Abstract
Glucocorticoid-induced TNF receptor ligand (GITRL), a recently identified member of the TNF superfamily, binds to its receptor, GITR, on both effector and regulatory T cells and generates positive costimulatory signals implicated in a wide range of T cell functions. In contrast to all previously characterized homotrimeric TNF family members, the mouse GITRL crystal structure reveals a previously unrecognized dimeric assembly that is stabilized via a unique “domain-swapping” interaction. Consistent with its crystal structure, mouse GITRL exists as a stable dimer in solution. Structure-guided mutagenesis studies confirmed the determinants responsible for dimerization and support a previously unrecognized receptor-recognition surface for mouse GITRL that has not been observed for any other TNF family members. Taken together, the unique structural and biochemical behavior of mouse GITRL, along with the unusual domain organization of murine GITR, support a previously unrecognized mechanism for signaling within the TNF superfamily.
Keywords: crystal structure, domain swap, oligomerization, T cell costimulation
The members of the TNF family and their receptors (TNFRs) modulate diverse biological functions including cell proliferation, differentiation, survival, and apoptosis (1). TNF family members are synthesized as type-II transmembrane proteins and function either in their transmembrane forms or as soluble ligands generated upon proteolytic shedding from the plasma membrane. Conventional TNF ligands self-assemble into homotrimers, in which three β-sandwich “jelly-roll” protomers noncovalently associate through interactions between hydrophobic surfaces. TNFRs are type-I transmembrane proteins characterized by pseudorepeats of one to four extracellular cysteine-rich domains (CRDs). Known structures of TNF:TNFR complexes, in general, display a common organization in which trimeric TNF ligands engage three receptor molecules resulting in an assembly with threefold symmetry and a 3:3 ligand:receptor stoichiometry. This organization results in the clustering of the receptor cytoplasmic tails that recruit and locally enrich signaling adaptor proteins, like the trimeric TNFR-associated factors (TRAFs), thereby leading to activation of the downstream signaling pathways (1, 2). The complete generalization of this model requires additional structural information because of the low sequence identity exhibited by the members of the TNF and TNFR families.
Glucocorticoid-induced TNFR (GITR) and its ligand, GITRL, are recently described members of the TNFR/ligand superfamilies and are implicated in a wide range of immune functions involving both effector and regulatory T cells (Tregs) (3–5). GITR is expressed at low levels on resting mouse and human T cells and is up-regulated upon activation of CD4+ and CD8+ T cells. A substantial level of GITR is constitutively expressed on CD4+CD25+ Tregs. GITR is activated by its ligand, GITRL, which is expressed in various antigen-presenting cells, including macrophages, B cells, and immature and mature dendritic cells, as well as multiple nonlymphoid tissues. GITR engagement in the presence of suboptimal T cell receptor stimulation generates a positive costimulatory signal leading to increased T cell proliferation and cytokine production (6–8). More importantly, GITR stimulation on effector T cells has been shown to reverse the suppressive effects by the Tregs in mice (7, 9, 10). The GITRL:GITR pathway thus provides a potential target for manipulating T cell responsiveness to clear infectious pathogens and tumors and to reverse globally suppressed immune responses resulting from chronic infections.
GITRL is an ≈20-kDa transmembrane protein that shares ≈20% sequence identity with other TNF ligands. GITR is an ≈26-kDa transmembrane protein that displays 14–28% sequence identity with other members of the TNFR superfamily (11). The degree of sequence identity between human and mouse orthologs of GITRL/GITR (≈50–60%) is similar to that which exists between human and mouse orthologs of other TNF/TNFR family members (8, 11). However, mouse GITRL does not recognize the human receptor, and human GITRL does not bind the mouse receptor (12), suggesting that these putative orthologs do not share a common recognition surface for their cognate binding partners. Interestingly, recent findings suggest that the GITRL:GITR signaling pathway plays different roles in mice and humans (13), because in humans the Treg-mediated suppression of effector T cell function is not inhibited by GITR stimulation (14). The mechanistic basis for these distinct costimulatory effects in the human and mouse GITRL:GITR pathways remains to be elucidated.
Our structural and biochemical studies have demonstrated that, like all previously characterized TNF family members, the human GITRL ectodomain self-assembles into a noncovalently associated homotrimer, in which solvent-exposed loops near the intersubunit clefts form the receptor-recognition surface (15). Notably, human GITRL differs from typical TNF family members in forming a less compact trimer, with considerably smaller intermonomer interfaces. Consistent with the sparse monomer–monomer interfaces observed in the crystalline state, human GITRL displays a unique monomer–trimer equilibrium in solution (Kd ≈ 10 μM), with the trimer being the biologically active species in terms of receptor binding (15). In the present study we report the crystal structure of the mouse GITRL ectodomain. Remarkably, in contrast to all previously characterized TNF family members, the mouse GITRL structure reveals a previously unrecognized dimeric assembly that is stabilized via a “domain-swapping” interaction (16). This unique structure in the crystalline state is consistent with our biochemical data demonstrating that mouse GITRL exists as a stable dimer in solution. In further support of the distinct functional properties exhibited by the mouse and human orthologs, mouse GITRL shows a weaker affinity for its cognate receptor mouse GITR, as compared with the human ligand and receptor. In addition, our structure-guided site-directed mutagenesis studies defined the determinants responsible for dimerization and suggest a receptor-recognition surface for mouse GITRL that has not been observed previously for any other TNF family members, including human GITRL. This work provides a structural and biochemical basis for the unique functional properties exhibited by mouse and human GITRL:GITR signaling pathways and highlights a mechanism for the evolution of immune function within the TNF superfamily.
Results
Overall Structure of the Mouse GITRL Ectodomain Protomer.
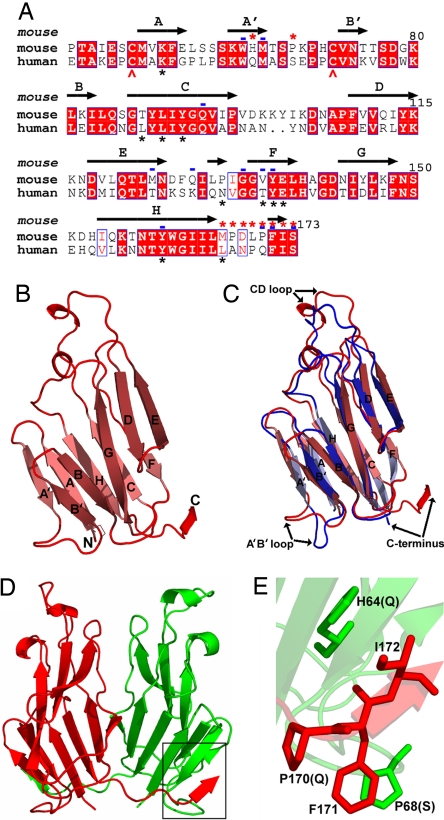
We have determined the crystal structure of the mouse GITRL ectodomain expressed in Escherichia coli (either purified from soluble fraction of the bacterial cell lysate or refolded from insoluble inclusion bodies; see Methods) [supporting information (SI) Table 1]. The mouse GITRL monomer exhibits the two-layer β-sandwich jelly-roll topology observed for other TNF family members, with inner and outer sheets composed of the A′AHCF and B′BGDE strands, respectively (Fig. 1 A and B). Consistent with the sequence identity of 55% (Fig. 1A), the TNF homology domains of mouse and human GITRL orthologs displayed considerable overall structural similarity (Fig. 1 B and C) (15), with an rms deviation of 1.5 Å (based on 108 equivalent Cα) (Fig. 1C). Other members in the TNF family also exhibit similar rms deviations when compared across the two species (0.97 Å for TNF-α and 1.8 Å for OX40L). The only pronounced differences between the mouse and human GITRL structures are present in the A′B′ and CD loops and at the C termini.
Fig. 1.
Structure of the mouse GITRL ectodomain shows a previously unrecognized dimeric assembly not observed in conventional TNF family members. (A) Amino acid sequence alignment of the mouse and human GITRL ectodomains. β-Strands identified in the mouse GITRL structure are indicated and labeled. Positions of the Cys residues that form the disulfide bond in both mouse and human GITRL are indicated with red arrows. Mouse GITRL residues mediating the domain swap and forming the potential hydrogen bonds at the monomer–monomer interface are marked with red asterisks and blue dashes, respectively, on the top of the alignment. Residues forming potential contacts at the intersubunit interfaces of human GITRL trimer are marked with black asterisks at the bottom of the alignment. (B) Ribbon diagram of the mouse GITRL monomer showing the classical jelly-roll fold of the TNF homology domains. The β-strands are labeled, and the N and C termini are marked. (C) Superposition of the mouse (red) and human (blue) GITRL shows significant differences in the arrangements of the A′B′ and CD loops and the C termini. (D) The mouse GITRL dimer is composed of two monomers, shown in red and green, that are associated with an angle of ≈40° with respect to each other. The C terminus of each subunit participates in a domain swap with the other subunit and is highlighted with a black box. (E) Detailed view of the mouse GITRL domain-swap interaction. Mouse GITRL residues, H64 and P68 from one monomer and P170, F171, and I172 from the other, are shown. P170, F171, and P68 are arranged to form a strong hydrophobic stacking interaction. The human GITRL residues at the corresponding positions (Q for H64, S for P68, and Q for P170) are indicated in the parentheses.
Mouse GITRL Ectodomain Adopts a Unique Dimeric Organization.
All previously characterized TNF family members conform to the paradigm that the ligands self-assemble into threefold symmetric homotrimers that represent the biologically active form of the molecule (2). Remarkably, in the crystalline state the mouse GITRL ectodomain is organized as a twofold symmetric dimer (Fig. 1D), an organization unique within the TNF family. Analysis of the mouse GITRL dimer shows that the two monomeric subunits are making an angle of ≈40° with respect to each other, while association of the inner β-strands from the two engaging monomers buries a total surface area of ≈2,998 Å2. Twelve residues (Trp-63, Met-65, Gln-94, Met-124, Gln-128, Gly-133, Val-135, Tyr-136, Tyr-160, Pro-170, Phe-171, and Ser-173) contribute 16 potential hydrogen bonds at this monomer–monomer interface (SI Table 2). Notably, eight of these 12 residues (all except Met-124, Gln-128, Val-35, and Pro-170) are conserved in both mouse and human GITRL (Fig. 1A), but only two of these conserved residues, Tyr-136 and Tyr-160, form part of the monomer–monomer interfaces in the human GITRL trimer (15).
Notably, the conformation of the C terminus of each mouse GITRL protomer differs remarkably from the analogous segment in the human ortholog. In mouse GITRL, the C terminus of each subunit adopts an unusual conformation such that it participates in a tight hydrophobic interaction with the A′ strand and the A′B′ loop of the other subunit (Fig. 1 D and E), thereby resulting in a classic example of domain swapping (16). The domain-swapped segment is stabilized by an array of hydrophobic associations, with Phe-171 and Ile-172 (at the C terminus) from one subunit interacting with His-64 (A′ strand) and Pro-68 (A′B′ loop) from the other subunit, respectively (Fig. 1E). Phe-171 and Ile-172 are conserved in both mouse and human proteins, whereas His-64 and Pro-68 are replaced by Gln and Ser in human GITRL sequence, respectively (Fig. 1A). As a consequence, the hydrophobic and packing interactions present in the mouse GITRL dimeric assembly are not available in the human structure. Although the three C-terminal residues 171Phe-Ile-Ser173 are conserved, the four linker residues (167Pro-Asp-Leu-Pro170) that connect them to the core of the ectodomain in mouse GITRL are replaced with an Ala-Asn-Pro-Gln segment in humans. The presence of Pro at positions 167 and 170 presumably biases the conformation of the mouse GITRL C-terminal segment, so that Phe-171 and Ile-172 from one monomer interact with His-64 and Pro-68 of the other monomer. It is therefore likely that the residues present exclusively in the mouse GITRL sequence are responsible for its dimeric organization.
Mouse GITRL Exists as a Stable Dimer in Solution.
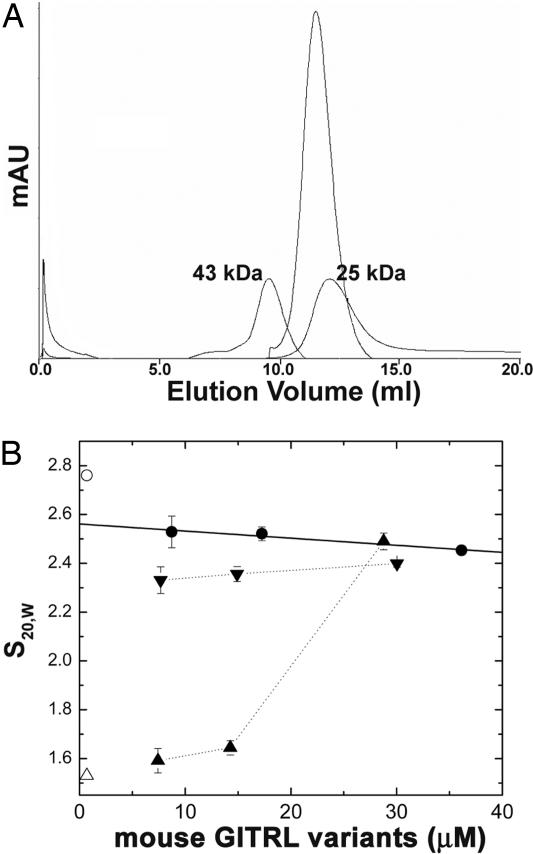
The unique dimeric assembly observed in the mouse GITRL crystal structure demanded further evaluation of the oligomeric state in solution. On a calibrated Superdex G-75 gel-filtration column (30 × 1.0), purified mouse GITRL (at a concentration of ≈40 μM) migrated as a symmetric monodisperse peak with an apparent molecular mass slightly higher than ≈25 kDa (Fig. 2A). Sedimentation velocity analysis demonstrated that the sedimentation constant is essentially invariant with increasing protein concentration (Fig. 2B), consistent with a stable stoichiometric assembly of the protein. Comparison of S20,w with the sedimentation coefficient calculated from the structure (HydroPro version 5a) (17) (Fig. 2B, open circle) suggests that mouse GITRL exists as a stable dimer in solution with a conformation comparable to that seen in the crystal. Equilibrium sedimentation analysis conducted as a function of solution temperature and salt concentration yielded a series of concentration distributions that was consistent with a single species with molecular masses (28–29 kDa) (SI Table 3) comparable to the molecular mass of 29.98 kDa calculated from the sequence of the mouse GITRL ectodomain dimer. Therefore, the solution and crystallographic data are consistent with a stable dimer.
Fig. 2.
Mouse GITRL shows a stable dimeric assembly in solution. (A) Elution profile of soluble mouse GITRL (refolded from inclusion bodies in E. coli) from Superdex G-75 gel-filtration column. The elution positions of size calibration standards of 25 kDa and 43 kDa are indicated. (B) The values of S20,w estimated for refolded mouse GITRL from sedimentation velocity analysis as a function of total monomer concentration at 5°C (●). The open circle and open triangle indicate the S values calculated from the crystal structure for dimer and monomer (top to bottom, respectively). ▴ and ▾ represent the values of S20,w estimated for the refolded humanized mouse GITRL and the murinized human GITRL, respectively, as a function of total monomer concentration at 25°C.
Domain-Swap Interactions Dictate the Dimeric Assembly of Mouse GITRL.
We used a structure-guided mutagenesis approach to determine whether the crystallographically observed domain swap was physiologically relevant. The mouse GITRL amino acid residues (His-64, P68, and 166Met-Ser173) that seem to favor this domain-swap interaction were systematically replaced with the corresponding residues from the human GITRL sequence (Fig. 1A). The rationale behind designing this “humanized” mouse GITRL mutant was to test whether disruption of the domain swap could perturb the stable dimeric assembly of the wild-type mouse GITRL. To test our hypotheses, the solution states of the wild-type and humanized mouse GITRL proteins were compared by sedimentation velocity analysis. In contrast to the wild-type mouse GITRL, S20,w increased with increasing concentrations of the humanized mouse GITRL, consistent with a reversible self-association process (Fig. 2B), as previously observed for human GITRL (15).
These data clearly suggest that mutations altering the putative domain-swap interactions in mouse GITRL dimer is sufficient to disrupt the dimeric organization of the wild-type protein. To further establish the critical role of the domain swap in mouse GITRL dimerization, a “murinized” human GITRL was generated by replacing the human GITRL sequence with the corresponding mouse GITRL residues (His-64, P68, and 166Met-Ser173 in mouse) (Fig. 1A). The S20,w values measured as a function of murinized human GITRL concentration more closely mimic those of the mouse GITRL nondissociating dimer than human GITRL, which exhibits a dynamic monomer–trimer equilibrium (Fig. 2B) (15). We conclude from these structure-guided mutagenesis experiments that the postulated domain-swapping interaction surface is the major determinant of the dimeric assembly of mouse GITRL.
Domain-Swapped C Termini of the Mouse GITRL Protomers Maintain Overall Stability of the Protein.
It is interesting to note that, although the mouse GITRL C-terminal segment (167Pro-Ser173) plays a critical role in the domain-swapped organization (hydrophobic association of the residues 171Phe-Ile-Ser173 with His-64 and P68 from the other subunit), it is not an integral part of the core of the TNF homology domain. To further examine the role of this segment in the overall structural integrity of the protein, a deletion construct of mouse GITRL lacking this region (167Pro-Ser173) was generated. Unlike the wild-type form of mouse GITRL, this deletion mutant failed to refold from the insoluble inclusion bodies expressed in E. coli and did not show any tendency to remain in the soluble fraction of the bacterial cell lysate, even when the cells were grown at 25°C. These observations demonstrate that the removal of this C-terminal segment from mouse GITRL dramatically affects overall stability of the protein and is consistent with the hypotheses that the domain-swap interaction is critical to the overall structural integrity of mouse GITRL. Notably, a similar deletion variant of human GITRL lacking the C-terminal seven residues could be refolded efficiently and was found to display chromatographic properties similar to those of wild-type human GITRL, suggesting that the homologous C-terminal segment of human GITRL has little, if any, impact on the stability and organization of the human protein.
Binding of Mouse GITRL to Mouse GITR.
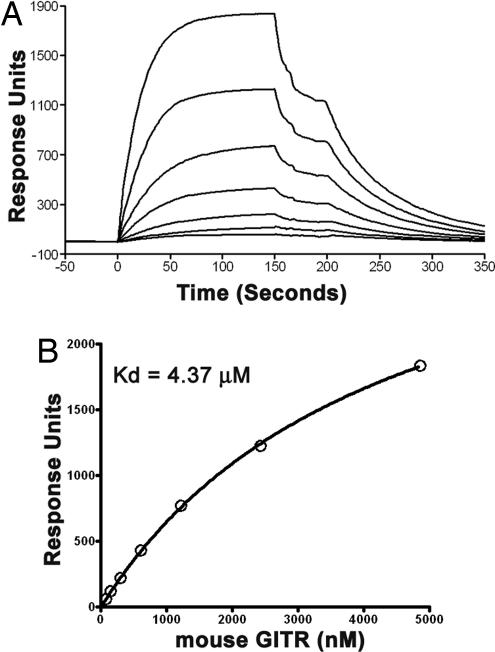
Our previous data demonstrated that the trimeric human GITRL binds to its receptor with a Kd value in the single-digit nanomolar range (15). Here, to test whether mouse GITRL expressed in E. coli and S2 cells retains receptor binding activity, the mouse GITRL:GITR interaction was measured by surface plasmon resonance (SPR) (Fig. 3 and SI Fig. 6; see also SI Results in SI Text). Fitting of the 1:1 Langmuir binding model to the steady-state plateaus reached after injection of mouse GITR/GITR-Ig yields a Kd value in the range of 4–15 μM for formation of the mouse GITRL:GITR complex. Consistent with earlier reports, mouse GITRL failed to bind to human GITR (data not shown) (12). Thus, the affinity of mouse GITRL for its receptor is significantly reduced compared with the human GITRL:GITR interaction (15). The reduced stability of the mouse GITRL:GITR complex is consistent with the dissociation reaction; koff is 1.5 × 10−2 s−1 compared with 1.1–1.5 × 10−4 s−1 measured for the human protein (15).
Fig. 3.
Receptor binding behavior of mouse GITRL. (A) Sensograms of the binding of mouse GITR (purified from HEK293T cells) at a range of concentrations (4,860 nM and 2-fold dilutions thereof) to immobilized mouse GITRL (expressed in E. coli). (B) Nonlinear 1:1 Langmuir fitting of the steady-state binding data yields a Kd of 4.37 ± 0.15 μM.
Site-Directed Mutagenesis Studies Suggest a Noncanonical Receptor Binding Surface for Mouse GITRL.
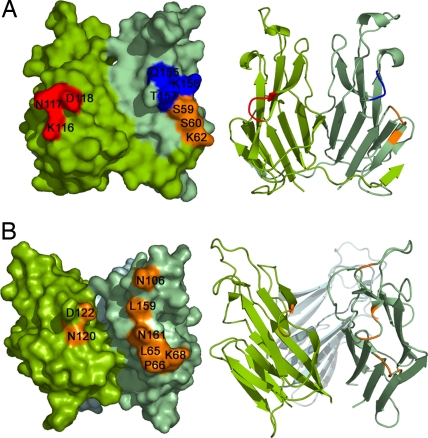
Based on existing structural and biochemical data of previously characterized TNF:TNFR family complexes, the receptor-recognition surfaces on the ligands are formed by the solvent-accessible loops (AA′, CD, DE, and GH) located at the clefts between neighboring monomers in the trimeric ligand (2). Based on the gross deviation of the mouse GITRL dimeric assembly from the canonical TNF family trimer, we investigated the surfaces that contribute to receptor recognition and binding. Three mutations were generated that targeted the surface-exposed loops AA′ (mutant S59A-S60-K62A), DE (mutant K116A-N117A-D118A), and GH (mutant Q155A-K156A-T157A), and they were tested for their receptor binding activity by SPR. Surprisingly, all of the mutants displayed receptor binding activity (Kd ≈ 4–6 μM) similar to that of the wild-type mouse GITRL (SI Fig. 7), whereas corresponding mutations in the human ortholog significantly altered the receptor binding efficacy of human GITRL (15). These data clearly indicate that, unlike the conventional TNF ligands, the AA′, DE, and GH loops in mouse GITRL do not make major contributions to receptor binding (Fig. 4), and suggest the presence of a noncanonical receptor-recognition surface for mouse GITRL.
Fig. 4.
Comparison of the receptor-recognition surfaces of mouse and human GITRL. Molecular surfaces of mouse (A) and human (B) GITRL show the arrangement of the surface exposed loops AA′, DE, and GH. Mutations of the loops AA′ (mutant S59A-S60-K62A; orange), DE (mutant K116A-N117A-D118A; red), and GH (mutant Q155A-K156A-T157A; blue) in mouse GITRL did not alter its receptor binding affinity, whereas corresponding mutations in the human ortholog significantly alter the receptor binding ability of the protein (15). Residues from these loops are closely clustered in the human GITRL, forming its receptor-recognition surface. In the mouse GITRL dimer, the corresponding residues are distantly located, suggesting a noncanonical receptor binding surface. Ribbon diagrams are shown on the right to indicate the orientation of the molecules.
Discussion
The extracellular events associated with signaling through the members of the TNF:TNFR superfamilies, in general, appear to involve the recruitment of three receptor ectodomains by the inherently trimeric TNF ligands. This “trimerization” of the ectodomains brings the receptor cytoplasmic tails into close proximity, resulting in the recruitment of signal adaptor molecules, TRAFs, which initiate the downstream signal transduction pathways (1, 2). In the majority of cases, TNF ligands self-assemble as “bell-shaped” homotrimers in which monomeric subunits are noncovalently associated through interactions between highly conserved hydrophobic surfaces (2, 18). Recently, the structures of OX40L (19) and human GITRL (15) revealed an expanded trimeric arrangement that more closely resemble an “open flower-like” assembly. Despite the somewhat altered organization, their gross overall quaternary features, in combination with the location of their receptor binding surfaces, clearly indicate that OX40L and human GITRL direct formation of a 3:3 receptor:ligand complex with overall similarity to other TNF:TNFR complexes. Taken together, these observations supported the notion that all members of the TNF/TNRF superfamilies use similar mechanisms for receptor–ligand engagement and signaling.
The present study demonstrates that mouse GITRL exhibits a dimeric quaternary structure not observed previously for any TNF ligand. Also, in contrast to all previously characterized TNF:TNFR interactions, including human GITRL:GITR system, mouse GITRL shows a strikingly weaker affinity (micromolar range) for its cognate receptor, mouse GITR. The mouse GITRL dimer is stabilized through a domain-swapping interaction mediated by a set of mostly hydrophobic residues (F171 and I172 at the C terminus from one subunit with H64 and P68 on the other subunit) present only in mouse GITRL. In the human ortholog, the analogous residues are incapable of participating in similar interactions and instead support the formation of a trimeric species. Humanization of mouse GITRL, via replacement of these residues with the human sequence, was predicted to disrupt the domain-swapping interaction and indeed resulted in the destabilization of the mouse GITRL dimeric assembly in favor of a species that exhibited “human-like” solution behavior. Similarly, murinization of the human GITRL sequence significantly altered the solution self-assembly behavior of the human protein to resemble the dimeric mouse GITRL. These results highlight the importance of the domain-swapped interface in stabilizing the atypical dimeric state of mouse GITRL. The mouse GITRL crystal structure demonstrates that the detailed conformation of the mouse GITRL C terminus not only stabilizes the dimeric assembly, but also sterically blocks inclusion of a third subunit into the mouse GITRL oligomeric assembly, thus preventing formation of the typical TNF family homotrimer.
A major concern is that these crystalline and solution properties are the consequence of the protocol used to express the protein in E. coli. Notably, recombinant mouse GITRL purified from the S2 insect cell culture medium showed similar chromatographic behavior consistent with a dimeric assembly of the protein (SI Fig. 6A) and displayed a receptor binding affinity similar to that observed with the E. coli expressed recombinant proteins (Fig. 3B and SI Fig. 6B; see also SI Results in SI Text). Secretion of the recombinant protein into the S2 cell culture medium maximizes the likelihood of obtaining correctly folded material. Thus, the similar physicochemical behaviors of mouse GITRL expressed in either E. coli or S2 expression system supports the notion that the features of mouse GITRL described here are the intrinsic properties of the molecule and are physiologically relevant.
Several existing structures of TNF:TNFR complexes demonstrate that two adjacent ligand monomers contribute solvent-exposed loops near the intersubunit clefts to form the receptor-recognition surface, and, as a consequence, each ligand trimer can bind three receptor molecules (2). The demonstration that dimeric mouse GITRL specifically binds mouse GITR suggests a unique receptor binding surface not exploited in other TNF family members. The unique receptor binding mode of mouse GITRL is supported by our demonstration that alterations of the solvent-accessible residues involved in receptor recognition of all previously characterized TNF family members do not affect its receptor binding activity.
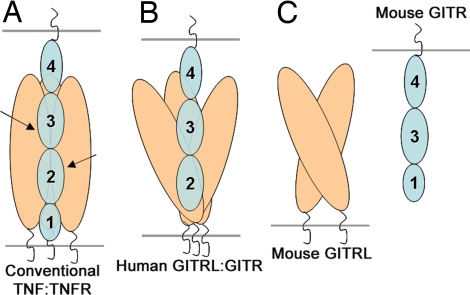
A distinct mode of interaction between mouse GITRL and its receptor is also suggested by the predicted domain organization of mouse GITR. Typical TNFR family members possess four extracellular cysteine-rich pseudorepeats (CRDs) that can be distinguished on the basis of primary sequence characteristics. Direct structural analysis of several TNF:TNFR family complexes demonstrates that the majority of the residues contacting the cognate ligands are contributed from CRD2 and CRD3, with CRD1 and CRD4 making few, if any, contacts (2). The mouse and human GITR are each composed of only three CRDs. The second and the third CRDs present in both the mouse and human GITR show considerable similarity to the canonical CRDs 3 and 4 of the classical TNFRs, respectively; however, there is a considerable divergence between the N-terminal CRDs. Based on sequence analysis, it has been suggested that the first CRD of human GITR conforms better with the characteristics of CRD2 of the conventional TNFRs, whereas that of the mouse protein exhibits a closer relationship to a canonical CRD1 (20, 21). The putative absence of CRD2 in mouse GITR is consistent with an atypical mode of interaction between mouse GITR and mouse GITRL, including the use of a unique ligand recognition surface and the unique dimeric organization of the mouse ligand. In contrast, human GITR appears to possess a canonical CRD2 and thus exhibits a more typical ligand binding interaction, including the use of a traditional binding surface that supports recognition of a trimeric ligand assembly. These differences in receptor sequence and ligand structure (Fig. 5) also correlate with the observed lack of interspecies cross-reactivity between GITRL and GITR (12).
Fig. 5.
Unique dimeric assembly and domain organization of mouse GITRL/GITR suggests a previously unrecognized mode of ligand:receptor interaction in the TNF/TNFR superfamily. (A) In conventional TNF:TNFR complexes, each receptor monomer (containing CRDs 1, 2, 3, and 4) binds at the cleft between the two adjacent subunits of the ligand homotrimer. TNFR CRDs 2 and 3, which typically contribute to ligand binding, are indicated with arrows. (B) Human GITR with CRDs 2, 3, and 4 is shown in complex with the human GITRL trimer. The human GITRL trimer, despite having an atypical expanded assembly, utilizes a canonical receptor binding surface similar to the conventional TNF ligands. (C) Dimeric mouse GITRL appears to possess a noncanonical receptor binding surface. This is consistent with the atypical domain organization of mouse GITR. Mouse GITR contains CRDs 1, 3, and 4 and lacks the signature of CRD 2, which critically contributes to the ligand binding surface(s) of the classical TNFRs.
Taken together, the unique dimeric structure of mouse GITRL, the biochemical demonstration of a previously uncharacterized receptor binding surface on mouse GITRL, and the unique properties of mouse GITR support a previously unrecognized mouse GITRL:GITR assembly. These differences may provide an explanation for the distinct biological functions associated with the mouse and human GITRL:GITR signaling pathways. Earlier reports demonstrated that mouse GITRL does not cross-react with human GITR receptor, and, analogously, human GITRL does not interact with the mouse receptor (12). These observations are consistent with our structural and biochemical data presented here. More interestingly, murine and human GITRL:GITR-mediated signaling appears to produce distinct costimulatory effects on Treg-mediated suppression of effector T cell function (14), which may be the consequence of the unique structural and biochemical properties. Notably, the recruitment of the trimeric signal adaptor molecules TRAFs to the TNFR cytoplasmic tails depends not only on the presence of a definite recruitment motif, but also on the distinct trimeric geometry of the receptor cytoplasmic tails (22, 23). The mouse GITR cytoplasmic tail possesses the TRAF recruitment motifs containing highly conserved acidic residues typical of other TNFR family members (24). However, because of the dimeric organization of murine GITRL, engagement of mouse GITRL:GITR complex would not be expected to support a typical interaction between the receptor cytoplasmic domain and the trimeric TRAFs. This unique behavior offers a potential mechanistic explanation for the distinct outcome of the GITRL:GITR interaction in mice and humans. Further work will be needed to define the exact nature of the mouse GITRL:GITR complex as well as its TRAF recruitment mechanism.
In summary, our studies suggest that the unique structural and biochemical properties exhibited by the mouse and human GITRL orthologs result in distinct signaling mechanisms that are responsible for their different biological functions. Comparison of the mouse GITRL structural features with those of its human ortholog demonstrates the evolution of previously unrecognized protein function through modest sequence divergence and represents a classic example of the sequence → structure → function paradigm.
Methods
Cloning, Expression, and Purification of the Extracellular Domains of Mouse GITRL and GITR.
The mouse GITRL extracellular domain (amino acids 46–173), with an N-terminal 6×His tag, was expressed in E. coli as insoluble inclusion body, refolded, and purified to homogeneity as described before (25). Alternatively, when grown at 25°C, a significant fraction of the protein remained soluble after bacterial cell lysis and was purified by Ni-NTA affinity chromatography and conventional gel-filtration chromatography. Gel-filtration profiles of the protein obtained by the two different purification methods superimposed on each other, indicating identical hydrodynamic properties. The N-terminal His tag was removed by thrombin cleavage. All of the mutants of mouse GITRL were generated by PCR mutagenesis, and the constructs were verified by DNA sequencing. The ectodomain of mouse GITRL, with an N-terminal 6×His tag, was also expressed in S2 insect cells and was purified from the culture medium by Ni-NTA affinity chromatography and gel-filtration chromatography. Extracellular domain of mouse GITR (amino acids 24–145) was expressed in HEK293T cells as Ig-fusion protein and was isolated from the Ig-fusion part by protease cleavage, as described previously (26).
Structure Determination and Analysis.
The crystal structures of mouse GITRL (either refolded from insoluble inclusion body expressed in E. coli or isolated from the soluble fraction of the bacterial cell lysate) were solved and analyzed as described in SI Experimental Procedures in SI Text.
Analytical Ultracentrifugation Analysis.
The analytical ultracentrifugation studies were conducted as described in SI Experimental Procedures in SI Text.
SPR Binding Assays.
Binding assays were performed with a BIAcore X optical biosensor at 25°C as described in SI Experimental Procedures in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We gratefully acknowledge the staff of the X4A beamline at the National Synchrotron Light Source; Drs. H. Wu and C. Terhorst for critical reading of the manuscript and insightful comments; Ya Wang and Dr. Charles Rubin for assistance with SPR experiments; and Rafael Toro for assistance with protein crystallization. This work was supported by National Institutes of Health Grant AI07289 (to S.G.N. and S.C.A.), Albert Einstein Cancer Center Grant (National Cancer Institute, P30CA13330), and a postdoctoral fellowship from the Cancer Research Institute (to K.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2QDN and 3B9I).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710529105/DC1.
References
- 1.Locksley RM, Killeen N, Lenardo MJ. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer JL, Schneider P, Tschopp J. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 3.Nocentini G, Ronchetti S, Cuzzocrea S, Riccardi C. Eur J Immunol. 2007;37:1165–1169. doi: 10.1002/eji.200636933. [DOI] [PubMed] [Google Scholar]
- 4.Shevach EM, Stephens GL. Nat Rev Immunol. 2006;6:613–618. doi: 10.1038/nri1867. [DOI] [PubMed] [Google Scholar]
- 5.Watts TH. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 6.Ronchetti S, Zollo O, Bruscoli S, Agostini M, Bianchini R, Nocentini G, Ayroldi E, Riccardi C. Eur J Immunol. 2004;34:613–622. doi: 10.1002/eji.200324804. [DOI] [PubMed] [Google Scholar]
- 7.Stephens GL, McHugh RS, Whitters MJ, Young DA, Luxenberg D, Carreno BM, Collins M, Shevach EM. J Immunol. 2004;173:5008–5020. doi: 10.4049/jimmunol.173.8.5008. [DOI] [PubMed] [Google Scholar]
- 8.Tone M, Tone Y, Adams E, Yates SF, Frewin MR, Cobbold SP, Waldmann H. Proc Natl Acad Sci USA. 2003;100:15059–15064. doi: 10.1073/pnas.2334901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHugh RS, Whitters MJ, Piccirillo CA, Young DA, Shevach EM, Collins M, Byrne MC. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu J, Yamazaki S, Takahashi T, Ishida Y, Sakaguchi S. Nat Immunol. 2002;3:135–142. doi: 10.1038/ni759. [DOI] [PubMed] [Google Scholar]
- 11.Gurney AL, Marsters SA, Huang RM, Pitti RM, Mark DT, Baldwin DT, Gray AM, Dowd AD, Brush AD, Heldens AD, et al. Curr Biol. 1999;9:215–218. doi: 10.1016/s0960-9822(99)80093-1. [DOI] [PubMed] [Google Scholar]
- 12.Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, Ambrose C, Tschopp J, Schneider P. J Biol Chem. 2006;281:13964–13971. doi: 10.1074/jbc.M601553200. [DOI] [PubMed] [Google Scholar]
- 13.Nocentini G, Riccardi C. Eur J Immunol. 2005;35:1016–1022. doi: 10.1002/eji.200425818. [DOI] [PubMed] [Google Scholar]
- 14.Levings MK, Sangregorio R, Sartirana C, Moschin AL, Battaglia M, Orban PC, Roncarolo MG. J Exp Med. 2002;196:1335–1346. doi: 10.1084/jem.20021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay K, Ramagopal UA, Mukhopadhaya A, Malashkevich VN, DiLorenzo TP, Brenowitz M, Nathenson SG, Almo SC. Proc Natl Acad Sci USA. 2007;104:19452–19457. doi: 10.1073/pnas.0709264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MJ, Schlunegger MP, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia De La Torre J, Huertas ML, Carrasco B. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eck MJ, Sprang SR. J Biol Chem. 1989;264:17595–17605. doi: 10.2210/pdb1tnf/pdb. [DOI] [PubMed] [Google Scholar]
- 19.Compaan DM, Hymowitz SG. Structure (London) 2006;14:1321–1330. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Kwon B, Yu KY, Ni J, Yu GL, Jang IK, Kim YJ, Xing L, Liu D, Wang SX, Kwon BS. J Biol Chem. 1999;274:6056–6061. doi: 10.1074/jbc.274.10.6056. [DOI] [PubMed] [Google Scholar]
- 21.Nocentini G, Giunchi L, Ronchetti S, Krausz LT, Bartoli A, Moraca R, Migliorati G, Riccardi C. Proc Natl Acad Sci USA. 1997;94:6216–6221. doi: 10.1073/pnas.94.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 23.Ye H, Park YC, Kreishman M, Kieff E, Wu H. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 24.Esparza EM, Arch RH. J Immunol. 2005;174:7875–7882. doi: 10.4049/jimmunol.174.12.7875. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Schwartz JC, Guo X, Bhatia S, Cao E, Lorenz M, Cammer M, Chen L, Zhang ZY, Edidin MA, et al. Immunity. 2004;20:337–347. doi: 10.1016/s1074-7613(04)00051-2. [DOI] [PubMed] [Google Scholar]
- 26.Chattopadhyay K, Bhatia S, Fiser A, Almo SC, Nathenson SG. J Immunol. 2006;177:3920–3929. doi: 10.4049/jimmunol.177.6.3920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.