenway S, Kaplan J. Proc Natl Acad Sci USA. 2002;99:12321–12326. doi: 10.1073/pnas.192449599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson PR, Kirby K, Hilliker A, Phillips JP. Hum Mol Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 18.Foury F. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- 19.Mühlenhoff U, Richhard N, Ristow M, Kispal G, Lill R. Hum Mol Gen. 2002;11:2025–2036. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- 20.Mühlenhoff U, Richhard N, Gerber J, Lill R. J Biol Chem. 2002;277:29810–29816. doi: 10.1074/jbc.M204675200. [DOI] [PubMed] [Google Scholar]
- 21.Mühlenhoff U, Gerber J, Richhard N, Lill R. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesuisse E, Santos R, Matzanke BF, Knight SAB, Camadro J, Dancis A. Hum Mol Genet. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 23.Bulteau AL, Lundberg KC, Ikeda-Saito M, Isaya G, Szweda LI. Proc Natl Acad Sci USA. 2005;102:5987–5991. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson RB, Roof DM. Nat Genet. 1997;16:252–257. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 25.Pandolfo M. Arch Neurol. 1999;56:1201–1208. doi: 10.1001/archneur.56.10.1201. [DOI] [PubMed] [Google Scholar]
- 26.Puccio H, Koenig M. Curr Opin Genet Dev. 2002;12:272–277. doi: 10.1016/s0959-437x(02)00298-8. [DOI] [PubMed] [Google Scholar]
- 27.Foury F, Cazzalini O. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 28.Wong A, Yang J, Cavadini P, Gellera C, Lonnerdal B, Taroni F, Cortopassi G. Hum Mol Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 29.Sturm B, Bistrich U, Schranzhofer M, Sarsero JP, Rauen U, Scheiber-Mojdehkar B, de Groot H, Ioannou P, Petrat F. J Biol Chem. 2005;280:6701–6708. doi: 10.1074/jbc.M408717200. [DOI] [PubMed] [Google Scholar]
- 30.Cañizares J, Blanca JM, Navarro JA, Monrós E, Palau F, Moltó MD. Gene. 2000;256:135–142. doi: 10.1016/s0378-1119(00)00343-7. [DOI] [PubMed] [Google Scholar]
- 31.Orr WC, Sohal RS. Arch Biochem Biophys. 1992;297:35–41. doi: 10.1016/0003-9861(92)90637-c. [DOI] [PubMed] [Google Scholar]
- 32.Llorens JV, Navarro JA, Martinez-Sebastian MJ, Baylies MK, Schneuwly S, Botella JA, Molto MD. FASEB J. 2007;21:333–344. doi: 10.1096/fj.05-5709com. [DOI] [PubMed] [Google Scholar]
- 33.Fridovich I. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 34.Kwong LK, Mockett RJ, Bayne AC, Orr WC Sohal RS. Arch Biochem Biophys. 2000;383:303–308. doi: 10.1006/abbi.2000.2093. [DOI] [PubMed] [Google Scholar]
- 35.Klichko VI, Radyuk SN, Orr WC. Arch Insect Biochem Physiol. 2004;56:34–50. doi: 10.1002/arch.10142. [DOI] [PubMed] [Google Scholar]
- 36.Radyuk SN, Sohal RS, Orr WC. Biochem J. 2003;371:743–752. doi: 10.1042/BJ20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missirlis F, Holmberg S, Georgieva T, Dunkov B, Rouault TA, Law JH. Proc Natl Acad Sci USA. 2006;103:5893–5898. doi: 10.1073/pnas.0601471103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner PR. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 39.Missirlis F, Hu J, Kirby K, Hilliker AJ, Rouault TA, Phillips JP. J Biol Chem. 2003;278:47365–47369. doi: 10.1074/jbc.M307700200. [DOI] [PubMed] [Google Scholar]
- 40.Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Nat Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 41.Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA. Hum Mol Genet. 2003;12:3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 42.Lu C, Cortopassi G. Arch Biochem Biophys. 2007;457:111–122. doi: 10.1016/j.abb.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mockett RJ, Bayne AC, Kwong LK, Orr WC, Sohal RS. Free Radic Biol Med. 2003;34:207–217. doi: 10.1016/s0891-5849(02)01190-5. [DOI] [PubMed] [Google Scholar]
- 44.Seznec H, Simon D, Bouton C, Reutenauer L, Hertzog A, Golik P, Procaccio V, Patel M, Drapier JC, Koenig M, Puccio H. Hum Mol Genet. 2005;14:463–474. doi: 10.1093/hmg/ddi042. [DOI] [PubMed] [Google Scholar]
- 45.Napoli E, Taroni F, Cortopassi GA. Antioxid Redox Signal. 2006;8:506–516. doi: 10.1089/ars.2006.8.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulteau AL, O'Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 47.Lodi R, Cooper JM, Bradley JL, Manners D, Styles P, Taylor DJ, Schapira AHV. Proc Natl Acad Sci USA. 1999;96:11492–11495. doi: 10.1073/pnas.96.20.11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson RB. Semin Pediatr Neurol. 2006;13:166–175. doi: 10.1016/j.spen.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. pp. 249–251. [Google Scholar]
- 50.Gustafson K, Boulianne GL. Genome. 1996;39:174–182. doi: 10.1139/g96-023. [DOI] [PubMed] [Google Scholar]
- 51.Kirby K, Hu J, Hilliker AJ, Phillips JP. Proc Natl Acad Sci USA. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griswold CM, Matthews AL, Bewley KE, Mahaffey JW. Genetics. 1993;134:781–788. doi: 10.1093/genetics/134.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]