Abstract
Promiscuous expression of tissue-restricted autoantigens in medullary thymic epithelial cells (mTECs) imposes central T cell tolerance. The molecular regulation of this unusual gene expression is not understood, in particular its delineation from cell lineage-specific gene expression control remains unclear. Here, we compared the expression profile of the casein gene locus in mTECs and mammary gland epithelial cells by single cell PCR. Mammary gland cells showed highly correlated intra- and interchromosomal coexpression of milk proteins (the casein genes, lactalbumin-α and whey acidic protein) and one of its transcriptional regulators (Elf5). In contrast, coexpression of these genes in mature CD80hi mTECs was rarely observed and no pattern of gene expression in individual mTECs was discernible. The apparent stochastic expression pattern of genes within the casein locus, the lower mRNA levels compared with mammary gland cells in conjunction with frequent coexpression of insulin in single mTECs clearly delineates the molecular mechanism(s) of promiscuous gene expression from cell lineage-specific gene control.
Keywords: central tolerance, single-cell PCR, tissue-restricted antigens, casein genes, autoimmune regulator
Expression of tissue-restricted antigens (TRA) in the thymus, termed promiscuous gene expression (pGE), extends the scope of central T cell tolerance to virtually all tissues of the body. pGE is foremost a physiological property of medullary thymic epithelial cells (mTECs) and is highly conserved between mouse and man. pGE appears necessary for preventing organ-specific autoimmune diseases in both species (1). Thus, a defect in pGE of numerous self-antigens leads to a multiorgan autoimmune disease in man, known as autoimmune polyglandular syndrome (APS1) (2), and lack of promiscuous expression of even a single TRA can precipitate spontaneous, organ-specific autoimmunity in mice (3, 4). Recessive (i.e., deletion) and dominant tolerance mechanisms (i.e., Treg induction/selection) cooperatively mediate tolerance toward TRAs (5, 6); thus, the immune system apparently does not discriminate between promiscuously and “conventionally” expressed self-antigens with respect to modes of tolerance induction. Interestingly, pGE is apparently not restricted to the thymus but also operates in stromal cells of peripheral lymph nodes and thus might constitute a conserved feature between central and peripheral tolerance (7).
pGE differs in basic aspects from tissue-specific gene regulation. Thus, the temporal regulation during pre- and postnatal development or sex-specific expression patterns as observed in particular cell lineages are abolished in mTECs (8). To date, the only known molecular factor identified to be involved in pGE is the autoimmune regulator (Aire), which controls (to varying degrees) the expression of a large proportion of TRAs in mTECs. Epigenetic mechanisms have been implicated but not yet directly addressed in detail. A role for epigenetic control is insinuated by the following observations (i) promiscuously expressed genes tend to localize in clusters, (ii) the imprinting status of Igf2 is lost in mTECs, (iii) expression of certain TRAs including cancer germ cell antigens correlates with promotor hypomethylation (ref. 1 and unpublished results).
Two models are currently discussed to explain this unorthodox gene regulation. The differentiation model holds that progressive differentiation of mTECs leads to a loss of repressive gene control mechanisms, which results in expression of diverse TRAs in an apparently stochastic manner. Thus, phenotypically mature mTEC, i.e., those expressing high levels of CD80 and MHC class II (6), display the highest degree and diversity of pGE (8). According to this model, single mTECs would express TRAs of mixed tissue origin rather than emulating cell lineage-affiliated patterns. An alternative explanation is the developmental or progressive restriction model, that assumes pGE to be a property of an immature, possibly pluripotent progenitor stage of mTECs. Differentiation of mTECs would progressively restrict pGE. Concomitant with this restriction, cell lineage-affiliated gene expression programs become activated in individual mTECs. In this way, individual mTECs would mimic particular cell lineages with respect to certain gene signatures and signaling cascades (9).
Because the two models predict different gene expression patterns at the single-cell level, expression analysis of selected TRAs in single mTECs should allow a distinction between alternative regulatory mechanisms underlying both models. Here, we applied this approach to genes colocalized in the mouse casein locus. Casein genes figure as prototypic TRAs, their expression is spatially restricted to mammary gland epithelial cells (MEC) and temporally tightly regulated during late pregnancy and the postpartum lactation period by well characterized hormone-dependent signaling pathways (10, 11). Our results clearly reveal distinct gene expression patterns in single mTECs and MECs and thus do not support the view that pGE parsimoniously adopts cell type-specific gene regulation patterns and circuits.
Results
MECs and mTECs Differ in Coexpression Patterns of Milk Protein Genes.
We previously reported that the casein gene family is contiguously expressed in MEC of lactating female mice and in the mature subset of mTEC of nonpregnant females (8). It was, however, unclear whether this read-through of the casein locus also extends to single cells. We applied the multiplex reverse-transcriptase PCR assay to address this issue (12). To validate this method we first performed efficiency and competition tests of primer sets with cDNA mixed from different tissues, and we obtained comparable amplification efficiencies with primer sets for the first and the second PCR. Second, the same gene was amplified separately or in multiplex in the first PCR round, which resulted in the same expression levels [supporting information (SI) Fig. 5].
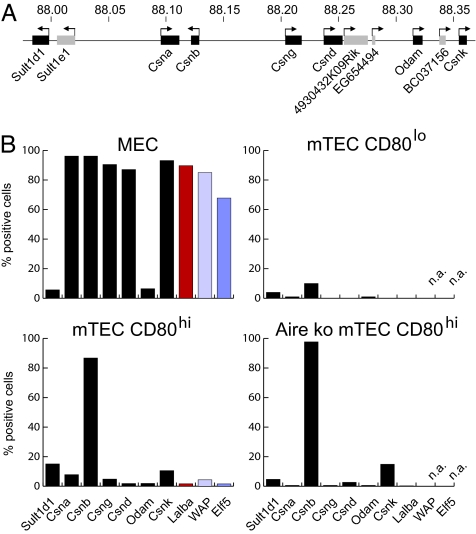
Single-cell PCR analysis was then performed on sorted mTECs and MECs. mTECs were purified from young adult female mice and terminally differentiated MECs from the glands of lactating mothers immediately after weaning. As shown in Fig. 1, the expression frequencies in both cell types were clearly distinct. Approximately 90% of all MECs analyzed coexpressed all casein genes (located on chromosome 5) and, in addition, the milk protein genes lactalbumin-α (Lalba) and the whey acidic protein (WAP) located on chromosomes 15 and 11, respectively (Fig. 1B). The transcription factor Elf5 (E74-like factor 5), implicated in the control of WAP expression (13–15), was expressed at somewhat lower frequency in 70% of cells. In contrast, the non-MEC-affiliated genes Sult1d1 (sulfotransferase family 1D, member 1) and Odam (odontogenic ameloblast associated) exhibited frequencies of expression <10%. Importantly, most MECs (220 of 263; 84%) coexpressed all casein genes and Lalba (Fig. 2A, Table 1, and SI Table 2). Although it was assumed that mature mammary gland cells coexpress all milk protein genes, this had not been verified at the single-cell level. A clear coexpression was also observed for WAP and Elf5 in a smaller sample size (Fig. 2A). The degree of coexpression of WAP and Elf5 was higher in MECs isolated from mothers 1 day after giving birth (coexpression was found in 91% of MEC, data not shown) than at day 28 of lactation (67% of MEC). This finding concurs with a previous report describing a critical role for Elf5-binding sites during late pregnancy but not throughout lactation (13).
Fig. 1.
Expression frequencies of mammary gland-specific genes differ between MEC and mTEC subsets. (A) Diagram of the casein gene region on chromosome 5. Genes analyzed in this study are shown in black, and genes not analyzed are shown in gray. Numbers indicate chromosomal position in megabases. (B) Gene expression was assessed by single-cell PCR in sorted MECs (263 cells), CD80hi mTECs of C57BL/6 (664 cells), or Aire KO mice (343 cells) and in CD80lo mTECs of C57BL/6 mice (100 cells). Genes of the casein gene locus are shown in chromosomal order in black and Lalba (chromosome 15) in red. The expression of Elf5 and its target gene WAP (chromosomes 11 and 2, respectively) was assessed in only approximately one-third of total cells analyzed (blue). The absolute number of positive cells was calculated by adding up all cells in which at least one of the genes analyzed was detectable. The surface molecule EpCAM, which served as sorting marker for all cell types, was detected in 94–99% of positive cells. n.a., not analyzed.
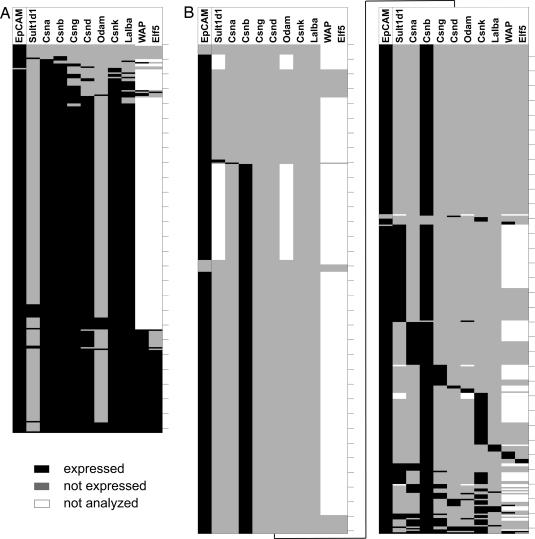
Fig. 2.
Distinct (co)expression patterns of TRAs in individual MECs and mTECs. Each row represents a single cell analyzed for expression of the genes listed above the columns; ticks indicate 10-cell intervals. Black denotes detected expression, gray lack of expression, and white expression not analyzed. Expression patterns were arranged from top to bottom according to increasing number of genes expressed per cell. (A) Expression pattern of mature MECs. Note the high number of cells coexpressing MEC specific genes including the transcription factor Elf5 and its regulated gene WAP. (B) CD80hi mTECs exhibit either expression of single genes or a varied selection of MEC-specific genes. Contiguous expression of the casein locus was found in only one cell (bottom row).
Table 1.
Correlation analysis of gene expression in mammary gland epithelial cells as detected by single-cell PCR
| Sult1d1 | Csna | Csnb | Csng | Csnd | Odam | Csnk | Lalba | WAP | Elf5 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sult1d1 | — | −0.0036* | 0.0048 | 0.0038 | −0.0006 | 0.0686 | 0.0002 | 0.0048 | −0.0072 | −0.0137 |
| Csna | — | 0.7921 | 0.5468 | 0.4205 | 0.0054 | 0.6996 | 0.5135 | 0.7929 | 0.3912 | |
| Csnb | — | 0.5468 | 0.4205 | 0.0054 | 0.6996 | 0.4563 | 0.6238 | 0.3517 | ||
| Csng | — | 0.6383 | 0.0144 | 0.6715 | 0.6586 | 0.8599 | 0.5043 | |||
| Csnd | — | 0.0111 | 0.6198 | 0.6112 | 0.679 | 0.5008 | ||||
| Odam | — | 0.0101 | 0.0157 | 0.0397 | 0.0321 | |||||
| Csnk | — | 0.6368 | 0.8069 | 0.4674 | ||||||
| Lalba | — | 0.8191 | 0.5403 | |||||||
| WAP | — | 0.479 | ||||||||
| Elf5 | — |
*κ < 0.1, no concordance; 0.10–0.40, weak concordance; 0.41–0.60, clear concordance; 0.61–0.80, strong concordance; 0.81–1.00, nearly complete concordance. κ Indices between 0.41 and 1.00 are highlighted in bold (see also SI Table 3).
A correlation analysis of this dataset confirms this conclusion, whereby coexpressed gene pairs show high values of the concordance index κ (0.42–0.86) (Table 1). A weak concordance was observed for Csna–Elf5 and Csnb–Elf5, Sult1d1, and Odam showed no correlation at all with the remaining genes analyzed. Thus, functionally related genes—the milk protein genes and the transcription factor Elf5—were coexpressed with high probability irrespective of genomic localization. This did not apply to colocalized but functionally unrelated genes.
The frequencies and coexpression patterns in single CD80hi mTECs were quite different (Fig. 1B). All genes analyzed except one exhibited expression frequencies between 2% and 15%, a frequency range well in accordance with data on protein expression of selected TRAs, which amount to 1–3% in unseparated mTECs (16–18). A notable exception was casein β (Csnb), which was expressed by 85% of cells. This unexpectedly high frequency is consistent with a previous study reporting invariable expression of Csnb in pools of 20–25 mTECs (19). In contrast to MECs, CD80hi mTECs did not show correlated expression among compared gene pairs. The different milk protein genes were either expressed alone (442 of 664 cells; 67%) or less frequently in various combination (141 of 664 cells; 21%) (Fig. 2B, SI Table 2, and SI Table 3). For instance, Csng and Csnk were found to be coexpressed in 10 cells. Of 664 single mTECs tested, only one cell expressed all but one milk protein gene (Lalba) and thus closely mimicked the MEC pattern. A clear correlation was only observed between the functionally nonrelated genes Csnd and Odam (SI Table 3). These data clearly show that mTECs do not emulate MEC-specific gene signatures.
Frequency of TRA-Expressing Cells Differs Among mTEC Subsets.
The up-regulation of pGE in whole populations of CD80hi versus CD80lo mTECs (8) could be due to a quantitative increase in mRNA expression levels per cell and/or an increase in the frequency of cells expressing these genes. Hence, we compared by single-cell PCR the frequency of CD80lo versus CD80hi mTECs for the same selected set of TRAs. The frequency for all eight TRAs was increased in CD80hi mTECs. Thus, of 100 CD80lo cells tested, not a single cell expressed Csng or Csnd, and the frequency of Csnb-expressing cells was increased approximately eightfold in the CD80hi population. These findings argue for de novo induction of pGE during mTEC maturation (Fig. 1B and SI Table 4). Note also that the rare antigen-positive cells among CD80lo mTECs express similar mRNA levels to those in the mature subset (see below).
Single Aire-Positive mTECs Coexpress both Aire-Dependent and -Independent Genes.
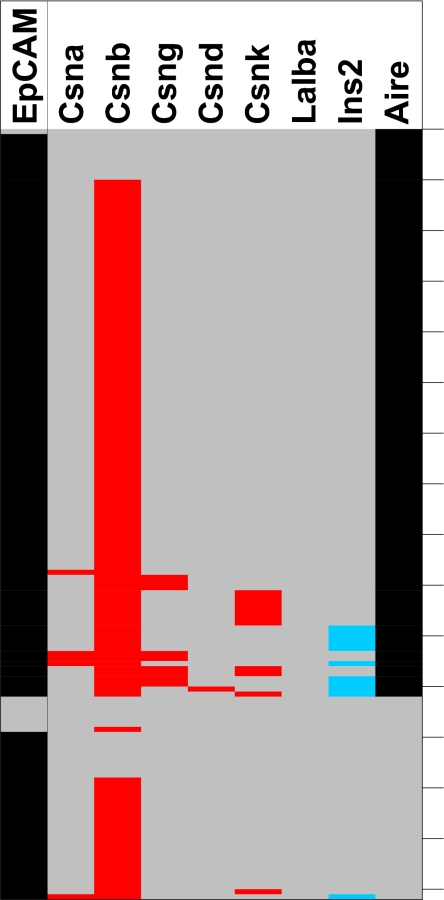
The delineation of Aire-dependent and -independent pools of promiscuously expressed TRAs (8, 20) and the heterogeneity of mTECs with regard to Aire expression (21, 22) raised the question whether there is a strict correlation between expression of Aire and the Aire-dependency of TRAs at the single-cell level. The casein locus encompasses Aire-dependent (casein α, δ, and γ) and -independent (casein β and κ) genes. At the single-cell level, Aire was expressed in approximately two-thirds of CD80hi mTECs, which was higher than reported (19) (Fig. 3). Although expression of Aire-dependent genes was restricted to Airepos cells (with one exception), the majority of Airepos mTECs did not (co)express Aire-dependent genes. Thus, Aire is necessary but not sufficient for expression of a particular Aire-dependent gene. We also note that 10 of 11 casein κ-positive cells segregated into the Airepos mTEC pool, whereas the more prevalent Aire-independent gene casein β was expressed in both Aire-positive and -negative mTECs (Fig. 3). The frequency of mTECs expressing Aire-dependent genes was drastically reduced in Aire KO mice, whereas there was a marginal increase in the frequencies of cells expressing Aire-independent genes (Fig. 1B). Thus, Csnb–mRNA-expressing cells increased from 87% to 98% and Csnk-positive cells from 11% to 15%, and there was no evidence for concordant regulation of these genes in the absence of Aire (SI Table 5).
Fig. 3.
(Co)expression of Aire, milk protein genes, and insulin2 in single CD80hi mTECs. Milk protein genes are marked in red and insulin in blue. Note that single CD80hi mTECs show coexpression of Aire-dependent and -independent milk protein genes together with insulin2. Black indicates expression, and gray indicates lack of expression.
Single mTECs Coexpress Genes Affiliated with Distinct Cell Lineages.
To address the question of whether single mTECs coexpress TRAs affiliated with distinct cell lineages, we analyzed the expression of the different casein genes in combination with insulin2, a gene that is selectively expressed in pancreatic β cells. We detected single mTECs coexpressing casein genes and insulin2 (Fig. 3 and data not shown). Overall, 18 of 236 cells analyzed expressed insulin2, and 9 of these cells coexpressed at least one casein gene in addition to Csnb. There was, however, no significant preference for coexpression of the Aire-dependent subset of casein genes with insulin. Thus, single mTECs do not strictly adhere to cell lineage-specific gene expression patterns but rather coexpress genes, the products of which serve dedicated functions in different terminally differentiated cell types.
Mature mTECs and MECs Express TRA at Different Levels.
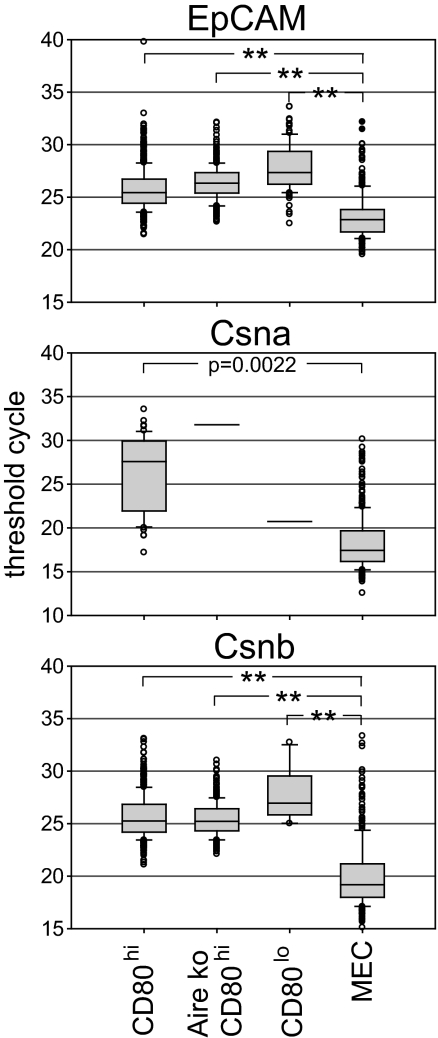
Comparison of mRNA expression levels of certain TRAs between whole thymus and the respective tissue revealed large differences, initially raising doubts as to whether such low expression levels would suffice to impose central tolerance. Thus, by quantitative RT-PCR analysis, levels of retinal S-antigen in human thymus were approximately five orders of magnitude lower than those in retina (23). Likewise, expression of Csna and Csnd differed between purified mTECs and mature MECs by four to five orders of magnitude (data not shown). This was, in part, explained by the low frequency of mTECs expressing a given TRA, as shown by protein staining either on cytospin preparations or on histological sections (24). However, it is important to establish at a single-cell level the amount of mRNA for TRAs in mTECs and compare this to individual cells of the specialized tissue. By using the threshold cycle number as a relative measure of quantitative differences in mRNA levels, we show that mTECs express 167-fold and 53-fold lower RNA levels for Csna and Csnb, respectively, when compared with MECs (Fig. 4). Thus, apart from differences in expression patterns, the cellular mRNA abundance constitutes a second distinguishing feature between promiscuous and tissue-specific gene expression. The low frequency of TRA-positive mTECs together with the low expression levels per cell attest to the high efficacy and sensitivity of the tolerance process (25).
Fig. 4.
Quantitative mRNA expression levels in MECs versus mTEC subsets. Threshold cycle (ct) numbers for all cells showing expression of EpCAM, Csna or Csnb are summarized as box plots including all outliers. MEC generally exhibited the lowest ct values, i.e., highest expression levels, whereas the mTEC subsets showed nearly similar values. Note that Csna expression was detectable in only two Aire KO mTEC CD80hi cells and a single WT mTEC CD80lo cell. **, P < 0.0001.
Discussion
Single-cell PCR analysis of mTECs and MECs reveals distinct intra- and interchromosomal coexpression patterns. The set of cells, in which expression of all casein genes and two additional milk proteins was assessed, revealed coexpression in 78% of MECs, whereas none of the mTECs displayed this signature. Expression patterns in mTECs varied from expression of a single gene and permutated combinations of coexpression to contiguous expression of the casein locus including the functionally unrelated Sult1d1 and the Odam gene. Thus, in rare mTECs, a genomic region spanning at least 0.4 Mb was accessible for transcription. The genealogy among these individual mTECs is unknown. Given the clonal origin of mTECs (26), it is conceivable that pGE becomes increasingly complex with clonal expansion. Accordingly, cells with broader expression patterns might have descended from those with a more limited expression pattern. We would like to point out that these findings do not contradict our previous observations that promiscuously expressed genes are clustered in the genome and that the casein locus is “apparently” contiguously transcribed at the population level (8).
Differences between mTECs and MECs included not only target genes but also regulatory circuits. WAP gene transcription is controlled by the factor Elf5 in MECs (13–15), and this is reflected by the highly concordant expression of both genes in these cells (Fig. 2 and Table 1). This concordance is not observed in mTECs (SI Table 3), suggesting that promiscuous expression of WAP is not contingent on Elf5. We, however, cannot formally exclude that such a dependency exists at the initiation of WAP transcription. Our conclusion that pGE differs from activation of cell lineage-specific gene expression patterns of a certain cell type is further corroborated by the lower mRNA expression levels in mTECs and the observation that single mTECs coexpress TRAs affiliated with distinct cell lineages, i.e., insulin and caseins.
Mature mTECs fall into an Airepos (70%) and an Airene subset (30%) consistent with a recent report (27). When correlating expression of Aire with Aire-dependent and Aire-independent genes at the single cell level, it became clear that Aire is necessary but not sufficient for expression of Aire-dependent genes (Fig. 3). The fact that cells expressing the Aire-independent gene Csnk also partitioned into the Airepos subset suggests that Aire expression rather than MHC/CD80 up-regulation marks the stage at which certain Aire-independent genes (with the obvious exception of Csnb) become accessible to expression (8). These data are compatible with the terminal differentiation, whereby CD80hi Airene and Airepos subsets represent sequential differentiation stages with the Airepos cells representing the most mature stage (28–30).
Our data do not exclude an alternative differentiation pathway, namely that mature CD80hi mTECs split into a major Airepos and a minor Airene sublineage. Such a subdivision concurs with the recent description of a mTEC sublineage marked by high expression of claudin 3 and 4 (22). This sublineage also displays high levels of MHC class II, CD80, and is largely Airepos when analyzed at the clonal level in situ. Our data argue against Aire functioning as a master switch for lineage-affiliated gene expression programs (31), at least as far as mammary epithelial cells are concerned, because certain milk proteins continued to be expressed in the absence of Aire, even at elevated frequencies.
The frequencies of mTECs expressing a particular TRA at the mRNA level are well in accordance with previous estimates based on protein detection, i.e., 1–3% of total mTECs. A tight correlation between protein and mRNA expression has also been confirmed by the finding that mTECs isolated on the basis of surface expression of a particular TRA are selectively enriched for the corresponding mRNA (16). This close correlation makes it unlikely that our single-cell PCR analysis grossly under estimates the frequency of mRNA-expressing cells because of levels below the detection threshold. Based on the observation that 2.9% of EpCAM-sorted CD80hi mTECs do not express the corresponding message, we estimate this potential error to be in a similar range (Fig. 2) for other genes. Following the same reasoning, it is unlikely that the slightly different protocols used for the isolation of the two cell types skew the expression frequencies in favor of MECs.
An obvious exception is Casein β mRNA, which is expressed in >85% of CD80hi and in ≈10% of CD80lo mTECs. This “overexpression” of casein β has also been noted in a previous analysis of pools of 20 to 25 mTECs (19). At present, the significance of this observation and the underlying mechanism are unclear. The Csnb gene and its promoter are evolutionarily closely related to Csn-α, -γ, and -δ, whereas Csnk is not related to this group (32). Expression of the casein genes in MEC is synergistically induced by prolactin and glucocorticoids, which activate composite response elements present in the proximal promoters of the casein genes and upstream enhancers of the Csna and Csnb genes. Three transcription factors, Stat5a (signal transducer and activator of transcription 5A), C/EBPβ (CCAAT/enhancer binding protein), and GR (glucocorticoid receptor), are required for activation of the casein genes, but none of these factors is mammary-specific, and no locus control region has been yet identified, which might control the expression of the whole region en bloc (11, 33). Interestingly, transgenes driven by the Csnb promoters either in mice or cell lines often led to higher expression levels than constructs driven by other casein promoters. Hence, it has been argued that the Csnb promoter might include additional elements that confer stronger promoter activity (33). Indeed, the first intron of the mouse Csnb gene does contain additional enhancer and promoter sequences, which increase the basal activity of this gene in MEC (34). This may also explain the elevated expression in mTECs. Irrespective of the underlying molecular mechanisms the expression of Csnb in virtually all mature mTECs might serve as an transcriptional entry site for this locus from which pGE spreads into either direction (35). It will be interesting to know whether this phenomenon is particular to the casein locus or applies also to other promiscuously expressed gene clusters.
Our PCR data only provide a static picture of gene expression. The varied expression pattern of the casein locus as observed in mTECs, however, might reflect oscillating rather than static expression at the level of individual genes. Thus, a gene may be transcribed only at a given point in time in each cell. Our snapshot analysis would not fully reveal the cumulative gene expression during the lifespan of an individual mTEC. Depending on the half-life of peptide/MHC complexes on mTECs (estimated to be ≈20 h; B.K., unpublished results), such a discontinuous transcription might still suffice to provide a steady supply of ligands for tolerance induction.
We also cannot exclude that mTECs and MEC differ in their usage of multiple transcription start sites and alternative promoters. There is precedence for cell type-specific usage of alternative promoters in spermatogenesis (36, 37). If this were the case, the PCR primers chosen might not have allowed detection of a fraction of these alternative transcripts in mTECs. It has only recently been appreciated that a significant proportion of the transcriptome is derived from overlapping and fusion transcripts generated from alternative promoters (38, 39).
Taken together, our data do not lend support to the view that mTECs emulate tissue-specific gene signatures, a central tenet of the developmental model (9). Instead the varied, apparently stochastic pattern of gene expression in single mTECs favors a model, in which gene clusters (also including functionally unrelated genes) become accessible for transcription in CD80hi mTECs. Conceivably, the particular stoichiometry of transcription factors, which may fluctuate in individual mTECs, then determines which pattern is turned on in a particular cell at a given time.
Materials and Methods
Animals.
C57BL/6 and C3H mice were obtained from Charles River Laboratories. Aire KO mice were generously provided by L. Peeltonen (University of Helsinki, Helsinki, Finland) and were bred on a mixed genetic background. All mice were kept under specific pathogen-free conditions at the animal facilities of the German Cancer Research Center (DKFZ).
Tissue Preparation and Single-Cell Sort.
mTECs were purified as described (8). Briefly, TECs were enriched on a discontinuous Percoll density gradient (densities 1.07, 1.045, and 1.0) from thymi sequentially digested by collagenase and collagenase/dispase. The TEC-enriched fraction was stained with FITC-anti-Ly51 (clone 6C3), PE-anti-CD80 (clone 16–10A1), PerCP-anti-CD45 (clone 30-F11), and Alexa Fluor 647-anti-EpCAM (clone G8.8). Staining was preceded by blocking with the anti-FcR mAb 2.4G2. mTECs were identified as CD45-Ly51-EpCAM+ and sorted according to their CD80 expression, as CD80hi or CD80lo, representing the top and bottom 30% of the population. For the isolation of MECs, all mammary glands from a mouse lactating for 4 weeks were excised, trimmed of connective tissue, and cut into little pieces. The tissue fragments were digested for two rounds with a collagenase/dispase solution followed by five rounds of trypsin digestion. All incubations were performed at 37°C with slow stirring for 20 min. After each step, the tissue slurry was filtered through 60-μm gauze, the tissue fragments were transferred back into the digest, and the flow-through was spun down. All cell pellets were combined, again filtered through 60-μm gauze, washed in PBS and the pellet was resuspended in 30 ml of Percoll solution (density 1.06). The solution was overlayered with 6 ml of PBS and the tube centrifuged for 30 min at 3,500 × g. The cells from the interphase were collected, washed, and stained with Alexa Fluor 647-anti-EpCAM (clone G8.8) and PI. EpCAM+PI− cells, i.e., MECs, were sorted. Cell sorting was performed with a FACSDiVa (Becton Dickinson) at 16 psi in single-cell mode by using the automatic cell deposition unit. Cells were collected in 5 μl of PBS-DEPC (0.1%) in 0.2-ml PCR 8-well stripes arranged in 96-well format and stored at −80°C. Reliable single-cell sorting into a volume of 5 μl required precise deposition of cells into the center of the tube. This was independently tested for each experiment by sorting fluorescent beads and visually inspecting their deposition in a target area corresponding to the surface area of the 5-μl cell collecting volume.
Single-Cell PCR.
Primer design, reverse transcription, first PCR amplification and real-time quantitative PCR were performed essentially as described (12) by using DNA engine Dyad (MJ Research) and 7300 real-time PCR System (Applied Biosystems) machines. The cycle number for the first PCR was lowered from 15 to 10, because we observed an improved correlation between input cDNA and resulting threshold cycle (Ct) values. Our data were analyzed in a qualitative fashion, because the PCR efficiency varied among cells for the same and different genes. In case of an atypical melting curve, the product from the respective well was reamplified by the appropriate primer combination and sequenced to verify the specificity. Primer sequences are available upon request.
Statistical Methods.
κ coefficients were calculated with SAS, Version 9.1 (SAS Institute). A linear mixed model (PROC MIXED in SAS) was used to determine the contrasts of the threshold cycle values between the different cell types. A P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS.
We are most grateful to Benedita Rocha and Marta Monteiro (Necker Institute, Paris) for teaching us the single-cell PCR method. We thank Annette Kopp-Schneider (German Cancer Research Center, Heidelberg) for assistance with the statistical analysis and Ludger Klein and Wolfgang Schmid for comments on the manuscript. This work was supported by the German Cancer Research Center (S.P., K.H., and B.K.) and the German Research Foundation (DFG) Sonderforschungsbereich 405 (J.D., S.R., and B.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707486105/DC1.
References
- 1.Kyewski B, Derbinski J. Nat Rev Immunol. 2004;4:688–698. doi: 10.1038/nri1436. [DOI] [PubMed] [Google Scholar]
- 2.Peterson P, Peltonen L. J Autoimmun. 2005;25(Suppl):49–55. doi: 10.1016/j.jaut.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 3.DeVoss J, Hou Y, Johannes K, Lu W, Liou GI, Rinn J, Chang H, Caspi RR, Fong L, Anderson MS. J Exp Med. 2006;203:2727–2735. doi: 10.1084/jem.20061864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavanescu I, Kessler B, Ploegh H, Benoist C, Mathis D. Proc Natl Acad Sci USA. 2007;104:4583–4587. doi: 10.1073/pnas.0700259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 6.Aschenbrenner K, D'Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, Rolink A, Klein L. Nat Immunol. 2007;8:351–358. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura AR, Heath JK, Turley SJ. Nat Immunol. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 8.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillard GO, Farr AG. J Exp Med. 2005;202:15–19. doi: 10.1084/jem.20050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennighausen L, Robinson GW. Nat Rev Mol Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 11.Kabotyanski EB, Huetter M, Xian W, Rijnkels M, Rosen JM. Mol Endocrinol. 2006;20:2355–2368. doi: 10.1210/me.2006-0160. [DOI] [PubMed] [Google Scholar]
- 12.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Genome Res. 2004;14:1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKnight RA, Spencer M, Dittmer J, Brady JN, Wall RJ, Hennighausen L. Mol Endocrinol. 1995;9:717–724. doi: 10.1210/mend.9.6.8592517. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D, et al. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris J, Stanford PM, Sutherland K, Oakes SR, Naylor MJ, Robertson FG, Blazek KD, Kazlauskas M, Hilton HN, Wittlin S, et al. Mol Endocrinol. 2006;20:1177–1187. doi: 10.1210/me.2005-0473. [DOI] [PubMed] [Google Scholar]
- 16.Cloosen S, Arnold J, Thio M, Bos GM, Kyewski B, Germeraad WT. Cancer Res. 2007;67:3919–3926. doi: 10.1158/0008-5472.CAN-06-2112. [DOI] [PubMed] [Google Scholar]
- 17.Derbinski J, Schulte A, Kyewski B, Klein L. Nat Immunol. 2001;2:1032–1039. [Google Scholar]
- 18.Taubert R, Schwendemann J, Kyewski B. Eur J Immunol. 2007;37:838–848. doi: 10.1002/eji.200636962. [DOI] [PubMed] [Google Scholar]
- 19.Gillard GO, Farr AG. J Immunol. 2006;176:5815–5824. doi: 10.4049/jimmunol.176.10.5815. [DOI] [PubMed] [Google Scholar]
- 20.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 21.Heino M, Peterson P, Sillanpaa N, Guerin S, Wu L, Anderson G, Scott HS, Antonarakis SE, Kudoh J, Shimizu N, et al. Eur J Immunol. 2000;30:1884–1893. doi: 10.1002/1521-4141(200007)30:7<1884::AID-IMMU1884>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 22.Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, Minato N. Nat Immunol. 2007;8:304–311. doi: 10.1038/ni1438. [DOI] [PubMed] [Google Scholar]
- 23.Takase H, Yu CR, Mahdi RM, Douek DC, Dirusso GB, Midgley FM, Dogra R, Allende G, Rosenkranz E, Pugliese A, et al. Int Immunol. 2005;17:1131–1140. doi: 10.1093/intimm/dxh275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith KM, Olson DC, Hirose R, Hanahan D. Int Immunol. 1997;9:1355–1365. doi: 10.1093/intimm/9.9.1355. [DOI] [PubMed] [Google Scholar]
- 25.Peterson DA, DiPaolo RJ, Kanagawa O, Unanue ER. J Immunol. 1999;162:3117–3120. [PubMed] [Google Scholar]
- 26.Rodewald HR, Paul S, Haller C, Bluethmann H, Blum C. Nature. 2001;414:763–768. doi: 10.1038/414763a. [DOI] [PubMed] [Google Scholar]
- 27.Venanzi ES, Gray DH, Benoist C, Mathis D. J Immunol. 2007;179:5693–5700. doi: 10.4049/jimmunol.179.9.5693. [DOI] [PubMed] [Google Scholar]
- 28.Gabler J, Arnold J, Kyewski B. Eur J Immunol. 2007;37:3363–3372. doi: 10.1002/eji.200737131. [DOI] [PubMed] [Google Scholar]
- 29.Gray D, Abramson J, Benoist C, Mathis D. J Exp Med. 2007;204:2521–2528. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, McConnell FM, Scott HS, Penninger JM, Jenkinson EJ, et al. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillard GO, Dooley J, Erickson M, Peltonen L, Farr AG. J Immunol. 2007;178:3007–3015. doi: 10.4049/jimmunol.178.5.3007. [DOI] [PubMed] [Google Scholar]
- 32.Rijnkels M, Elnitski L, Miller W, Rosen JM. Genomics. 2003;82:417–432. doi: 10.1016/s0888-7543(03)00114-9. [DOI] [PubMed] [Google Scholar]
- 33.Kolb AF. Biochim Biophys Acta. 2002;1579:101–116. doi: 10.1016/s0167-4781(02)00533-x. [DOI] [PubMed] [Google Scholar]
- 34.Kolb A. Biochem Biophys Res Commun. 2003;306:1099–1105. doi: 10.1016/s0006-291x(03)01104-5. [DOI] [PubMed] [Google Scholar]
- 35.Oliver B, Parisi M, Clark D. J Biol. 2002;1:4. doi: 10.1186/1475-4924-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecht NB. Dev Genet. 1995;16:95–103. doi: 10.1002/dvg.1020160202. [DOI] [PubMed] [Google Scholar]
- 37.Sassone-Corsi P. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 38.Kapranov P, Willingham AT, Gingeras TR. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 39.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.