Abstract
By transfer of T cell receptor (TCR) genes, antigen specificity of T cells can be redirected to target any antigen. Adoptive transfer of TCR-redirected T cells into patients has shown promising results. However, this immunotherapy bears the risk of autoreactive side effects if the TCR recognizes antigens on self-tissue. Here, we introduce a safeguard based on a TCR-intrinsic depletion mechanism to eliminate autoreactive TCR-redirected T cells in vivo. By the introduction of a 10-aa tag of the human c-myc protein into murine (OT-I, P14) and human (gp100) TCR sequences, we were able to deplete T cells that were transduced with these myc-tagged TCRs with a tag-specific antibody in vitro. T cells transduced with the modified TCR maintained equal properties compared with cells transduced with the wild-type receptor concerning antigen binding and effector function. More importantly, therapeutic in vivo depletion of adoptively transferred T cells rescued mice showing severe signs of autoimmune insulitis from lethal diabetes. This safeguard allows termination of adoptive therapy in case of severe side effects.
Keywords: adoptive T cell therapy, antibody depletion, myc-tag, suicide, TCR
Cytotoxic T lymphocytes specifically recognize antigens presented on target cells via MHC class I molecules. Their specificity is conferred by the T cell antigen receptor (TCR), a heterodimer composed of one α-chain and one β-chain (TCRαβ). Adoptive transfer of T cells specific for viral, tumor, or minor histocompatibility antigens into patients has been successfully applied in the treatment of malignant and viral diseases (1, 2). Yet, isolation and in vitro expansion of antigen-specific T cell clones remains time consuming and laborious and will only be available for a limited number of patients. Alternatively, T cells can be genetically modified with TCR genes in vitro, making it possible to redirect them to any target antigen. It has been shown that TCR-modified T cells secrete cytokines and lyse antigen-presenting target cells (3–5). Adoptive transfer of redirected T cells into melanoma patients led to partial remission in some patients in a clinical trial (6).
However, therapy using TCR gene-modified cells is associated with potential risks. First, many tumor antigens are not tumor-specific, but also are expressed in normal tissue (7). Recognition of such tumor-associated antigens presented on nontumor cells by TCR-redirected T cells may lead to autoreactivity. Second, the introduced TCR chains can form mixed heterodimers with the α- and β-chains of the endogenous TCR (8, 9). If a polyclonal pool of T cells is TCR-modified, the specificity of these mispaired TCRs cannot be predicted, and they might have autoreactive capacity. Third, it has been shown that the activation of the introduced TCR also may induce a response of the endogenous receptor (10). Although clonal deletion in the thymus eliminates the majority of high-affinity autoreactive T cells, low-affinity autoreactive T lymphocytes escape central tolerance mechanisms (11, 12). If these T cells are transduced with a second TCR and become activated, they may react against self-tissue. Finally, stable TCR expression in transduced T cells requires integration into the host genome that bears the risk of insertional mutagenesis. Therefore, it is desirable to equip TCR gene-modified T cells with a safeguard that allows the specific and efficient elimination of these cells if severe side effects occur.
Suicide genes such as HSV thymidine kinase (HSV-TK) or apoptosis-inducing fusion constructs in combination with activating prodrugs have been tested as safety modalities in adoptive cell transfer (13–18). Also, the use of the CD20 molecule that can be targeted by a CD20-specific antibody has been used to eliminate specific T cell populations (19, 20).
HSV-TK was already applied as a safety modality in patients with graft-versus-host disease after donor lymphocyte infusion; however, the safety mechanism has not been successful in all patients (13). Major limitations of the HSV-TK system are (i) immunogenicity of the HSV-TK gene product resulting in elimination of the transferred T cells (15, 21, 22), (ii) silencing or inhomogeneous expression of the transgene leading to ganciclovir-resistant cells (23), and (iii) that the prodrug ganciclovir cannot be used to treat upcoming viral infections (e.g., CMV, EBV) in patients. Apoptosis-inducing fusion genes and the use of the CD20 molecule also rely on high-level expression in transduced cells (24), and administration of a CD20-specific antibody in clinical application also will lead to the elimination of B cells. Furthermore, all of these strategies require the introduction of an additional gene into T cells. Retroviral vectors most commonly used to transduce T cells only have a limited transgene capacity. Considering the size of the TCRαβ genes, it is unlikely that vectors carrying an additional gene can efficiently transduce T cells. Hence, peripheral blood lymphocytes (PBLs) will have to be independently transduced with a TCR and a second gene-encoding vector, increasing the number of retroviral integrations into the host cell genome and, thus, the risk of insertional mutagenesis (25). Also, the purification and analysis steps needed to ensure that all TCR-redirected cells express the safety modality will prolong the in vitro culture time and decrease their functionality (26).
Therefore, we used a TCR-intrinsic safety mechanism for the elimination of TCR gene-modified T cells. This mechanism does not rely on the introduction of a second gene, and safety is not hampered by the down-regulation or low expression of the transgene.
Results
Expression of myc-Tagged TCRs in Murine T Cells.
We sought to introduce the amino acids 410–419 of the human c-myc protein (myc-tag) into the structure of a TCR in a position where it can be recognized by a myc-specific antibody without interfering with TCR function. The myc-tag was chosen because of its expected low immunogenicity in humans. Based on crystal structures of human and murine TCRs (27), we selected different sites in the murine P14 TCR (recognizing gp33, a peptide of lymphocytic choriomeningitis virus) α- and β-chains for tag insertion. We generated nine retroviral constructs, in which either one or two tags were inserted in a specific position or parts of the original TCR were substituted by one or two tags (Fig. 1). Interestingly, all TCRs were expressed in the TCRαβ-deficient murine T cell line 58 and recognized the antigen as shown by specific tetramer staining (E.K., unpublished data). However, only one construct carrying a double myc-tag at the N terminus of the variable region of the TCRα-chain allowed efficient in vitro depletion of T cells. Therefore, this site was chosen for myc-tag insertion into different TCRs.
Fig. 1.
Positions of myc-tag insertion in the murine P14 TCR. (A and B) The 10-aa myc-tag sequence was incorporated into the sequence of the TCRα-chain (A) or the TCRβ-chain (B). Four constructs were generated, in which one or two myc-tags were inserted (marked with a “1”). Numbers indicate the amino acid position behind which the tag was introduced. In five constructs, the sequences of the original TCR were substituted with one or two myc-tag sequences (marked with a “2”). Here, numbers indicate the first and last amino acid position of the original sequence that was replaced. The position selected for the modification of other human and murine TCRs is marked by an asterisk. S, signal peptide; V, variable region; D, diversity region; J, joining region; C, constant region.
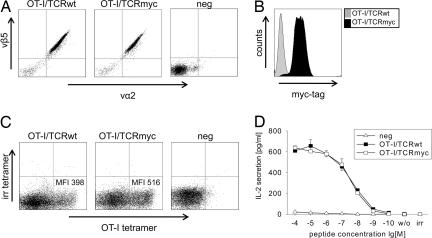
We generated retroviral constructs encoding the P14 TCR, as well as constructs encoding the OT-I TCR, which recognizes the ovalbumin-derived peptide ova. For each of these TCRs, either myc-tagged (P14/TCRmyc and OT-I/TCRmyc) or wild-type (P14/TCRwt and OT-I/TCRwt) TCR-encoding retroviruses were produced and used to transduce 58 cells. The cells were enriched with a TCRβ-specific antibody and then analyzed by flow cytometry. TCRα- and TCRβ-chain specific antibodies were used to confirm similar expression levels of OT-I/TCRwt and OT-I/TCRmyc (Fig. 2A). Incubation with a myc-specific antibody revealed only binding to OT-I/TCRmyc-modified cells, but not to OT-I/TCRwt-transduced cells (Fig. 2B). Similar results were obtained with the P14 TCR [supporting information (SI) Fig. 7 A and B].
Fig. 2.
OT-I/TCRmyc is expressed and functions comparably to OT-I/TCRwt. (A) The TCR-deficient 58 cells were transduced with the indicated OT-I TCR constructs. Cells were enriched with vβ-chain-specific antibodies and analyzed by flow cytometry with vα2- and vβ5-specific antibodies. Untransduced cells (neg) served as a negative control. (B) The 58 cells transduced with the OT-I/TCRmyc were stained with an antibody specific for the myc-tag sequence. Cells transduced with the wild-type receptor were used as a control. (C) The B6 splenocytes were transduced with either OT-I/TCRwt or OT-I/TCRmyc and after 72 h were stained with a CD8-specific antibody, an OT-I specific MHC-tetramer, and an irrelevant tetramer (irr). Cells are gated on CD8 expression. Numbers indicate the MFI of the tetramer staining. (D) The 1 × 105 OT-I/TCRwt- or OT-I/TCRmyc-transduced 58 (CD8α+) cells or untransduced cells (neg) were stimulated for 24 h with 1 × 105 T2-Kb cells pulsed with ova peptide. IL-2 concentration of the culture supernatant was analyzed by ELISA. Unloaded T2-Kb cells (w/o) or T2-Kb cells loaded with irrelevant peptide (irr) served as negative controls. Data represent the mean values of duplicates, and error bars indicate SD.
T Cells Transduced with myc-Tagged and Wild-Type TCRs Exhibit Comparable Functionality.
For the potential clinical application of myc-tagged TCRs, it is essential that the receptor function is not impaired by the insertion of the tag. For functional characterization of myc-tagged TCRs, splenocytes of C57BL/6J (B6) mice were transduced with OT-I/TCRmyc or OT-I/TCRwt retroviruses and were stained with an OT-I-specific and irrelevant tetramer. Both TCRs similarly bound the OT-I tetramer, as shown by comparable mean fluorescence intensity (MFI) in flow cytometry (Fig. 2C). For detection of cytokine secretion, CD8α-positive 58 cells were transduced with OT-I/TCRmyc or OT-I/TCRwt retroviruses with a similar efficiency (≈90%) (data not shown) and were stimulated with ova-peptide-loaded T2 cells transfected to express murine MHC-I H-2Kb molecules. The secretion of IL-2 was detected in a peptide concentration-dependent manner and was similar for cells transduced with either construct (Fig. 2D). These findings could be reproduced for P14/TCRmyc compared with P14/TCRwt (SI Fig. 7 C and D).
T Cells Expressing myc-Tagged TCRs Can Be Depleted in Vitro and in Vivo.
To analyze whether TCRmyc-expressing T cells can be depleted with a myc-specific antibody, TCR-enriched 58 cells (>95% purity) were subjected to complement-mediated lysis by incubation with a myc-specific antibody and complement factors. Cell viability was determined by using 7-amino-actinomycin D (7-AAD) staining. OT-I/TCRmyc-positive cells showed specific lysis of 78%, whereas OT-I/TCRwt-transduced cells were not lysed (Fig. 3A). Similar results were obtained by using 58 cells transduced with either P14/TCRmyc or P14/TCRwt (SI Fig. 7E).
Fig. 3.
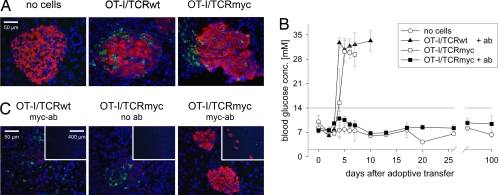
TCRmyc-transduced T cells can be depleted in vitro and in vivo with a myc-specific antibody. (A) The 58 cells were transduced with OT-I/TCRwt or OT-I/TCRmyc and were enriched with vβ5-specific antibodies. For depletion, cells were incubated with a myc-specific antibody and rabbit complement factors. 7-AAD was used to discriminate between living and dead cells. Numbers indicate the calculated specific lysis. Results show data from one of two independent experiments with comparable results. (B) Splenocytes of B6 mice were transduced with either OT-I/TCRwt or OT-I/TCRmyc. The 5 × 106 TCR-transduced cells were adoptively transferred i.v. into Rag-1−/− recipients. After 13 days, blood was stained for CD8- and myc-positive cells. One group of mice received 500 μg of a myc-specific antibody i.p. for depletion. One day after antibody injection, blood samples were collected and stained with CD8- and myc-specific antibodies for the TCRmyc construct or with CD8-, vα2- and vβ5-specific antibodies for the TCRwt construct. Stainings show cells gated on CD8 expression and represent one of two treated animals.
To demonstrate that TCRmyc-modified T cells also can be depleted in vivo, splenocytes of B6 mice were transduced with either OT-I/TCRwt or OT-I/TCRmyc retroviruses, and cells were injected into Rag-1−/− mice. Flow-cytometry analysis of blood samples taken 13 days after injection demonstrated the presence of the adoptively transferred cells in the blood (Fig. 3B). Five hundred micrograms of myc-specific antibody was injected, and blood samples were analyzed 1 day later. In mice that received OT-I/TCRmyc-transduced T cells, no myc-positive cells could be detected, indicating that TCRmyc-transduced cells were completely depleted. In contrast, in mice that received OT-I/TCRwt T cells or did not receive antibody treatment, the population of adoptively transferred cells remained unchanged (Fig. 3B).
Depletion of myc-Tagged T Cells Rescues Mice from Lethal Autoimmune Diabetic Disease After Adoptive Transfer.
To analyze whether myc-tagged TCRs can be used to prevent autoimmune disease, we used RIP-mOVA mice that express ovalbumin under the control of the rat insulin promoter in the β-islet cells of the pancreas (28). If OT-I/TCR T cells are transferred into these mice, they develop autoimmune diabetes due to the destruction of insulin-producing cells by the T cells. In this model, disease onset is extremely rapid: As early as day 2 after adoptive transfer, insulitis (defined by the infiltration of islets with lymphocytes) can be detected. Blood glucose values increase from normal to highly glycemic within 24 h at day 4 or 5 after adoptive transfer, and mice have to be killed at days 6–10 because of severity of symptoms.
Splenocytes of B6 mice were transduced with either OT-I/TCRmyc or OT-I/TCRwt retroviruses, and cells were injected into sublethally irradiated RIP-mOVA mice. Two days later, injected mice (but not control animals) showed infiltration of pancreatic islets, with CD8-positive T cells demonstrating onset of insulitis (Fig. 4A). For treatment, 500 μg of myc-specific antibody was injected i.p. None of the animals that received OT-I/TCRmyc T cells and antibody treatment developed diabetes, as measured by blood glucose concentration, until the end of the observation period on day 100. In contrast, all animals in the control groups receiving either OT-I/TCRwt T cells plus antibody or OT-I/TCRmyc T cells but no antibody became glycemic within 4–5 days after adoptive T cell transfer (Fig. 4B) and had to be killed 2–6 days later. Immunohistochemistry (IHC) staining of pancreatic sections of severely sick diabetic mice on day 6 after transfer showed some remaining CD8-positive T cells in the tissue and an almost complete lack of ovalbumin-expressing cells due to destruction by OT-I T cells. In striking contrast, animals that had received TCRmyc-transduced T cells and antibody treatment exhibited intact islet structure and a lack of T cells in the pancreas (Fig. 4C).
Fig. 4.
Treatment of autoimmune insulitis mediated by myc-specific antibody depletion. B6 splenocytes were transduced with either OT-I/TCRwt or OT-I/TCRmyc and injected i.v. into sublethally irradiated RIP-mOVA mice. Mice that were irradiated but received no cells served as a negative control. (A) Two days after adoptive transfer, pancreases of mice from each group were analyzed by IHC with ovalbumin- (red) and CD8- (green) specific antibodies. Nuclei were stained with DAPI (blue). (B) Two days after cell transfer, 500 μg of a myc-specific antibody was administered i.p. into all mice that had received T cells harboring the TCRwt (n = 5) and half of the mice (n = 5) that had received T cells carrying the TCRmyc. The blood glucose concentration was determined. Depicted are mean values of all animals in one group; error bars indicate SD. If measurement exceeded the upper detection limit of 33.3 mM, values were set as 35 mM to allow the calculation of mean blood glucose levels. (C) Pancreatic sections from diabetic and antibody-treated mice were stained on day 6 with ovalbumin- (red) and CD8- (green) specific antibodies and DAPI (blue). (Insets) Larger parts of the tissue at a lower magnification.
Human TCRmyc T Cells Function Comparably to TCRwt Cells and Can Be Depleted in Vitro.
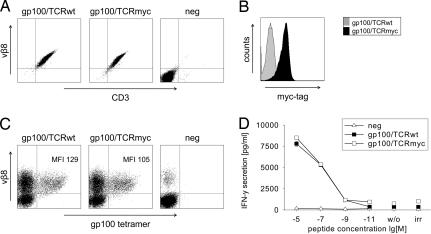
To assess whether the myc-tag also can be applied as a safeguard to eliminate human TCR-redirected T cells, we modified a human TCR that is reactive against the melanoma antigen gp100 with two myc-tags at the N terminus of the α-chain, corresponding to the same position modified in the murine TCRs. Retroviral constructs were generated and used to transduce the TCR-deficient human T cell line Jurkat76. The cells were enriched with β-chain-specific antibodies and subcloned by limiting dilution. Both the modified and the wild-type TCR were expressed on Jurkat76 cells as detected with a vβ8-specific antibody (Fig. 5A). Because no antibodies are available for the detection of the gp100α-chain, this staining could not be performed. Jurkat76 cells transduced with the gp100/TCRmyc, but not cells transduced with gp100/TCRwt, could be stained with a myc-specific antibody as analyzed by flow cytometry (Fig. 5B). For functional comparison, PBLs were transduced with the TCR constructs and analyzed by tetramer staining and peptide-specific IFN-γ secretion. Both TCRs similarly bound the gp100-tetramer demonstrated by comparable MFI in flow cytometry (Fig. 5C). Upon stimulation with gp100 peptide-pulsed T2 cells, PBLs transduced with either TCR secreted comparable amounts of IFN-γ (Fig. 5D).
Fig. 5.
The human gp100/TCRmyc is expressed and functions comparably to gp100/TCRwt. (A) The human T cell line Jurkat76 was transduced with gp100/TCRwt or gp100/TCRmyc, enriched with vβ8-chain specific antibodies, and subcloned. TCR expression was analyzed by flow-cytometry staining with a vβ8-specific antibody. Untransduced cells (neg) served as a negative control. (B) Jurkat76 cells transduced with gp100/TCRmyc were stained with a myc-specific antibody and analyzed by flow cytometry. Cells transduced with the wild-type receptor served as a control. (C) PBLs were transduced with gp100/TCRwt or gp100/TCRmyc vectors and were stained with a vβ8-specific antibody and a gp100-specific tetramer. Untransduced PBLs (neg) show the endogenous vβ8-positive T cells. Numbers indicate the MFI of the tetramer staining. (D) gp100/TCRwt- or gp100/TCRmyc-transduced PBLs were cocultured with T2 cells pulsed with gp100 peptide for 24 h. Untransduced PBLs were used as a negative control (neg). Culture supernatant was analyzed for IFN-γ concentration by ELISA. Unloaded T2 cells (w/o) or T2 cells loaded with irrelevant peptide (irr) served as negative controls. The data represent mean values of duplicates, and error bars indicate SD. The results were reproduced in two independent experiments and with two different donors.
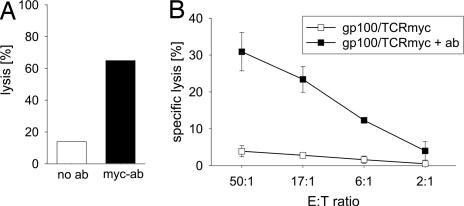
To show that gp100/TCRmyc-modified T cells can be eliminated, PBLs transduced with gp100/TCRmyc were enriched with a myc-specific antibody and were restimulated. PBLs that were 85–99% positive for myc expression were subjected to complement-mediated lysis (Fig. 6A) or antibody-dependent cell-mediated cytotoxicity (ADCC) (Fig. 6B). Depending on the assay, 31–65% of the gp100/TCRmyc-transduced cells were depleted in the presence of a myc-specific antibody, whereas cells incubated without antibody showed only a little unspecific lysis.
Fig. 6.
T cells transduced with gp100/TCRmyc can be depleted in vitro by complement- and cell-mediated lysis. PBLs were transduced with gp100/TCRmyc, sorted, and restimulated. (A) For complement-mediated depletion, cells were incubated with a myc-specific antibody and rabbit complement factors. 7-AAD was used to discriminate between living and dead cells. Cells incubated without antibody served as a control. (B) For cell-mediated lysis, autologous NK cells were used as effectors. 51Cr-labeled TCRmyc-positive PBLs were incubated with effector cells at different E/T ratios. A myc-specific antibody and a secondary rabbit anti-mouse IgG1 antibody were added, and lysis was measured in a standard 4-h chromium release assay. Samples without antibody served as a control. The data represent mean values of duplicates, and error bars indicate the SD. Similar results were obtained in an independent experiment with a different donor.
Discussion
We demonstrated that T cells transduced with murine and human TCRs, which were modified with a myc-tag, functioned comparably to cells transduced with the wild-type receptor. Myc-tagged T cells could be eliminated in vitro. Moreover, in vivo depletion of adoptively transferred myc-tagged T cells rescued mice showing severe signs of insulitis from the development of lethal autoimmune diabetes.
The therapeutic potential of TCR-redirected T cells has been demonstrated in a recent clinical trial in which a tumor antigen-reactive TCR was transduced into autologous T cells which were then adoptively transferred into patients with metastatic melanoma (6). However, some clinical data demonstrate that adoptive therapy with nonmodified or gene-modified T cells bears the risk of autoreactive side effects (29–31). Autoreactivity of transferred T cells can hardly be predicted and may vary from patient to patient. Hence, including a safeguard in TCR gene-modified T cells that allows the termination of therapy by the elimination of autoreactive cells is desirable. Such modality has to meet several criteria: (i) it should not interfere with TCR function; (ii) it should be specific, efficient, and rapid; and (iii) the implementation in a clinical setting should be feasible. So far, different safety modalities such as suicide genes (13–16, 32), apoptosis-inducing fusion genes (17, 18, 23), or cell-surface proteins (e.g., CD20) that are targeted by specific antibodies (19, 20) have been proposed for the elimination of transduced T cells. However, these strategies comprise some disadvantages, rendering them less feasible for adoptive therapy with TCR gene-modified T cells (15, 21–24).
In this article, we used a safeguard for the elimination of TCR gene-modified T cells that is based on a TCR-intrinsic mechanism. This safeguard differs in several important features from other safety modalities. First, the safety of the system does not depend on high-level expression of the transgene. myc-tag TCR-redirected T cells that are not eliminated because of down-regulation or low expression of the TCR most probably are not autoreactive. Second, the antibody depletion mechanism that is used for the elimination of TCR-redirected T cells is specific. C-myc is a nuclear protein and is not expressed on the surface of normal cells (33). Hence, application of a myc-specific antibody is not toxic for other cells. Also, antibody therapy has been successfully and safely applied in the clinic to treat different tumor types (34, 35). Third, being derived from a human protein, the myc-tag is unlikely to be immunogenic in humans. However, we currently cannot exclude that an immunogenic peptide is generated at the fusion site of the myc-tag and the TCRα-chain. Fourth, redirection of T cell antigen specificity by TCR gene transfer and implementation of the safeguard are provided in one step by using only one vector. Thus, the introduction of an additional gene and purification steps are not required, and the risk of insertional mutagenesis is reduced.
In summary, we have developed a safeguard that efficiently eliminates TCR gene-modified autoreactive T cells after adoptive transfer. This safeguard relies on a TCR-intrinsic mechanism and improves the safety of adoptive T cell transfer by the specific depletion of transferred T cells without side effects if this is necessary. Whether this strategy can treat an established autoimmune disease in a clinical setting is currently not known and may depend on the nature of the side effects that occur.
We envisage that this approach also can be used as a safety modality for T cells carrying other therapeutic proteins, including single-chain antibody chimeric receptors or cytokine-receptor common γ chain.
Methods
Mice.
B6 mice were purchased from Charles River Laboratories. Rag-1−/− mice (B6.129S7-Rag1tm1Mom) were obtained from The Jackson Laboratory. RIP-mOVA mice were a gift from M. Canarille (Ludwig-Maximillians-Universität, Munich, Germany). Animal experiments were approved by the responsible institution and were performed according to national and regional regulations.
Molecular Cloning of myc-Tagged TCRs and Retroviral Vectors.
The OT-I TCR (vα2/vβ5, recognizing the ovalbumin257–264 peptide SIINFEKL) was isolated from the cDNA of splenocytes of OT-I/Rag-1−/− mice (36) and cloned into the MP71 retroviral vector (37) via NotI and BsrGI restriction sites. The construction of P14 TCR (vα2/vβ8, recognizing the lymphocytic choriomeningitis virus glycoprotein33–41 peptide KAVYNFATM) and gp100 TCR (vα13/vβ8, recognizing the human gp100209–217 peptide IMDQVPFSV) vectors has been described previously (8, 38, 39). In bicistronic constructs, the α- and β-chains, were linked via the P2A peptide (40) to obtain MP71–TCRα–P2A–TCRβ.
Only the generation of the construct carrying two myc-tags at the N terminus of the TCRα-chain is described. Insertion of the first myc-tag (amino acid sequence EQKLISEEDL) into the TCRα-chain was facilitated with pairs of overlapping primers (sequences provided in SI Table 1). First, the signal peptide was amplified by using a 5′ primer (P1) with a NotI restriction site and a 3′ primer encoding the myc-tag sequence (P2). The variable and constant regions of the α-chain were amplified by using a 5′ primer containing the myc-tag sequence (P3) and a 3′ primer with a BsrGI restriction site (P4). Second, the fragments were combined in a separate PCR by using the primers P1 and P4 and cloned into the MP71 vector. The second myc-tag was inserted by annealing the oligonucleotides M1 and M2 to a double-stranded fragment that was subsequently cloned into the TCRmyc via the BclI restriction site. Oligonucleotides were obtained from TIB MOLBIOL.
Cell Culture and Transduction.
T2, T2-Kb (a gift from H. Schreiber, University of Chicago, Chicago), RPMI 8866 cells (kindly provided by G. Trinchieri (Wistar Institute, Philadelphia), Jurkat76 cells (41), and 58 cells (42) were grown in RPMI medium 1640 (Invitrogen) supplemented with 10% FCS (Biochrom), 10 mM Hepes, and 100 units/ml penicillin/streptomycin (T cell medium). 293T cells (ATCC CRL-11268; American Type Culture Collection) and the ecotropic packaging cell line Plat-E (43) were cultured in DMEM (Invitrogen) with 10% FCS and 100/per ml penicillin/streptomycin. The isolation of murine splenocytes and human peripheral blood mononuclear cells (PBMCs) was performed as described previously (37). To produce ecotropic retrovirus for the transduction of murine T cells, Plat-E cells were transiently transfected with the vector construct by using CaPO4 precipitation. Amphotropic virus supernatant for the transduction of human T cells was produced by triple transfection of 293T cells with the vector construct, pcDNA3.1MLVg/p (C. Baum, MH-Hannover, Hannover, Germany) encoding the MLV gag and pol genes and pALF-10A1GaV (44) encoding the MLV 10A1 env gene. A 48-h culture supernatant of the packaging cells was harvested and filtered through 0.45-μm pore-size filters. Transduction of T cells was performed as described previously (37).
Peptides, Antibodies, and Tetramers.
Ova, gp33, and gp100 peptides were obtained from Biosyntan. mAbs directed against murine vα2, vβ5, vβ8, CD3, CD8, and human vβ8 were obtained from BD Caltag or Immunotech. Tetramers were used to stain gp100 TCR (Immunomics), P14 TCR (Immunotech), and OT-I TCR (Dirk Busch, Technische Universität, Munich, Germany). The myc-specific antibodies 3A7 and 9E10 were obtained from U.S. Biologicals or purified from hybridoma supernatant (ATCC CRL-1729), respectively. For FACS staining, a rabbit anti-myc antibody with a secondary goat anti-rabbit antibody (Santa Cruz Biotechnology) was used. Fluorescence intensity was measured by using a FACSCalibur flow cytometer and CellQuestPro Ver. 9 software (BD Biosciences). Data analysis was performed with FlowJo Ver. 5.7.2 software (Tree Star).
Cytokine Release Assay.
Peptide-presenting target cells were incubated for 2 h at 37°C with different amounts of peptide in serum-free medium and washed twice. Per well, 1 × 105 effector cells were cocultured with peptide-loaded targets in a 1:1 ratio in 96-well round-bottom plates (Corning Costar) for 24 h at 37°C. The supernatant was tested for human IFN-γ or murine IL-2 amount by ELISA (sensitivity 4 or 2 pg/ml, respectively; eBioscience).
Complement-Mediated Depletion Assay.
Exponentially growing 58 cells or Ficoll-Hypaque-purified PBLs, respectively, were seeded in a 96-well plate (Corning Costar) with 1 × 105 cells per well in RPMI 1640 medium plus 25 mM Hepes and 0.3% BSA. Cells were labeled with 1 μg of myc-specific antibody per well (clone 3A7) for 1 h at 4°C, washed, and incubated with rabbit complement (for 58 cells, LOW-TOX-M; for PBLs, rabbit complement MA; Cedarlane) diluted 1:6 or 1:9 for 2 h at 37°C. For live and dead cell discrimination, cells were stained with 7-AAD (BD Biosciences) for 10 min and analyzed by flow cytometry. Cells incubated with antibody or complement alone served as controls. Percentage of specific depletion was calculated as [% cytotoxicity (antibody plus complement) − % cytotoxicity (complement alone)]/[100% − % cytotoxicity (complement alone)] × 100.
Antibody-Dependent Cell-Mediated Cytotoxicity Assay.
Target cells were prepared by enriching TCR-transduced PBLs by using myc-specific magnetic beads and MACS separator columns (Miltenyi Biotec) and specifically restimulating them with irradiated peptide-loaded T2 cells. As effector cells, autologous NK cells were used that were prepared from freshly isolated PBMC by centrifugation on Ficoll-Hypaque, depletion of monocytes by adhesion to cell culture plastics, and cocultivation for 6 days with RPMI 8866 cells irradiated with 64 Gy. For stimulation, 300 units/ml Proleukin were added to the effector cells 1 day before the ADCC assay. Lysis was performed by incubating 5 × 103 51Cr-labeled (Amersham) target cells [100 μCi (1 Ci = 37 GBq) per sample] with effector cells in effector-to-target cell (E/T) ratios from 50:1 to 2:1 for 4 h in the presence of 1 μg of myc-specific antibody (clone 9E10) and 1 μg of rabbit anti-mouse IgG1-Fc polyclonal antibody (Jackson ImmunoResearch).
Adoptive T Cell Transfer.
RIP-mOVA mice were sublethally irradiated with 4 Gy 1 day before adoptive transfer. Age- and sex-matched recipient mice were injected in the tail vein with 2 × 107 (RIP-mOVA mice) or 5 × 106 (Rag-1−/− mice) TCR-positive splenocytes 1 day after the second transduction. For depletion of adoptively transferred cells, 500 μg of myc-specific antibody (clone 9E10) were injected i.p. 2 (RIP-mOVA mice) or 13 (Rag-1−/− mice) days after adoptive transfer. The expansion and depletion of cells were monitored by flow cytometry of blood samples. Diabetes development in RIP-mOVA mice was followed by measuring blood glucose levels with Ascensia ELITE SENSOR strips (Bayer). Mice with blood glucose levels >14 mM at 2 consecutive days were considered diabetic.
Immunohistochemical Staining.
Cryosections of the pancreas of killed mice were fixed with acetone and preincubated subsequently with protein block (Immunotech) and PBS/1% BSA/1% donkey serum. Ovalbumin was stained with a polyclonal rabbit anti-ova antibody (Acris) and secondary donkey anti-rabbit coupled to Alexa594 (Molecular Probes). CD8-positive cells were detected with rat anti-CD8α antibody (clone 53–6.7; BD) and secondary donkey anti-rat antibody coupled to Alexa488 (Molecular Probes). Nuclei were visualized with DAPI. Images were obtained with an Axiowert 200 microscope and Axiovision Rel. 4.5 software.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Martina Grabbert, Irmgard Küttner, and Kordelia Hummel for excellent technical assistance, Shabnam Shalapour for great help with immunohistology, T. Brocker for providing the RIP-mOVA mice, H. Schreiber for providing T2-kb cells, and G. Trincheri for providing RPMI 8866 cells. This work was supported by Wilhelm Sander-Stiftung Grant 2004.106.1 and Deutsche Forschungsgemeinschaft Grant SFB-TR36.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710198105/DC1.
References
- 1.Blattman JN, Greenberg PD. Science. 2004;305:200–205. doi: 10.1126/science.1100369. [DOI] [PubMed] [Google Scholar]
- 2.Dudley ME, Rosenberg SA. Nat Rev Cancer. 2003;3:666–675. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engels B, Uckert W. Mol Aspects Med. 2007;28:115–142. doi: 10.1016/j.mam.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Xue S, Gillmore R, Downs A, Tsallios A, Holler A, Gao L, Wong V, Morris E, Stauss HJ. Clin Exp Immunol. 2005;139:167–172. doi: 10.1111/j.1365-2249.2005.02715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tey SK, Bollard CM, Heslop HE. Immunol Cell Biol. 2006;84:281–289. doi: 10.1111/j.1440-1711.2006.01441.x. [DOI] [PubMed] [Google Scholar]
- 6.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, et al. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novellino L, Castelli C, Parmiani G. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommermeyer D, Neudorfer J, Weinhold M, Leisegang M, Engels B, Noessner E, Heemskerk MH, Charo J, Schendel DJ, Blankenstein T, et al. Eur J Immunol. 2006;36:3052–3059. doi: 10.1002/eji.200636539. [DOI] [PubMed] [Google Scholar]
- 9.Heemskerk MH, Hagedoorn RS, van der Hoorn MA, van der Veken LT, Hoogeboom M, Kester MG, Willemze R, Falkenburg JH. Blood. 2007;109:235–243. doi: 10.1182/blood-2006-03-013318. [DOI] [PubMed] [Google Scholar]
- 10.Gladow M, Uckert W, Blankenstein T. Eur J Immunol. 2004;34:1882–1891. doi: 10.1002/eji.200425041. [DOI] [PubMed] [Google Scholar]
- 11.Hafler DA, Weiner HL. Immunol Rev. 1995;144:75–107. doi: 10.1111/j.1600-065x.1995.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 12.Bach JF, Chatenoud L. Annu Rev Immunol. 2001;19:131–161. doi: 10.1146/annurev.immunol.19.1.131. [DOI] [PubMed] [Google Scholar]
- 13.Bonini C, Ferrari G, Verzeletti S, Servida P, Zappone E, Ruggieri L, Ponzoni M, Rossini S, Mavilio F, Traversari C, Bordignon C. Science. 1997;276:1719–1724. doi: 10.1126/science.276.5319.1719. [DOI] [PubMed] [Google Scholar]
- 14.Ciceri F, Bonini C, Marktel S, Zappone E, Servida P, Bernardi M, Pescarollo A, Bondanza A, Peccatori J, Rossini S, et al. Blood. 2007;109:4698–4707. doi: 10.1182/blood-2006-05-023416. [DOI] [PubMed] [Google Scholar]
- 15.Riddell SR, Elliott M, Lewinsohn DA, Gilbert MJ, Wilson L, Manley SA, Lupton SD, Overell RW, Reynolds TC, Corey L, Greenberg PD. Nat Med. 1996;2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 16.Tiberghien P, Ferrand C, Lioure B, Milpied N, Angonin R, Deconinck E, Certoux JM, Robinet E, Saas P, Petracca B, et al. Blood. 2001;97:63–72. doi: 10.1182/blood.v97.1.63. [DOI] [PubMed] [Google Scholar]
- 17.Straathof KC, Pule MA, Yotnda P, Dotti G, Vanin EF, Brenner MK, Heslop HE, Spencer DM, Rooney CM. Blood. 2005;105:4247–4254. doi: 10.1182/blood-2004-11-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomis DC, Marktel S, Bonini C, Traversari C, Gilman M, Bordignon C, Clackson T. Blood. 2001;97:1249–1257. doi: 10.1182/blood.v97.5.1249. [DOI] [PubMed] [Google Scholar]
- 19.Introna M, Barbui AM, Bambacioni F, Casati C, Gaipa G, Borleri G, Bernasconi S, Barbui T, Golay J, Biondi A, Rambaldi A. Hum Gene Ther. 2000;11:611–620. doi: 10.1089/10430340050015798. [DOI] [PubMed] [Google Scholar]
- 20.Serafini M, Manganini M, Borleri G, Bonamino M, Imberti L, Biondi A, Golay J, Rambaldi A, Introna M. Hum Gene Ther. 2004;15:63–76. doi: 10.1089/10430340460732463. [DOI] [PubMed] [Google Scholar]
- 21.Jung D, Jaeger E, Cayeux S, Blankenstein T, Hilmes C, Karbach J, Moebius U, Knuth A, Huber C, Seliger B. Hum Gene Ther. 1998;9:53–62. doi: 10.1089/hum.1998.9.1-53. [DOI] [PubMed] [Google Scholar]
- 22.Berger C, Flowers ME, Warren EH, Riddell SR. Blood. 2006;107:2294–2302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank O, Rudolph C, Heberlein C, von Neuhoff N, Schrock E, Schambach A, Schlegelberger B, Fehse B, Ostertag W, Stocking C, Baum C. Blood. 2004;104:3543–3549. doi: 10.1182/blood-2004-03-0852. [DOI] [PubMed] [Google Scholar]
- 24.van Meerten T, van Rijn RS, Hol S, Hagenbeek A, Ebeling SB. Clin Cancer Res. 2006;12:4027–4035. doi: 10.1158/1078-0432.CCR-06-0066. [DOI] [PubMed] [Google Scholar]
- 25.Woods NB, Muessig A, Schmidt M, Flygare J, Olsson K, Salmon P, Trono D, von Kalle C, Karlsson S. Blood. 2003;101:1284–1289. doi: 10.1182/blood-2002-07-2238. [DOI] [PubMed] [Google Scholar]
- 26.Sussman JJ, Parihar R, Winstead K, Finkelman FD. Cancer Res. 2004;64:9124–9130. doi: 10.1158/0008-5472.CAN-03-0376. [DOI] [PubMed] [Google Scholar]
- 27.Rudolph MG, Stanfield RL, Wilson IA. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 28.Kurts C, Heath WR, Carbone FR, Allison J, Miller JF, Kosaka H. J Exp Med. 1996;184:923–930. doi: 10.1084/jem.184.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, et al. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yee C, Thompson JA, Roche P, Byrd DR, Lee PP, Piepkorn M, Kenyon K, Davis MM, Riddell SR, Greenberg PD. J Exp Med. 2000;192:1637–1644. doi: 10.1084/jem.192.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamers CH, Sleijfer S, Vulto AG, Kruit WH, Kliffen M, Debets R, Gratama JW, Stoter G, Oosterwijk E. J Clin Oncol. 2006;24:e20–e22. doi: 10.1200/JCO.2006.05.9964. [DOI] [PubMed] [Google Scholar]
- 32.Junker K, Koehl U, Zimmerman S, Stein S, Schwabe D, Klingebiel T, Grez M. Gene Ther. 2003;10:1189–1197. doi: 10.1038/sj.gt.3301977. [DOI] [PubMed] [Google Scholar]
- 33.Evan GI, Hancock DC, Littlewood T, Pauza CD. Ciba Found Symp. 1986;119:245–263. doi: 10.1002/9780470513286.ch14. [DOI] [PubMed] [Google Scholar]
- 34.Adams GP, Weiner LM. Nat Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 35.Carter P. Nat Rev Cancer. 2001;1:118–129. doi: 10.1038/35101072. [DOI] [PubMed] [Google Scholar]
- 36.Schuler T, Blankenstein T. J Immunol. 2003;170:4427–4431. doi: 10.4049/jimmunol.170.9.4427. [DOI] [PubMed] [Google Scholar]
- 37.Engels B, Cam H, Schuler T, Indraccolo S, Gladow M, Baum C, Blankenstein T, Uckert W. Hum Gene Ther. 2003;14:1155–1168. doi: 10.1089/104303403322167993. [DOI] [PubMed] [Google Scholar]
- 38.Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR, Wunderlich JR, Hughes MS, Restifo NP, Rosenberg SA. J Immunol. 2003;171:3287–3295. doi: 10.4049/jimmunol.171.6.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 40.Arnold PY, Burton AR, Vignali DA. J Immunol. 2004;173:3103–3111. doi: 10.4049/jimmunol.173.5.3103. [DOI] [PubMed] [Google Scholar]
- 41.Heemskerk MH, Hoogeboom M, de Paus RA, Kester MG, van der Hoorn MA, Goulmy E, Willemze R, Falkenburg JH. Blood. 2003;102:3530–3540. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 42.Letourneur F, Malissen B. Eur J Immunol. 1989;19:2269–2274. doi: 10.1002/eji.1830191214. [DOI] [PubMed] [Google Scholar]
- 43.Morita S, Kojima T, Kitamura T. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 44.Stitz J, Buchholz CJ, Engelstadter M, Uckert W, Bloemer U, Schmitt I, Cichutek K. Virology. 2000;273:16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.