Abstract
In the present report, a method based on chip-based nanoelectrospray mass spectrometry (nanoESI-MS) is described to detect noncovalent ligand binding to the human estrogen receptor α ligand-binding domain (hERα LBD). This system represents an important environmental interest, because a wide variety of molecules, known as endocrine disruptors, can bind to the estrogen receptor (ER) and induce adverse health effects in wildlife and humans. Using proper experimental conditions, the nanoESI-MS approach allowed for the detection of specific ligand interactions with hERα LBD. The relative gas-phase stability of selected hERα LBD–ligand complexes did not mirror the binding affinity in solution, a result that demonstrates the prominent role of hydrophobic contacts for stabilizing ER–ligand complexes in solution. The best approach to evaluate relative solution-binding affinity by nanoESI-MS was to perform competitive binding experiments with 17β-estradiol (E2) used as a reference ligand. Among the ligands tested, the relative binding affinity for hERα LBD measured by nanoESI-MS was 4-hydroxtamoxifen ≈ diethylstilbestrol > E2 >> genistein >> bisphenol A, consistent with the order of the binding affinities in solution. The limited reproducibility of the bound to free protein ratio measured by nanoESI-MS for this system only allowed the binding constants (Kd) to be estimated (low nanomolar range for E2). The specificity of nanoESI-MS combined with its speed (1 min/ligand), low sample consumption (90 pmol protein/ligand), and its sensitivity for ligand (30 ng/mL) demonstrates that this technique is a promising method for screening suspected endocrine disrupting compounds and to qualitatively evaluate their binding affinity.
Keywords: electrospray ionization mass spectrometry, noncovalent, nuclear receptor, estrogen receptor, endocrine disruptors, solution affinity
The estrogen receptor (ER) belongs to the nuclear receptor (NR) superfamily of ligand-activated transcription regulators, which are involved in many processes such as growth, organ differentiation, and development of reproductive tissues. The ER, which comprises a DNA-binding domain (DBD) and a ligand-binding domain (LBD), activates the transcription of target genes in response to the binding of estrogens, a group of steroid compounds, to the LBD. Once bound by hormones, the ER undergoes a conformational change that facilitates dimerization and subsequent interactions with the specific DNA sequence (Kumar and Chambon 1988; Steinmetz et al. 2001). The large hydrophobic cavity of the ER LBD allows the binding of a wide variety of nonsteroidal compounds through hydrophobic interactions. In the last decades, a variety of biologically active synthetic chemicals have been released into the environment that can disrupt normal endocrine functions in wildlife and humans (Colborn et al. 1993; Harrison et al. 1997). It is now recognized that some of these environmental contaminants disrupt the endocrine system by binding to the ER LBD with varying affinities. Due to the ability of these hormone-like compounds to interfere with endocrine systems, they are called endocrine disruptors.
The primary question in the field of endocrine disruptors remains whether or not a given chemical has an endocrine disruptor activity. Many in vivo and in vitro assays have been used to investigate estrogenic activity, and they possess their own advantages and drawbacks (Sonnenschein and Soto 1998). It is usually recognized that in vitro assays have advantages in terms of cost effectiveness and screening efficiency of a large number of compounds, but for a definitive demonstration of endocrine-disrupting activity it is necessary to perform in vivo tests (Beresford et al. 2000). For many chemicals, solution affinity has been determined by radiolabeled assays (Kuiper et al. 1998; Blair et al. 2000) and fluorescence polarization methods (Bolger et al. 1998; Nikov et al. 2000; Ohno et al. 2002, 2003). To investigate the bioactivity of a large number of compounds suspected as endocrine disruptors, the method of choice needs to be simple, robust, and easily automated in order to obtain high sample throughput.
The method proposed here is electrospray ionization mass spectrometry (ESI-MS), well known as an important tool for primary structure determination and characterization of purified proteins. Several reports have shown the potential of ESI-MS to characterize the binding of NR ligands to their receptor in terms of stochiometry, specificity, and stability (Greschik et al. 2002; Lengqvist et al. 2002, 2004, 2005; Bitsch et al. 2003; Potier et al. 2003; Stehlin-Gaon et al. 2003; Sanglier et al. 2004). ESI-MS has been successfully used to assess noncovalent complex stability in the vacuum of the mass spectrometer by analyzing their resistance to dissociation (Loo et al. 1997; Wu et al. 1997; van der Kerk-van Hoof and Heck 1999) and for measuring solution binding constants (Jorgensen et al. 1998; Sannes-Lowery et al. 2000; Daniel et al. 2003; Gabelica et al. 2003; Wendt et al. 2003; Tjernberg et al. 2004; Wortmann et al. 2005; De Vriendt et al. 2006). By using correction factors or assumptions to derive solution concentrations from the ion intensities of the different species, a number of these studies have demonstrated that binding affinities determined by ESI are in good agreement with solution-phase data. With regard to its speed, sensitivity, and ability to directly determine binding stochiometry, ESI-MS has the potential to become a superior screening method for suspected endocrine-disrupting compounds. The recent development of an automated chip-based nanoflow electrospray (nanoESI) adds other important advantages to ESI-MS for studying protein–ligand complexes, i.e., high sensitivity and low sample consumption combined with high-throughput capability (Keetch et al. 2003; Zhang et al. 2003).
ESI-MS still faces challenges to characterize native proteins for two main reasons. First, it is necessary to use an aqueous environment to maintain intact complexes in solution. This requires harsher MS transfer/desolvation conditions compared with organic solvents, under which the complex may be destroyed. Therefore, a compromise between sufficient desolvation and intact complex detection has to be found. This can result in peak broadening due to adduct formation with solvent, salt, and buffer molecules present in the spray solution. Secondly, the removal of the solvent environment during the ESI process influences the strength of the noncovalent interaction. Thus, the survival of a noncovalent complex in the gas phase is largely dictated by the nature of the interactions. Ionic and hydrogen bonds are strengthened (Wu et al. 1997), while hydrophobic interactions are weakened (Robinson et al. 1996).
In the present work, a chip-based nanoESI-MS study of the triple mutant hERα LBD is reported for which a crystal structure has been shown previously (Gangloff et al. 2001). The triple mutant hERα LBD (for simplification, hERα LBD) has been demonstrated to bind 17β estradiol (E2) as strongly as the wild type and to have limited transcriptional capacity due to an antagonist conformation of helix H12. These protein characteristics and stability allowed us to use the triple mutant as a model for developing an efficient ligand screening method based on nanoESI-MS. The application of nanoESI-MS will be demonstrated to investigate the gas-phase stability of hERα LBD complexed with a natural hormone, drug molecules, an environmental contaminant, and a phytoestrogen. Using competitive binding experiments with a reference ligand, we demonstrate here the ability of nanoESI-MS to probe the relative solution-binding affinity of different ligands. The method can be easily applied for screening a library of suspected endocrine-disrupting compounds. In addition, application of nanoESI-MS for calculating solution-phase equilibrium constants will be discussed for hERα LBD incubated with small hydrophobic ligands.
Results and Discussion
Detection of native hERα LBD by nanoESI-MS
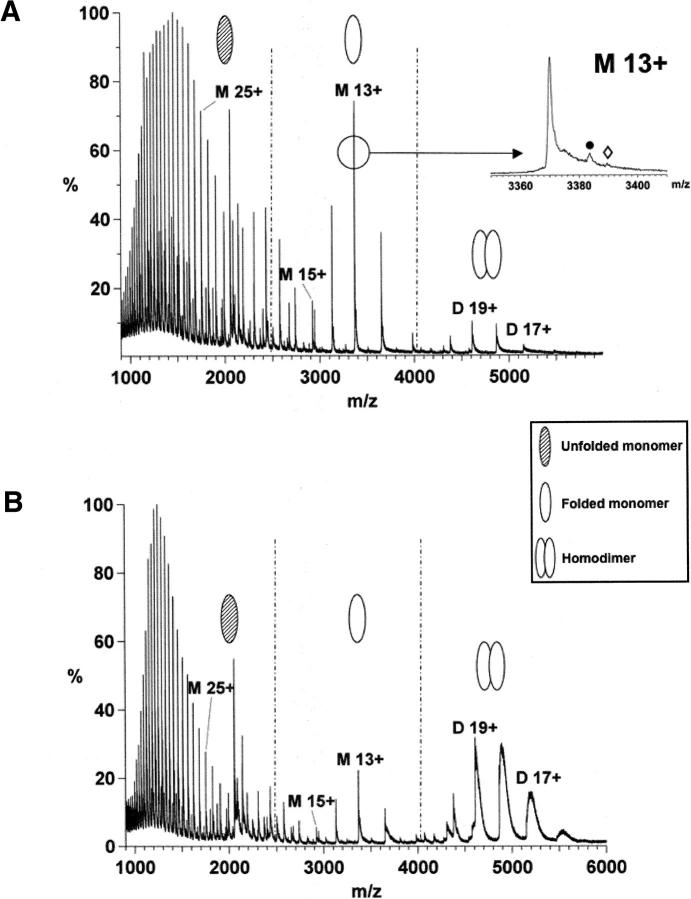
The ability to detect noncovalent complexes using ESI-MS depends on both instrumental and solution conditions. As a first step, the solution conditions (e.g., pH and buffer concentration) were optimized to simulate a near-native environment for hERα LBD in order to preserve noncovalent interactions, including ligand binding. Under denaturing conditions, a broad distribution of charge states (21+–60+) consistent with ions of the unfolded hERα LBD monomer was measured (data not shown). Each protein peak showed satellite peaks at higher m/z that correspond to the addition of 178 Da and 258 Da. They are assigned to the gluconoylation (+C6H10O6, MW 178.14) and phosphogluconoylation (+C6H11O9P, MW 258.12), respectively, of the His6-affinity tag (Geoghegan et al. 1999). Such post-translational modifications were previously observed for nuclear receptor proteins overexpressed in Escherichia coli (Lengqvist et al. 2002, 2004, 2005; Stehlin-Gaon et al. 2003). The deconvoluted molecular mass of 43,814 Da did not agree with the molecular mass based on the known amino acid sequence (43,947 Da). This 133 Da difference was attributed to the loss of the N-terminal methionine. With nondenaturing conditions (Fig. 1), the resulting spectra show three charge states envelopes, which were assigned to the hERα LBD homodimer (labeled D n+, m/z range 4000–5600), the folded monomer (labeled M n+, m/z range 2500–4000), and the unfolded monomer (m/z range 900–2500). Increasing the pH from 6 to 7.4 enhanced the relative homodimer abundance and decreased the homodimer and monomer average charge state (from +18.4 to +18 and +13.1 to +12.7, respectively). This suggests that a higher pH better simulates a native environment for the protein (native solution pH of this protein is 7–8). Increasing the ammonium acetate concentration from 1 to 50 mM had little influence on the homodimer relative abundance (data not shown), but resulted in a decrease of the homodimer and folded monomer average charge states (from +19.3 to +18.4 and from +14 to +13.1, respectively). This suggests a more folded conformation at high ionic strength (Donald et al. 2001; Kapur et al. 2002). These distinct molecular ion envelopes were observed for the hERα LBD (303–553) in fusion with ubiquitin (Witkowska et al. 1996).
Figure 1.
Positive nanoESI mass spectra of hERα LBD acquired under different conditions. (A) hERα LBD in 50 mM NH4OAc, pH 6. (B) hERα LBD in 50 mM NH4OAc, pH 7.4. The broad homodimer peak under B indicates that more water/salt molecules are trapped in the folded structure. The M 13+ charge state expansion for pH 6 shows the post-translational modifications: (•) His6-tag gluconolyation, (⋄) His6-tag phosphogluconoylation.
The detection of noncovalent complexes by ESI-MS depends on the collision frequency of the ions with the residual gas along the path from atmosphere to vacuum and on the center-of-mass energy. Therefore, a careful selection of the MS transfer conditions, i.e., the transfer voltages and the gas pressure, is required where the internal energy of the ions is below the dissociation threshold of the complex (Sobott et al. 2005). Experimentally, this is controlled by reducing the transfer voltages, which control the kinetic energy of the ions, and/or increasing the source pressure (Schmidt et al. 2001; Tahallah et al. 2001; Sanglier et al. 2002; Tjernberg et al. 2004). Thus, the detection of the folded hERα LBD monomer at a native pH could in theory be due to homodimer dissociation during the MS transfer. However, using “softer” MS transfer conditions, the homodimer abundance was not affected (data not shown), an observation that is fully consistent with the strong monomer–monomer interactions stabilized by hydrogen bonds (Shiau et al. 1998). Therefore, the nanoESI-MS data suggest a solution equilibrium between the folded monomer and the homodimer for the triple mutant hERα LBD. The purpose of the fusion protein (see Materials and Methods) for the triple mutant hERα LBD overexpressed in E. coli is to stabilize and allow a correct folding of the protein. Therefore, it is concluded that the unfolded hERα LBD monomer observed under nondenaturing conditions (Fig. 1) was the result of a partial misfolding during the overexpression or protein denaturation due to the freezing/thawing process before sample preparation.
Control experiments were needed to exclude protein aggregation during the ESI process and correlate the ESI mass spectrum with higher-order structure and specific noncovalent protein–protein complexes. The disappearance of the homodimer envelope under denaturing conditions is consistent with specific interactions, but is not sufficient to exclude aggregation of hERα LBD monomer during the ionization process. The nanoESI-MS data obtained after incubation with E2, i.e., the detection of folded monomer and homodimer bound to one and two E2 molecules, respectively, clearly support the assignment for folded hERα LBD monomer and homodimer charge state distributions (for details, see next paragraph). This demonstrates that nanoESI-MS is very well suited to characterize protein homogeneity and to provide protein structural information before performing further structural studies.
Detection of hERα LBD–ligand complexes using nanoESI-MS
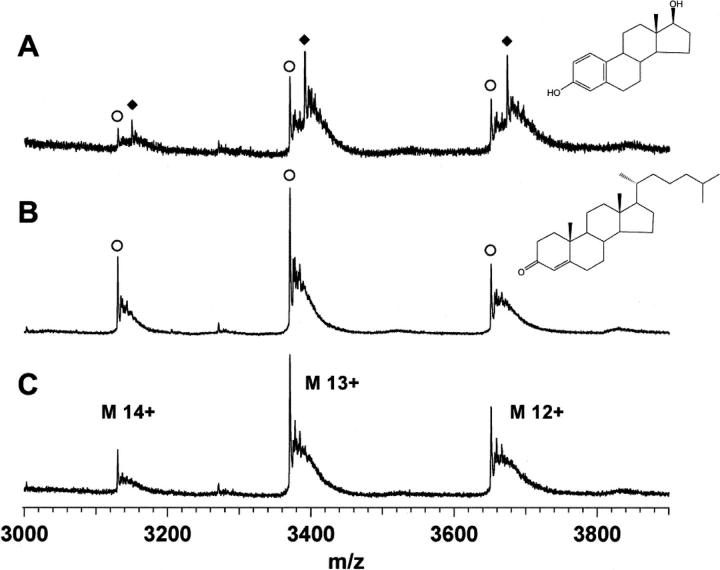
The width of the peaks measured at pH 7.4 (Fig. 1B) indicates more trapped water/salt molecules in the folded structure, which may complicate the detection of low molecular weight endocrine disruptors (MW 200–300 Da) at the mass range of the hERα LBD (MW 44 kDa). A slightly acidic pH was therefore selected for the analysis of hERα LBD–ligand complexes to improve the sensitivity and decrease the peak width observed in Figure 1B. By selecting MS transfer conditions carefully, intact hERα LBD monomer and homodimer bound to one and two E2 molecules, respectively, were detected (Fig. 2). The source pressure was raised to 4.5 mbar to improve the detection of ligand-bound protein. Under the conditions of high pressure, the ion collisions with the residual gas are less energetic and allow proper tuning over a wider range of transfer potentials while preserving the intact noncovalent complexes (Schmidt et al. 2001). Subjecting the ions to harsher MS transfer conditions (i.e., higher transfer voltages) helped to decluster them and produced narrower peaks with higher signal-to-noise ratio, but also led to the dissociation of hERα LBD bound with E2 (Fig. 2). The heavier homodimer ions require more collisions than the lighter monomer ions for building up sufficient internal energy for dissociation to take place. Therefore, the transfer potential should be selected carefully to find a balance between good ion desolvation and complex dissociation. Since nonspecific protein–ligand aggregation cannot generally be excluded during the ionization process (Loo 1997), the experiments were repeated by incubating hERα LBD with an inactive cholesterol derivative (4-cholesten-3-one). Under the same instrumental conditions, the absence of binding with 4-cholesten-3-one validates the detection of a specific hERα LBD–E2 complex by nanoESI-MS (Fig. 3). In the case of NRs, it has been claimed that the location of the ligand-binding site deep inside the LBD allows the detection of intact complexes by ESI-MS even if the complex formation in solution is mainly driven by the hydrophobic effect (Potier et al. 2003). The observed 1:1 monomer:E2 and 1:2 homodimer:E2 stochiometry of the gas-phase complexes further supports the complex specificity.
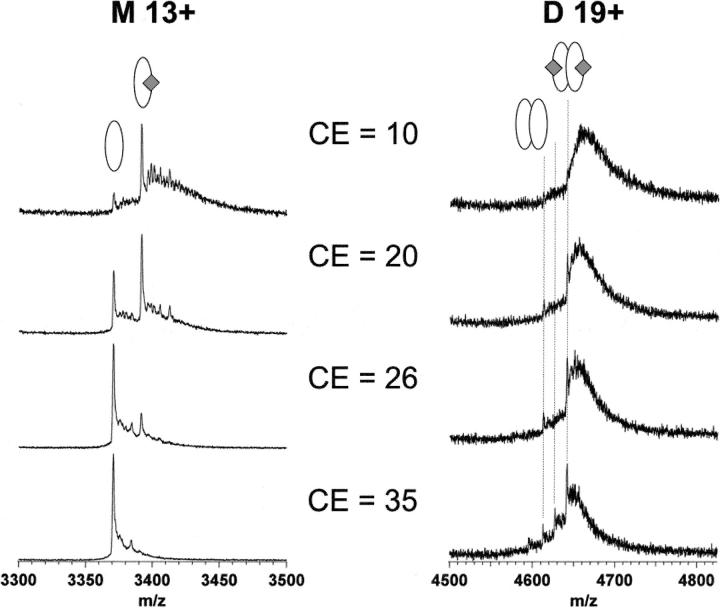
Figure 2.
Gas-phase stability analysis of hERα LBD-E2 complex (in 50 mM NH4OAc, pH 6) at different collision energies. Only the monomer charge state +13 and the homodimer charge state +19 are shown. The complex gradually dissociates when the CE voltage is increased, the result of the more energetic ion collisions with the collision cell gas.
Figure 3.
NanoESI mass spectra of hERα LBD monomer charge states acquired after incubation with E2 (A) and 4-cholest-3-one (B) in 50 mM NH4OAc at pH 6. (C) NanoESI mass spectrum of hERα LBD alone. These results demonstrate the specificity of hERα LBD–E2 complex. (○) Unbound hERα LBD, (♦) hERα LBD complexed with E2.
Collision-induced dissociation of hERα LBD–ligand complexes
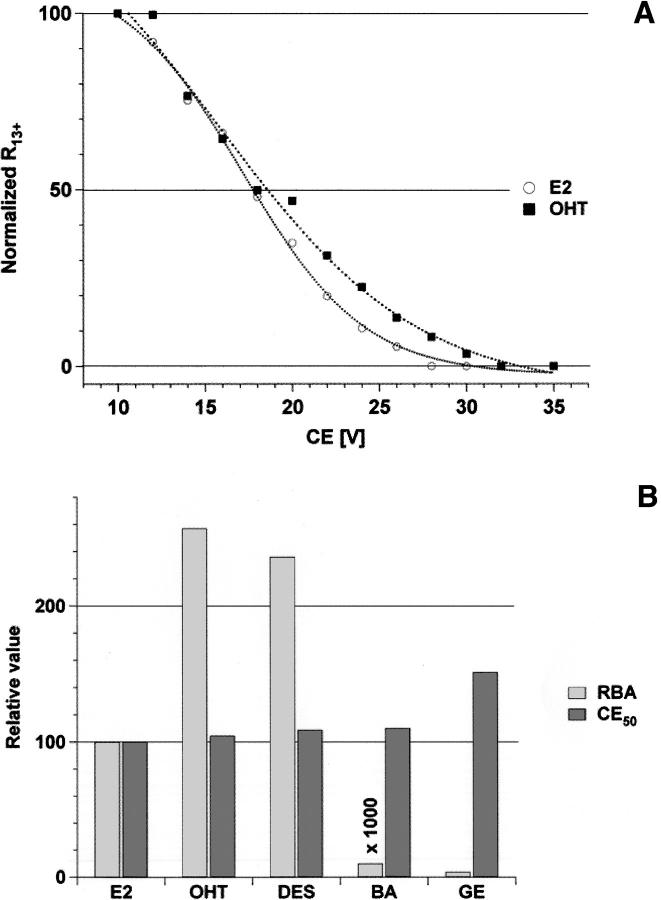
The above results demonstrate the specificity of the hERα LBD–ligand complexes detected by nanoESI-MS. This complex is stabilized by a number of hydrophobic contacts in the binding pocket. The high accessible volume of the binding pocket allows it to accept a wide variety of nonsteroidal compounds. Thus, solution and gas-phase behavior were compared to understand how solvation affects the stability of hERα LBD–ligand complexes. Subjecting the ions to collision-induced dissociation (CID), e.g., by increasing the collision energy (CE) voltage, results in dissociation of the complex due to elevated collision energy of the ions with the collision gas. The effect of the charge state on complex stability has been previously discussed (Nesatyy 2001). CID analysis on the +13 charged complex of the hERα LBD monomer ion is reported with two pharmaceutical compounds, 4-hydroxytamoxifen (OHT) and diethylstilbestrol (DES); with an environmental estrogenic compound, bisphenol A (BA); and with a phytoestrogen, genistein (GE). These ligands were selected because these have a wide range of affinities for hERα LBD and represent the different classes of interest for endocrine disruptors. The dissociation curves for E2 and OHT complexes are displayed in Figure 4A. The acceleration CE voltage required to dissociate 50% of the complex (CE50) is a measure of the complex gas-phase stability. The data in this work were compared with the relative binding affinities (RBAs) determined by competition binding assays of radiolabeled E2 for the hERα (Kuiper et al. 1998). Every CE50 value was normalized to a reference ligand, E2 (CE50[E2] = 100%). As shown by Figure 4B, the gas-phase stability data clearly indicate that measurements of relative gas-phase stability for hERα LBD–ligand complexes do not reflect the binding affinity in solution, in agreement with other ESI studies (Wu et al. 1997; van der Kerk-van Hoof and Heck 1999; Nesatyy 2001; Pan et al. 2006). These results confirm the prominent role of hydrophobic contacts for stabilizing ER–ligand complexes in solution (Shiau et al. 1998), an interaction that disappears during the removal of water molecules in the ESI process (Robinson et al. 1996).
Figure 4.
(A) Normalized bound to free protein intensity ratio R of the noncovalent complexes between hERαLBD and its target ligand E2 and OHT as a function of the collision energy (CE). CID analysis was performed on the hERα LBD monomer charged state +13. (B) Comparison of the relative CE50 and the relative binding affinity (RBA) in solution (Kuiper et al. 1998) for hERα LBD and its target ligand.
The crystal structures of hERα LBD with different ligands show that each ligand has a specific interaction network within the binding pocket (Brzozowski et al. 1997; Shiau et al. 1998; Manas et al. 2004). The H-bond interactions of DES with the ERα resemble that of E2, but DES forms more hydrophobic contacts with the binding pocket. OHT possesses one H-bond less than E2 and DES, but is more stabilized by hydrophobic contacts. The additional hydrophobic interactions present in OHT and DES complexes may account for the higher solution affinity, as suggested by Shiau et al. (1998). Based on the less extensive H-bonding network of OHT, one would thus expect a lower CE50 for this compound relative to E2 and DES. However, this is not supported by our data. The slightly acidic pH used for the experiments can disrupt a H-bond in E2 or DES complexes, which can be reflected by similar CE50 values relative to OHT. These similar CE50 values for OHT, DES, and E2 could also be the result of the OHT being more deeply packed into the binding pocket (Shiau et al. 1998), a structural feature that could enhance the OHT complex stability during CID experiments. In this case, even if OHT possesses fewer H-bonds than E2 and DES, similar CE50 values will be measured. GE, a low-affinity ligand, is a good example to illustrate the influence of the binding pocket flexibility on the complex stability. As for E2 and DES, GE is stabilized by three H-bonds (Manas et al. 2004). However, some binding pocket side chains form unfavorable interactions with the ligand, which lower the binding affinity. Therefore, the different crystal structures demonstrate the importance of hydrophobic contacts and binding pocket flexibility for stabilizing such complexes in solution and reveal why CID is not adapted to evaluate the relative solution binding affinity of a ligand.
An important finding is that nanoESI-MS is well suited to detect specific hERα LBD–ligand complexes rapidly and with high sensitivity. With the chip-based nanoESI system, an analysis time of 1 min/ligand could be obtained, resulting in consumption of <90 pmol of protein (i.e., 5 μL of protein at 0.6 mg/mL). The limit of detection of a bound ligand is in the range of 30 ng/mL. Therefore, the speed, specificity, low sample consumption, and capability for automation of the nanoESI robot demonstrate that nanoESI-MS is a fast and efficient screening method for the identification of suspected endocrine disruptors. With the present level of clustering, and given the accuracy of the Q-ToF instrument, it is possible to detect a mass difference of 0.4 Da for the monomer charge state +12. Thus, the method currently allows resolving the mass of an unknown ligand bound to the hERα LBD with a 5-Da accuracy.
Binding constant measurements by nanoESI-MS titration
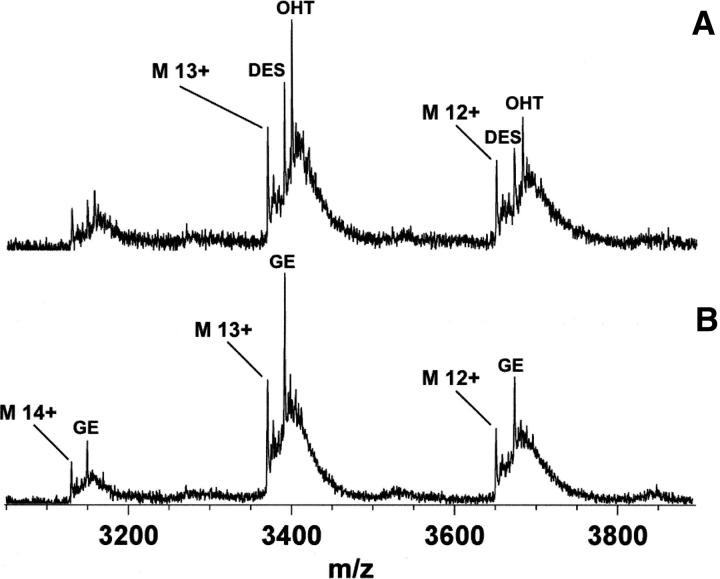
If one considers ESI-MS as a way to determine a solution-phase equilibrium constant (Kd), a key question that should be answered is whether the mass spectrum quantitatively reflects the solution composition. In other words, can the free protein and protein–ligand ion intensities be used to derive concentration in solution? Due to the small mass of ligand relative to the protein, a similar ionization efficiency was assumed for the free and bound protein (Peschke et al. 2004). ESI-MS has shown its capability to provide relative solution-binding affinities by competition experiments and direct solution affinities by titration experiments in a number of studies (Jorgensen et al. 1998; Sannes-Lowery et al. 2000; Daniel et al. 2003; Gabelica et al. 2003; Wang et al. 2003; Wendt et al. 2003; Tjernberg et al. 2004). Qualitative analysis was performed here using competitive binding experiments in which hERα LBD was incubated with a reference ligand (E2) and the target ligand. The peak height of the different protein–ligand complexes was taken as proportional to their relative solution affinity. When DES or OHT was added with E2 at an equimolar concentration (i.e., 0.5 μM each) to hERα LBD, only DES or OHT complex was observed, while only E2 complex was detected when incubation was done with BA and GE. Therefore, OHT and DES have a stronger solution affinity for hERα LBD relative to E2; BA and GE must have a lower affinity. The ability of a high-affinity ligand to displace a low-affinity ligand from the hERα LBD is another demonstration of the specificity of the protein–ligand interactions detected by nanoESI-MS. Subsequent competition experiments were performed between the stronger (OHT and DES) and lower (BA and GE) affinity ligands. The resulting nanoESI mass spectra are presented in Figure 5 and suggest the following relative solution affinity of hERα LBD–ligand complexes: OHT ≈ DES > E2 >> GE >> BA, which is in agreement with the binding affinity measured in solution (i.e., RBA values presented in Fig. 4B). Thus, nanoESI-MS can qualitatively estimate the solution affinity of potential endocrine disruptors.
Figure 5.
Competitive binding experiments analysis of hERα LBD with a mixture of OHT and DES (A) or GE and BA (B). NanoESI mass spectra of hERα LBD were acquired after incubation with an equimolar mixture of ligand (i.e., 0.5 μM of each) in 50 mM NH4OAc (pH 6).
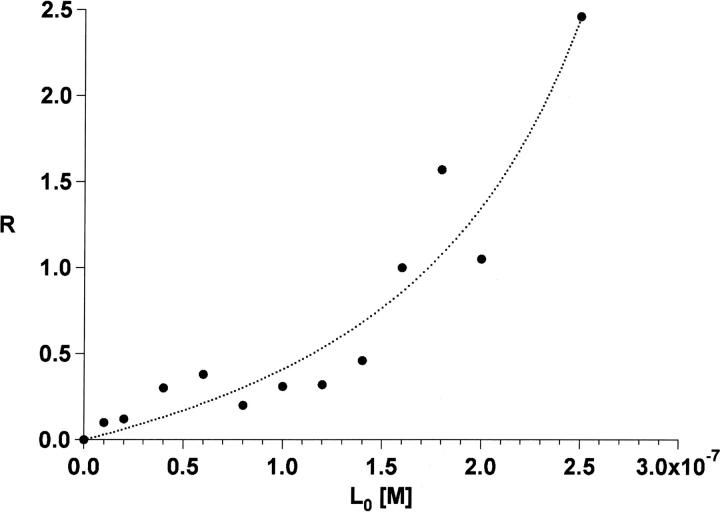
The assumption that the ratio of the bound to free protein (R) ion intensities is the same as the concentrations in solution at equilibrium allows us to compute absolute solution equilibrium constants by measuring the amount of bound and free protein as a function of ligand concentration. In this work, hERα LBD was titrated with E2 and analyzed with the automated nanoESI-MS setup. For calculating Kd, the bound to free protein ratio R was derived from the deconvoluted spectrum to take into account the variation of the R values with charge state, as shown in Figure 3. Lower charge states show more complexation, a variation that may be attributed to two factors: (1) Lower charge states represent more folded conformations of the protein, and thus induce more ligand binding; and (2) ions bearing lower charges experience lower acceleration and collide with less energy with the residual gas, resulting in the survival of more complex. Figure 6 shows the resulting titration curve for E2 in which the experimental data were fitted according to an expression described in “data processing” below (Daniel et al. 2003). From the nanoESI titration data, hERα LBD was estimated to have a Kd in the low nanomolar range for E2, a value in reasonable agreement with the 0.92 nM reported in the literature (Gangloff et al. 2001).
Figure 6.
NanoESI-MS titration graph showing the relative amount of hERα LBD-E2 complex formed with increasing the amount of E2 added to hERα LBD protein in 50 mM NH4OAc (pH 6).
We give only a rough estimation of Kd for the following reasons: (1) The protein concentration after purification is not known; therefore, Kd and the initial protein concentration were used as variable parameters; and (2) to have meaningful MS-derived values, the bound to free protein ratio R initially present in the solution must be conserved throughout the ES process and reflected in the mass spectrum. Considering the first point, the protein concentration found by fitting the data was 0.35 μM. If it is assumed that there is a similar ionization and mass spectrometer transfer efficiencies for the different hERα LBD ions, the folded monomer was found to represent 15% of the total protein concentration based on the nanoESI mass spectra, which correspond to 10%–15% recovery for the purification step. As described by Gabelica et al. (2002), when species have different m/z values, such for the different hERα LBD ions, discrimination can occur during the ES process, the transmission into the mass spectrometer, and in the detector. Considering the actual knowledge in this field, such discrimination is difficult to predict for this system.
Secondly, it rapidly became clear during the measurements that the reproducibility of the bound to free protein ratio was poor. Substantial variation of the R values was found with the distance of the spray needle from the sample orifice (i.e., a decrease by a factor close to three was found while moving away from the cone axis) and also for individual measurements (i.e., nozzle-to-nozzle variation by a factor of three). These effects are not well documented for nanoESI and only a few studies report a similar dependence on the cone-to-needle distance (Gabelica et al. 2002; Benkestock et al. 2004). Benkestock et al. (2004) demonstrate in their study that if the ligand had a hydrophilic or hydrophobic character, then a number of spray parameters (e.g., distance, tip diameter, polarity) affect the measured ratio R. According to these investigators, the understanding of this process was related to the ESI mechanism, which is still subject to many debates.
In conclusion, it is shown here that automated nanoESI-MS is an efficient screening method for suspected endocrine-disrupting compounds in terms of specificity, sample consumption possibility, speed, and automation. Ligand binding was clearly observed, even for low-affinity ligands. It is demonstrated that the best approach to evaluate qualitatively solution-binding affinity of ligand to hERα LBD by nanoESI-MS was a competition experiment with a reference ligand. Sampling the complex and free-protein solution composition by nanoESI should be done carefully by considering the physicochemical properties of the protein and ligand, especially if a low-nanomolar equilibrium constant should be determined. The capability of using the natural affinity of the receptor protein with nanoESI-MS for ligand demonstrates that this analytical method can be used routinely to identify new endocrine-disrupting compounds.
Materials and Methods
Protein purification and incubation
The triple mutant hERα LBD was overexpressed in E. coli and purified as previously described (Gangloff et al. 2001). Amino acid sequence 302–553 of hERα corresponding to the LBD was generated with cysteine to serine mutations at positions 381, 417, and 530 in fusion with thioredoxin, six histidine residues, and a thrombin cleavage sequence (MW 43,947 Da). Prior to ESI-MS measurements, the protein stock solution (4.5 mg/mL in 10% [v/v] glycerol, 50 mM NaCl, and 100 mM Tris/HCl, pH 8) was desalted and buffer exchanged against 50 mM NH4OAc (pH 6) using Micro Bio-spin 6 columns (Bio-Rad Laboratories). Fifty microliters of the protein at 30 μM diluted in 50 mM NH4OAc (pH 6) were applied. As the recovery of the purification step is unknown, the hERα LBD concentration indicated throughout this study is based on a 100% recovery, an approximation that likely overestimates the protein concentration used during the experiments.
The ligand was dissolved in ethanol and added to 6 μL of the purified protein solution (15 μM), the final ethanol concentration being 2%. hERα LBD and ligand were mixed and incubated for 30 min at room temperature before analysis. 17β-estradiol, 4-hydroxytamoxifen, and diethylstilbestrol were obtained from Sigma, bisphenol A from Aldrich, and genistein from Fluka.
Electrospray ionization mass spectrometry
ESI mass spectra were acquired on a quadrupole time-of-flight mass spectrometer (Q-ToF ULTIMA) equipped with an automated chip-based nanoESI system (Nanomate 100, Advion Biosciences). Calibration was performed by using the multiply charged ions produced by a mixture of 1 μM myoglobin and trypsinogen dissolved in MeOH:H2O (1:1, v/v) with 1% (v/v) acetic acid.
The source block was heated only to 40°C to prevent dissociation of the noncovalent complexes. The mass spectrometer was tuned with gentle desolvation parameters to maintain hERα LBD–ligand noncovalent complexes intact during their transfer from the solution phase to the mass spectrometer vacuum. The cone and first ion tunnel RF1 voltages, the parameters that control the kinetic energy of the ions in the source region of the Q-ToF ULTIMA mass spectrometer, were optimized at 80 V and 60 V, respectively. The pressure in the source region was increased at 4.5 mbar with a Speedivalve (BOC Edwards) to enhance the transmission of high m/z ions (Tito et al. 2001; Sobott et al. 2002; Chernushevich and Thomson 2004) and to preserve the noncovalent complexes in the gas phase (Schmidt et al. 2001; Tahallah et al. 2001; Sanglier et al. 2002; Tjernberg et al. 2004). After passing the cone and the ion tunnels, the ion beam was transmitted to the quadrupole used in RF-only mode and passed through a hexapole collision cell pressurized with argon (Purity 5.0, PanGas). Collision-induced dissociation (CID) experiments were performed by varying the acceleration CE voltage, which determines the kinetic energy of the ions when they collide with the collision cell gas. For competitive binding assays and titration experiments, CE voltage was optimized at 10 V. Ions were detected with a multichannel plate (MCP) detector set at 2250 V.
Data processing
Before data processing, each spectrum was background subtracted (fifth-order polynomial, 25% below curve) and smoothed (Savitzky-Gollay smooth, 10 × 5 channels) with the MassLynx software (version 4.0). For CID experiments, the CE voltage giving a 50% complex dissociation was calculated by fitting the dissociation curve with a sigmoid curve. The abundance of the free and the ligand-bound protein cannot be directly correlated from their respective maximum intensities, because the peak position of the ligand-bound protein matches those of the adduct and post-translational modification of the free protein. It was assumed that the peaks of the ligand-bound protein differ only in their position on the mass scale and their amplitude, but not in their shape. To calculate the bound to free protein ratio R, a reference nanoESI mass spectra of hERα LBD alone was measured under the same MS conditions as hERα LBD incubated with a ligand. The corresponding ion peak was duplicated and shifted by the respective m/z ligand value. The abundances of the free and bound protein were evaluated by adjusting the peak heights of the reference mass spectra with the mass spectra of hERα LBD incubated with the ligand. For the Kd measurement, R was calculated after deconvolution with MaxEnt 1 software on m/z range 2500–4000 with the same procedure.
The Kd determination from a titration experiment was based on the equations already described in the literature (Daniel et al. 2003). The complex formation of the protein P with its ligands L can be described by the following equations:
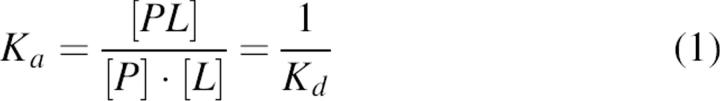 |
Solving the above equations for R, the concentration ratio of the bound to free protein measured by nanoESI-MS, yields Equation 4, where Ka and the initial protein concentration [P]0 are adjustable parameters. The initial protein concentration was used as an adjustable parameter, because the yield of the protein purification step was unknown. The experimental R versus known [L]0 values were fitted by using Equation 4, which give a value for Ka, respectively Kd.
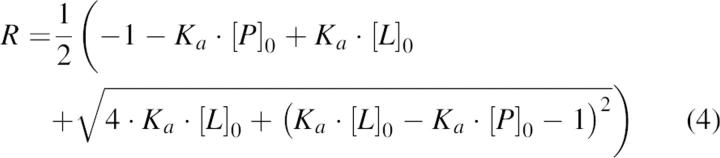 |
Acknowledgments
This work was supported by the Swiss National Science Foundation (project no. 4050-104373) and in part by Novartis AG. We are grateful to the Functional Genomics Center Zurich, in particular Dorothea Rutishauser, and to Reinaldo Almeida from Advion Biosciences for providing technical support.
Footnotes
Reprint requests to: Renato Zenobi, Department of Chemistry and Applied Biosciences, ETH Zurich, 8093 Zurich, Switzerland; e-mail: zenobi@org.chem.ethz.ch; fax: 41 44 632 12 92.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062664107.
References
- Benkestock K., Sundqvist, G., Edlund, P.O., and Roeraade, J. 2004. Influence of droplet size, capillary-cone distance and selected instrumental parameters for the analysis of noncovalent protein–ligand complexes by nano-electrospray ionization mass spectrometry. J. Mass Spectrom. 39: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Beresford N., Routledge, E.J., Harris, C.A., and Sumpter, J.P. 2000. Issues arising when interpreting results from an in vitro assay for estrogenic activity. Toxicol. Appl. Pharmacol. 162: 22–33. [DOI] [PubMed] [Google Scholar]
- Bitsch F., Aichholz, R., Kallen, J., Geisse, S., Fournier, B., and Schlaeppi, J.M. 2003. Identification of natural ligands of retinoic acid receptor-related orphan receptor α ligand-binding domain expressed in Sf9 cells—A mass spectrometry approach. Anal. Biochem. 323: 139–149. [DOI] [PubMed] [Google Scholar]
- Blair R.M., Fang, H., Branham, W.S., Hass, B.S., Dial, S.L., Moland, C.L., Tong, W.D., Shi, L.M., Perkins, R., and Sheehan, D.M. 2000. The estrogen receptor relative binding affinities of 188 natural and xenochemicals: Structural diversity of ligands. Toxicol. Sci. 54: 138–153. [DOI] [PubMed] [Google Scholar]
- Bolger R., Wiese, T.E., Ervin, K., Nestich, S., and Checovich, W. 1998. Rapid screening of environmental chemicals for estrogen receptor binding capacity. Environ. Health Perspect. 106: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski A.M., Pike, A.C.W., Dauter, Z., Hubbard, R.E., Bonn, T., Engstrom, O., Ohman, L., Greene, G.L., Gustafsson, J.A., and Carlquist, M. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389: 753–758. [DOI] [PubMed] [Google Scholar]
- Chernushevich I.V. and Thomson, B.A. 2004. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 76: 1754–1760. [DOI] [PubMed] [Google Scholar]
- Colborn T., Saal, F.S.V., and Soto, A.M. 1993. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ. Health Perspect. 101: 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.M., McCombie, G., Wendt, S., and Zenobi, R. 2003. Mass spectrometric determination of association constants of adenylate kinase with two noncovalent inhibitors. J. Am. Soc. Mass Spectrom. 14: 442–448. [DOI] [PubMed] [Google Scholar]
- De Vriendt K., Van Driessche, G., Devreese, B., Bebrone, C., Anne, C., Frere, J.M., Galleni, M., and Van Beeumen, J. 2006. Monitoring the zinc affinity of the metallo-β-lactamase CphA by automated nanoESI-MS. J. Am. Soc. Mass Spectrom. 17: 180–188. [DOI] [PubMed] [Google Scholar]
- Donald L.J., Hosfield, D.J., Cuvelier, S.L., Ens, W., Standing, K.G., and Duckworth, H.W. 2001. Mass spectrometric study of the Escherichia coli repressor proteins, Iclr and GclR, and their complexes with DNA. Protein Sci. 10: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabelica V., Vreuls, C., Filee, P., Duval, V., Joris, B., and De Pauw, E. 2002. Rapid Commun. Mass Spectrom. 16: 1723–1728. [DOI] [PubMed] [Google Scholar]
- Gabelica V., Galic, N., Rosu, F., Houssier, C., and De Pauw, E. 2003. Influence of response factors on determining equilibrium association constants of noncovalent complexes by electrospray ionization mass spectrometry. J. Mass Spectrom. 38: 491–501. [DOI] [PubMed] [Google Scholar]
- Gangloff M., Ruff, M., Eiler, S., Duclaud, S., Wurtz, J.M., and Moras, D. 2001. Crystal structure of a mutant hER α ligand-binding domain reveals key structural features for the mechanism of partial agonism. J. Biol. Chem. 276: 15059–15065. [DOI] [PubMed] [Google Scholar]
- Geoghegan K.F., Dixon, H.B., Rosner, P.J., Hoth, L.R., Lanzetti, A.J., Borzilleri, K.A., Marr, E.S., Pezzullo, L.H., Martin, L.B., LeMotte, P.K., et al. 1999. Spontaneous α-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: The cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 267: 169–184. [DOI] [PubMed] [Google Scholar]
- Greschik H., Wurtz, J.M., Sanglier, S., Bourguet, W., van Dorsselaer, A., Moras, D., and Renaud, J.P. 2002. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell 9: 303–313. [DOI] [PubMed] [Google Scholar]
- Harrison P.T.C., Holmes, P., and Humfrey, C.D.N. 1997. Reproductive health in humans and wildlife: Are adverse trends associated with environmental chemical exposure? Sci. Total Environ. 205: 97–106. [DOI] [PubMed] [Google Scholar]
- Jorgensen T.J.D., Roepstorff, P., and Heck, A.J.R. 1998. Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogues and vancomycin group antibiotics by electrospray ionization mass spectrometry. Anal. Chem. 70: 4427–4432. [Google Scholar]
- Kapur A., Beck, J.L., Brown, S.E., Dixon, N.E., and Sheil, M.M. 2002. Use of electrospray ionization mass spectrometry to study binding interactions between a replication terminator protein and DNA. Protein Sci. 11: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keetch C.A., Hernandez, H., Sterling, A., Baumert, M., Allen, M.H., and Robinson, C.V. 2003. Use of a microchip device coupled with mass spectrometry for ligand screening of a multi-protein target. Anal. Chem. 75: 4937–4941. [DOI] [PubMed] [Google Scholar]
- Kuiper G.G.J.M., Lemmen, J.G., Carlsson, B., Corton, J.C., Safe, S.H., van der Saag, P.T., van der Burg, P., and Gustafsson, J.A. 1998. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology 139: 4252–4263. [DOI] [PubMed] [Google Scholar]
- Kumar V. and Chambon, P. 1988. The estrogen-receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell 55: 145–156. [DOI] [PubMed] [Google Scholar]
- Lengqvist J., Griffiths, W.J., Perlmann, T., and Sjovall, J. 2002. Detection of a receptor–ligand noncovalent complex using a triple quadrupole mass spectrometer. Rapid Commun. Mass Spectrom. 16: 2003–2006. [DOI] [PubMed] [Google Scholar]
- Lengqvist J., de Urquiza, A.M., Bergman, A.C., Willson, T.M., Sjovall, J., Perlmann, T., and Griffiths, W.J. 2004. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor α ligand-binding domain. Mol. Cell. Proteomics 3: 692–703. [DOI] [PubMed] [Google Scholar]
- Lengqvist J., Alvelius, G., Jornvall, H., Sjovall, J., Perlmann, T., and Griffiths, W.J. 2005. Electrospray mass spectrometry for the direct accurate mass measurement of ligands in complex with the retinoid X receptor α ligand binding domain. J. Am. Soc. Mass Spectrom. 16: 1631–1640. [DOI] [PubMed] [Google Scholar]
- Loo J.A. 1997. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 16: 1–23. [DOI] [PubMed] [Google Scholar]
- Loo J.A., Hu, P.F., McConnell, P., Mueller, W.T., Sawyer, T.K., and Thanabal, V. 1997. A study of Src SH2 domain protein–phosphopeptide binding interactions by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 8: 234–243. [Google Scholar]
- Manas E.S., Xu, Z.B., Unwalla, R.J., and Somers, W.S. 2004. Understanding the selectivity of genistein for human estrogen receptor-β using X-ray crystallography and computational methods. Structure 12: 2197–2207. [DOI] [PubMed] [Google Scholar]
- Nesatyy V.J. 2001. Gas-phase binding of noncovalent protein complexes between bovine pancreatic trypsin inhibitor and its target enzymes studied by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 36: 950–959. [DOI] [PubMed] [Google Scholar]
- Nikov G.N., Hopkins, N.E., Boue, S., and Alworth, W.L. 2000. Interactions of dietary estrogens with human estrogen receptors and the effect on estrogen receptor–estrogen response element complex formation. Environ. Health Perspect. 108: 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Fukushima, T., Santa, T., Waizumi, N., Tokuyama, H., Maeda, M., and Imai, K. 2002. Estrogen receptor binding assay method for endocrine disruptors using fluorescence polarization. Anal. Chem. 74: 4391–4396. [DOI] [PubMed] [Google Scholar]
- Ohno K., Suzuki, S., Fukushima, T., Maeda, M., Santa, T., and Imai, K. 2003. Study on interactions of endocrine disruptors with estrogen receptor using fluorescence polarization. Analyst 128: 1091–1096. [DOI] [PubMed] [Google Scholar]
- Pan S., Sun, X.J., and Lee, J.K. 2006. Stability of complementary and mismatched DNA duplexes: Comparison and contrast in gas versus solution phases. Int. J. Mass Spectrom. 253: 238–248. [Google Scholar]
- Peschke M., Verkerk, U.H., and Kebarle, P. 2004. Features of the ESI mechanism that affect the observation of multiply charged noncovalent protein complexes and the determination of the association constant by the titration method. J. Am. Soc. Mass Spectrom. 15: 1424–1434. [DOI] [PubMed] [Google Scholar]
- Potier N., Billas, I.M.L., Steinmetz, A., Schaeffer, C., Van Dorsselaer, A., Moras, D., and Renaud, J.P. 2003. Using nondenaturing mass spectrometry to detect fortuitous ligands in orphan nuclear receptors. Protein Sci. 12: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.V., Chung, E.W., Kragelund, B.B., Knudsen, J., Aplin, R.T., Poulsen, F.M., and Dobson, C.M. 1996. Probing the nature of noncovalent interactions by mass spectrometry. A study of protein-CoA ligand binding and assembly. J. Am. Chem. Soc. 118: 8646–8653. [Google Scholar]
- Sanglier S., Ramstrom, H., Haiech, J., Leize, E., and Van Dorsselaer, A. 2002. Electrospray ionization mass spectrometry analysis revealed a similar to 310 kDa noncovalent hexamer of HPr kinase/phosphatase from Bacillus subtilis. Int. J. Mass Spectrom. 219: 681–696. [Google Scholar]
- Sanglier S., Bourguet, W., Germain, P., Chavant, V., Moras, D., Gronemeyer, H., Potier, N., and Van Dorsselaer, A. 2004. Monitoring ligand-mediated nuclear receptor–coregulator interactions by noncovalent mass spectrometry. Eur. J. Biochem. 271: 4958–4967. [DOI] [PubMed] [Google Scholar]
- Sannes-Lowery K.A., Griffey, R.H., and Hofstadler, S.A. 2000. Measuring dissociation constants of RNA and aminoglycoside antibiotics by electrospray ionization mass spectrometry. Anal. Biochem. 280: 264–271. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Bahr, U., and Karas, M. 2001. Influence of pressure in the first pumping stage on analyte desolvation and fragmentation in nano-ESI MS. Anal. Chem. 73: 6040–6046. [DOI] [PubMed] [Google Scholar]
- Shiau A.K., Barstad, D., Loria, P.M., Cheng, L., Kushner, P.J., Agard, D.A., and Greene, G.L. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95: 927–937. [DOI] [PubMed] [Google Scholar]
- Sobott F., Hernandez, H., McCammon, M.G., Tito, M.A., and Robinson, C.V. 2002. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74: 1402–1407. [DOI] [PubMed] [Google Scholar]
- Sobott F., McCammon, M.G., Hernandez, H., and Robinson, C.V. 2005. The flight of macromolecular complexes in a mass spectrometer. Philos. Trans. R. Soc. London, Ser. A 363: 379–389. [DOI] [PubMed] [Google Scholar]
- Sonnenschein C. and Soto, A.M. 1998. An updated review of environmental estrogen and androgen mimics and antagonists. J. Steroid Biochem. Mol. Biol. 65: 143–150. [DOI] [PubMed] [Google Scholar]
- Stehlin-Gaon C., Willmann, D., Zeyer, D., Sanglier, S., Van Dorsselaer, A., Renaud, J.P., Moras, D., and Schule, R. 2003. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR β. Nat. Struct. Biol. 10: 820–825. [DOI] [PubMed] [Google Scholar]
- Steinmetz A.C.U., Renaud, J.P., and Moras, D. 2001. Binding of ligands and activation of transcription by nuclear receptors. Annu. Rev. Biophys. Biomol. Struct. 30: 329–359. [DOI] [PubMed] [Google Scholar]
- Tahallah N., Pinkse, M., Maier, C.S., and Heck, A.J.R. 2001. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 15: 596–601. [DOI] [PubMed] [Google Scholar]
- Tito M.A., Miller, J., Walker, N., Griffin, K.F., Williamson, E.D., Despeyroux-Hill, D., Titball, R.W., and Robinson, C.V. 2001. Probing molecular interactions in intact antibody: Antigen complexes, an electrospray time-of-flight mass spectrometry approach. Biophys. J. 81: 3503–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjernberg A., Carno, S., Oliv, F., Benkestock, K., Edlund, P.O., Griffiths, W.J., and Hallen, D. 2004. Determination of dissociation constants for protein–ligand complexes by electrospray ionization mass spectrometry. Anal. Chem. 76: 4325–4331. [DOI] [PubMed] [Google Scholar]
- van der Kerk-van Hoof A. and Heck, A.J.R. 1999. Covalent and noncovalent dissociations of gas-phase complexes of avoparcin and bacterial receptor mimicking precursor peptides studied by collisionally activated decomposition mass spectrometry. J. Mass Spectrom. 34: 813–819. [DOI] [PubMed] [Google Scholar]
- Wang W.J., Kitova, E.N., and Klassen, J.S. 2003. Influence of solution and gas phase processes on protein–carbohydrate binding affinities determined by nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 75: 4945–4955. [DOI] [PubMed] [Google Scholar]
- Wendt S., McCombie, G., Daniel, J., Kienhofer, A., Hilvert, D., and Zenobi, R. 2003. Quantitative evaluation of noncovalent chorismate mutase-inhibitor binding by ESI-MS. J. Am. Soc. Mass Spectrom. 14: 1470–1476. [DOI] [PubMed] [Google Scholar]
- Witkowska H.E., Green, B.N., Carlquist, M., and Shackleton, C.H.L. 1996. Intact noncovalent dimer of estrogen receptor ligand-binding domain can be detected by electrospray ionization mass spectrometry. Steroids 61: 433–438. [DOI] [PubMed] [Google Scholar]
- Wortmann A., Rossi, F., Lelais, G., and Zenobi, R. 2005. Determination of zinc to β-peptide binding constants with electrospray ionization mass spectrometry. J. Mass Spectrom. 40: 777–784. [DOI] [PubMed] [Google Scholar]
- Wu Q.Y., Gao, J.M., Joseph-McCarthy, D., Sigal, G.B., Bruce, J.E., Whitesides, G.M., and Smith, R.D. 1997. Carbonic anhydrase-inhibitor binding: From solution to the gas phase. J. Am. Chem. Soc. 119: 1157–1158. [Google Scholar]
- Zhang S., Van Pelt, C.K., and Wilson, D.B. 2003. Quantitative determination of noncovalent binding interactions using automated nanoelectrospray mass spectrometry. Anal. Chem. 75: 3010–3018. [DOI] [PubMed] [Google Scholar]