Abstract
The bacterial flagellum is a highly complex prokaryotic organelle. It is the motor that drives bacterial motility, and despite the large amount of energy required to make and operate flagella, motile organisms have a strong adaptive advantage. Flagellar biogenesis is both complex and highly coordinated and it typically involves at least three two-component systems. Part of the flagellum is a type III secretion system, and it is via this structure that flagellar components are exported. The assembly of a flagellum occurs in a number of stages, and the “checkpoint control” protein FliK functions in this process by detecting when the flagellar hook substructure has reached its optimal length. FliK then terminates hook export and assembly and transmits a signal to begin filament export, the final stage in flagellar biosynthesis. As yet the exact mechanism of how FliK achieves this is not known. Here we review what is known of the FliK protein and discuss the evidence for and against the various hypotheses that have been proposed in recent years to explain how FliK controls hook length, FliK as a molecular ruler, the measuring cup theory, the role of the FliK N terminus, the infrequent molecular ruler theory, and the molecular clock theory.
Keywords: FliK, bacteria, motility, flagella, type III secretion
Motility and chemotaxis are the processes by which bacteria can move away from stressful environments, or toward more favorable conditions (Amsler et al. 1993). The bacterial motor is the flagellum, which, although requiring a significant amount of energy to construct and run, offers the cell a strong adaptive advantage over nonmotile organisms. The significance and conservation of flagellar proteins is illustrated by the fact that plants and animals have evolved a subtype of Toll-like innate immune receptor (i.e., TLR-5) specifically dedicated to the recognition of this complex organelle (Gomez-Gomez and Boller 2002; Smith and Ozinsky 2002). In addition to being the driving force of translational motion, the flagellum also can act as an adhesin (Inglis et al. 2003; Kirov et al. 2004), and has been used by scientists as a tool for peptide display (Westerlund-Wikstrom 2000). In the present review we will discuss one particular protein which has a key role in regulating the complex process of flagellar biogenesis. FliK acts as an essential checkpoint controller; it detects when one process is complete (i.e., hook assembly), it terminates this stage, and triggers the next stage in the biosynthetic pathway (i.e., filament export). We will concentrate on discussing the role of FliK as a hook-length controlling protein. For more information on the switch in export specificity mediated by FliK we refer you to this recent excellent review (Ferris and Minamino 2006).
Flagellar biosynthesis has been most studied in Escherichia coli and Salmonella enterica (Pallen et al. 2005). To avoid unnecessary repetition, the data and concepts discussed herein will explicitly refer to S. enterica, unless otherwise stated. E. coli and S. enterica have 5–10 peritrichous flagella (that is flagella distributed over the whole cell) per cell, which together can rotate counterclockwise, causing filaments to form a bundle that produces translational motion, or clockwise, causing the cell to tumble and reorient (Silverman and Simon 1974; Macnab 1977). Some bacteria such as Helicobacter pylori have lophotrichous flagella (a tuft of flagella at one pole), while others such as Vibrio parahaemolyticus are monotrichous and have only one polar flagellum. Coupled with chemotaxis, the alternating pattern of translational motion and tumbling is a biased random walk that brings the bacterium away from stressful environments (Silverman and Simon 1974; Macnab 1977).
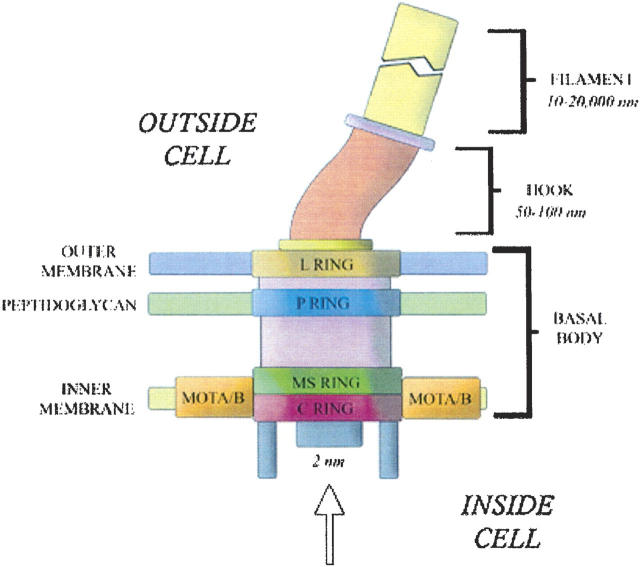
The bacterial flagellum consists of three protein substructures: the basal body complex, the hook, and the filament (Fig. 1). The whip-like filament is made up of repeating units of flagellin protein (Abram et al. 1970; DePamphilis and Adler 1971). The morphological pathway of flagellar biogenesis proceeds from cell-proximal to cell-distal structures, i.e., from the basal-body to the flagellar hook and finally to the filament (Suzuki et al. 1978; Kubori et al. 1992); and the flagellum uses a type III secretion system to export these proteins (Komoriya et al. 1999). This is a mechanism distinct from the Sec-dependent export pathway, and is similar to that employed for many virulence factors. Both flagellar proteins and virulence factors pass the cytoplasmic and outer membranes either through the flagella or the injectisome, respectively. These two secretory macrostructures resemble each other, and their components share many similarities (Ghosh 2004; Tampakaki et al. 2004; Ferris and Minamino 2006). Flagellar proteins cross the cytoplasmic membrane by an ATP-driven mechanism, diffuse down a channel in the nascent flagellar structure (indicated by an arrow in Fig. 1), and assemble at its distal end (Macnab 1999).
Figure 1.
Simplified schematic representation of a bacterial flagellum. This complex structure can be thought of consisting of three substructures: the basal body, hook, and filament. Export of flagellar components is through a putative pore as indicated by the arrow.
The flagellar export apparatus consists of six membrane proteins: FlhA, FlhB, FliP, FliO, FliQ, FliR, and three cytoplasmic proteins FliH, FliI, FliJ (Minamino and Macnab 1999, 2000c; Minamino et al. 2000; Macnab 2003). All the membrane components of the export apparatus are thought to lie within a putative central pore of the basal body MS ring (Fig. 1; Fan et al. 1997; Kihara et al. 2001), though the pore has not been identified by electron microscopy (Francis et al. 1994; Suzuki et al. 2004). FliI is the ATPase that provides the energy for the translocation of proteins across the cytoplasmic membrane (Fan and Macnab 1996). FliH acts as an ATPase regulator, coupling the energy of ATP hydrolysis to flagellar protein export (Minamino and Macnab 2000b; Auvray et al. 2002; Zhu et al. 2002; Minamino et al. 2003). FliJ functions as a general chaperone to prevent export substrates from premature aggregation in the cytoplasm (Minamino et al. 2000).
In total, at least 40 proteins are involved in flagellar formation and function, encoded by at least 13 different operons in the Enterobacteriaceae (Kutsukake et al. 1990). The genes for flagellar synthesis are termed flg, flh, fli; for flagellar rotation they are termed mot; for chemotactic membrane receptors they are termed tar, tsr, tap, trg; and for chemotactic signal transduction they are termed che. All are part of the same flagellar flhCD regulon in the Enterobacteriaceae (Macnab 1992) and are clustered together in several regions on the chromosome (Ohnishi et al. 1994). Operons in S. enterica can be grouped into three classes. Class I consists only of the flhCD operon, containing the genes flhC and flhD, which is regulated by a variety of external stimuli including the cAMP–CAP complex (Kutsukake et al. 1990). Its products are required for the expression of all other operons. Class II consists of the following operons: flaG, flgB, flhB, fliA, fliE (of which fliK is a member), fliF, and fliL; these are under the control of the class I operon and are required for subsequent expression of class III genes (Kutsukake et al. 1990). The class III operons are flgK, fliD, fliC, motA, and tar. The product of the fliD operon represses transcription of the class III operons (Kutsukake et al. 1990). The genes in the class II operons, except fliA, are all involved in the hook–basal body complex. The class III gene products are primarily responsible for filament formation (Kutsukake et al. 1990).
The flagellar hook: A historical perspective
The flagellar hook is the substructure that connects the long whip-like filament to the membrane anchoring basal body. It acts like a flexible universal joint that transmits the rotational movement generated by the basal body to the filament, which in turn, propels the cell forward (Kagawa et al. 1976; Kutsukake et al. 1979). The flagellar hook is composed of ∼120 subunits of the FlgE protein in S. enterica (Kagawa et al. 1976, 1979; Komeda et al. 1978). FlgE polymerizes to form a hook in the presence of FlgD, a scaffolding protein which associates with the tip of the elongating hook, whereas purified FlgE protein in the absence of FlgD polymerizes but not productively (Ohnishi et al. 1994). In flgD mutants, flagellar formation is arrested after completion of the basal body as the hook is not correctly assembled (Suzuki et al. 1978; Suzuki and Komeda 1981).
It has been postulated that the hook would need to be of a particular length; too short a hook would not be able to generate a sufficient bend angle, while too long a hook would no longer be efficient in the transmission of torque (Kawagishi et al. 1996; Williams et al. 1996). Hence, controlling hook length is essential for proper function and behavior of the cell. The lengths of hooks from wild-type S. enterica were first measured after straightening the hooks with the use of a low-pH, low-temperature staining solution, and the average length was found to be 55.0 ± 5.9 nm (Hirano et al. 1994). Compared to other biological structures of controlled length, this deviation in hook length of about 10% of the mean is much greater than for the tobacco mosaic virus, whose deviation is only 2% of the mean (Caspar 1963), and for the deviation of the bacteriophage tail, which is 5% of the mean (Katsura and Hendrix 1984).
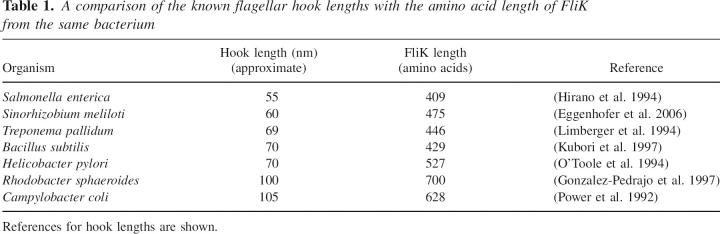
The hook length is similarly regulated in other flagellated bacteria (Table 1). Hook length for each species is controlled in order to optimize the transmission of torque to the filament. Monotrichous flagellated bacteria, such as Campylobacter, Caulobacter, and Rhodobacter, have significantly longer hooks than multiflagellated bacteria (Gonzalez-Pedrajo et al. 1997). The longer hooks were suggested to be related to the need for greater torque that must be applied to the single flagellum (Power et al. 1992).
Table 1.
A comparison of the known flagellar hook lengths with the amino acid length of FliK from the same bacterium
It appears that there is nothing intrinsic about the hook structure that controls its length, and purified FlgE can polymerize indefinitely in vitro (Kato et al. 1982). In an attempt to elucidate how hook length is regulated, bacterial mutants that were defective in hook-length control were generated and screened (Patterson-Delafield et al. 1973; Suzuki and Iino 1981). Mutations were mapped to the fliK gene, and mutants had hooks of indeterminate length with no filament attached. These mutants were nonmotile, and thus FliK was implicated as the protein responsible for controlling hook length. The long hooks of unregulated length were first termed superhooks (Patterson-Delafield et al. 1973) and later polyhooks, and ranged in length from 40 to 1000 nm (Hirano et al. 1994). The name “polyhook” is rather misleading though, as it implies a hook composed of multiples of a hook structure of a defined length, whereas in reality they are simply very long hooks of polymerized FlgE subunits (Hirano et al. 1994). The wavelength of the undulations of an H. pylori polyhook structure tended to be largely uniform, measuring 100 nm (Ryan et al. 2005). This unusual H. pylori polyhook uniformity is most likely due to the stabilizing presence of the flagellar sheath which is normally absent from polyhooks of other species. Even though the hook is composed of FlgE subunits (Ohnishi et al. 1994), no flgE mutant, until recently (Moriya et al. 2006), had been found to cause abnormal hook lengths.
FliK: Structure and bioinformatic analysis
Most flagellar ortholog pairs of S. enterica and E. coli are quite similar, with a sequence identity level >80% (Kawagishi et al. 1996). The identity level between the FliK proteins, however, is only ∼50%. A diagram of the conserved predicted structure of FliK is shown in Figure 2. The N terminus and central segments of FliK are the least conserved parts of the protein. Comparing FliK from S. enterica and E.coli, the first 251 residues are only 36% identical, with the longest stretch of identity being a mere 12 residues long (Kawagishi et al. 1996). The C-terminal portion, which is rich in glutamine residues, is the most conserved, with 71% identity in the last 154 residues (Kawagishi et al. 1996). The central part of FliK is rich in proline residues; more specifically within a 96-amino acid stretch between residues 139 and 234 in FliK of E. coli K-12, there are 17 proline residues which is 18 mol%. In the remainder of the sequence, the proline content is 5 mol% (Kawagishi et al. 1996).
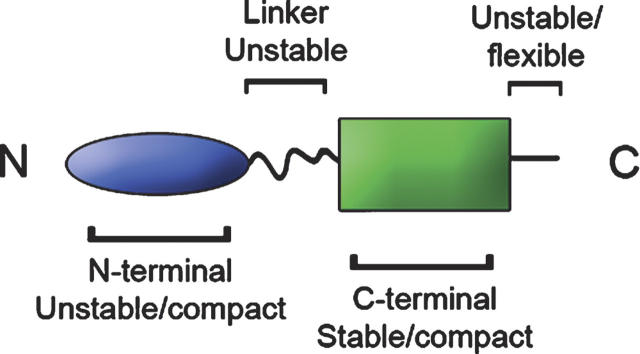
Figure 2.
Domain organization of the FliK protein. FliK typically consists of four regions: (1) the N-terminal region, relatively unstable but still compact; (2) an unstable linker region; (3) an extremely stable and compactly folded C-terminal region; and (4) an unstable and flexible region (Minamino et al. 2004).
An alignment of the FliK protein from 13 different bacteria is shown in Figure 3. Also included in the alignment is the YscP injectisome protein from Yersinia pestis, which will be discussed later. As expected, the greatest degree of sequence conservation can clearly be seen in the C-terminal portion of the protein. FliK from Campylobacter jejuni and Campylobacter coli contain six short insertions, making them the largest proteins. As discussed earlier, this may be a consequence of these cells having polar flagella, although it is worth noting that Pseudomonas syringae and Xanthomonas campestris, which also have polar flagella, do have shorter FliK proteins (Fig. 3). To our knowledge, the length of the flagellar hook is not known for these two bacteria. In reality, the environment in which an organism lives and swims will determine how much force is required by a flagellum to propel the cell and thus influence hook length. Our own analysis of the amino acid sequences of FliK from the two sequenced H. pylori strains shows the proteins have an overall identity of 89%. Interestingly, identity in the first 250 amino acids is only 83% compared to 96% for the last 150.
Figure 3.

Multiple alignment of amino acid sequences of FliK from 13 bacteria. Accession numbers for fliK from each organism are as follows: Bacillus licheniformis, YP_079020; Bacillus subtilis, NP_389509; Campylobacter jejuni, CAL34221; Campylobacter coli, ZP_00368165; Escherichia coli, NP_416453; Shigella flexneri, NP_707828; Salmonella enterica, NP_456535; Yersinia pestis, NP_405391; Bordetella bronchiseptica, NP_889120; Helicobacter pylori, NP_207698; Pseudomonas syringae, NP_791789; Xanthomonas campestris, YP_363724; Treponema phagedenis, AAB61249. Also included in the alignment for comparison purposes is yscP from Yersinis pestis, NP_395176. The alignment was created with ClustalW (Thompson et al. 1994).
In Salmonella, FliK is consistently found in two forms: one with a molecular mass of 44 kDa, the other of 43 kDa (Homma et al. 1988; Kawagishi et al. 1996). The 43-kDa form of FliK is due to C-terminal truncation of the 44-kDa form and is not functional (Muramoto et al. 1998). The most C-terminal 35 residues of FliK are highly susceptible to digestion by endogenous protease, degrading it to the 43-kDa form (Minamino et al. 2004), suggesting high conformational flexibility in the region (Kawagishi et al. 1996; Muramoto et al. 1998). In solution, FliK is monomeric and highly elongated (Minamino et al. 2004).
A FliK crystal structure has not yet been solved, so what is known about the structure and domain organization of the protein is mainly the result of studies on various fliK mutants. Williams et al. (1996) analyzed mutations in fliK that give rise to the polyhook phenotype. Most were deletions which produced a frame-shifted sequence. A few were nonsense mutations toward the 3′ end of the gene. There was only one example of an insertion, occurring at nucleotide position 1003, and this resulted in the addition of two amino acids. Taken together, it was apparent that disruption of the more stable C-terminus of FliK was responsible for the polyhook phenotype (Williams et al. 1996).
To test the effect of FliK concentration on hook length, wild-type fliK was either under- or overexpressed in a fliK mutant strain (Muramoto et al. 1998). For underexpression, a fliK allele was made in which the sole UGG codon at position 271 was replaced by UGA (Muramoto et al. 1998). The stop codon UGA is recognized at a low frequency by tRNATrp (Hirsh and Gold 1971; Eggertsson and Soll 1988). The FliK variant with this opal codon, when overexpressed, was estimated to be present at levels of 10–20 molecules per cell; it produced the polyhook-filament phenotype, yet with a filament attached, providing it with variably weak motility. Wild-type FliK levels were estimated to be around 40–80 molecules per cell. Overproduced wild-type FliK was estimated to be at a concentration of 2000–6000 molecules per cell, and these produced slightly shorter hooks, averaging 46 ± 7 nm. Hence, as the amount of FliK is increased, the phenotype progresses from polyhook, to polyhook filament, to wild-type hook filament, to short hook filament (Muramoto et al. 1998). It was found that C-terminally truncated FliK was nonfunctional no matter how much it was overproduced (Muramoto et al. 1998). With such low wild-type levels of FliK, a single event is probably responsible for the substrate switch. Furthermore, since both polyhooks and polyhook filaments can exist in the same cell, the event probably occurs at the level of the individual flagellum (Muramoto et al. 1998).
Hook-length control is intimately linked with filament assembly. A fliK mutant with the capacity to regulate hook length but not to initiate filament assembly has not yet been described (Williams et al. 1996). It has been hypothesized that FliK may not so much control hook length as it may measure it and then send a signal to FlhB to switch substrate specificity (Williams et al. 1996). FlhB is a membrane protein, the gene for which is part of the flhCD regulon. It responds to a signal from FliK that the correct hook length has been reached by triggering export of the antisigma factor FlgM, which in turn, allows for the subsequent expression of class III genes. This results in a switch of export substrate specificity from rod/hook-type to filament-type, consequently initiating filament assembly (Williams et al. 1996). Interactions between FlhB and FliK have been recently reviewed elsewhere (Ferris and Minamino 2006). Using in-frame linker-insertion mutagenesis, mutant alleles of the E. coli fliK gene were constructed (Kawagishi et al. 1996). The allele with an insertion in the N-terminal region (at position 98) could complement a fliK null mutant to wild-type motility levels. The allele with an insertion in the central region (at position 208) gave 50% wild-type motility levels, while the allele with an insertion in the C terminus at position 324 was nonmotile (Kawagishi et al. 1996). Deletion of residues 63–157 in FliK did not abolish filament assembly, showing that the N terminus is capable of withstanding significant disruption without hindering function to a great degree (Williams et al. 1996). Hence, the C-terminus of FliK is more highly conserved.
In S. enterica, the 44-kDa full-length FliK is exported into the culture medium (Minamino et al. 1999). It was detected in the culture supernatant from the wild-type strain and from flgD, flgE, and flgK mutants, but not in the flgB and fliK mutants (Minamino et al. 1999). Filament-type proteins, like flagellin (Kuwajima et al. 1989; Kornacker and Newton 1994) and FlgM (Iyoda and Kutsukake 1995) can still be exported when their C terminus has been truncated. Muramoto showed that FliK can also be exported when C-terminally truncated at residues 319 and 380 (Muramoto et al. 1998). From deletion analysis, it appears that only the first 40 amino acids are absolutely necessary for FliK export (Minamino et al. 1999). As the ability of FliK to be exported decreased, the corresponding phenotype changed from wild-type hook filament to polyhook filament to polyhook (Minamino et al. 1999). As FliK is a rod/hook-type substrate, once the substrate specificity switch occurs, FliK is no longer exported (Minamino et al. 1999).
When a fragment of FliK consisting of amino acids 199–405 (i.e., His-FliK199–405), which is stable and not exported, was overproduced in a fliK mutant, the amounts of FlgD and FliC (flagellin) exported into the culture were the same as in the vector control. Therefore, despite increasing the amount of His-FliK199–405 in the cell, substrate specificity switching still did not occur, indicating that FliK export is necessary for its proper functioning (Minamino et al. 2004). Using affinity blots, full-length FliK and FliK1–147 bound strongly to His-FLAG-FlhBC (the cytoplasmic domain of FlhB), while FliK265–405 did not bind at all (Minamino et al. 2004). This binding pattern demonstrates the recognition of FliK as an export substrate. One possible explanation for the lack of observed binding between FliK265–405 and FlhBC could be the probable regulation of the interaction between FliKC and FlhBC so that it only occurs during FliK export. This would also explain why when this domain is overproduced, it still cannot switch the substrate specificity of the export apparatus (Minamino et al. 2004). On the other hand, FliK265–405 most likely does not include the entire C-terminal domain, which was found to consist of the region from residues 205–405 (Minamino et al. 2004) so the above results cannot be viewed as definitive evidence that the purified FliK C-terminal domain will not bind to FlhBC. Thus, it appears that FliK1–147 is involved in the export of FliK, while FliKC is involved in signaling the substrate specificity switch (Williams et al. 1996; Minamino et al. 2004). In fliK mutants, therefore, this signal does not occur, and the export apparatus is locked in a state specific for rod/hook-type substrates (Kutsukake et al. 1994; Williams et al. 1996; Kutsukake 1997). Recent deletion analysis showed that removing residues in the Salmonella FliK C-terminal region from amino acid residues 265–405 abolished substrate specificity switching (Minamino et al. 2006). This region was subsequently dubbed the type III secretion substrate specificity switch, or T3S4 domain (Minamino et al. 2006).
Our studies with H. pylori have shown that a FliK polyhook mutant which cannot control hook length also has overexpression of the class II genes flaB and flgE while the class III gene flaA was underexpressed (Ryan et al. 2005). Hence, as expected, the switch in transcriptional specificity mediated by FliA (σ28) did not occur in the absence of a functional FliK, and the cell continues to produce class II proteins.
In summary, FliK typically consists of four regions (Fig. 2): (1) an N-terminal region, relatively unstable but still compact; (2) an unstable linker region; (3) an extremely stable and compactly folded C-terminal region; and (4) a short unstable and flexible region (Minamino et al. 2004).
Possible mechanisms of flagellar hook-length control
As discussed above, much is known about the kinetics and biochemistry of hook and filament assembly. The exact mechanism of how FliK measures and controls flagellar hook length, however, is still largely unknown. A number of diverse and sometimes contradictory hypotheses have been proposed to try to explain this. These are discussed along with supporting and opposing arguments in the next section.
FliK as a molecular ruler
The length of a number of biological structures is controlled by means of a molecular ruler. Some examples are the tail of the phage λ, where the GpH protein is the ruler (Katsura and Hendrix 1984; Katsura 1987), and the thin (F-actin) filaments of muscle, where nebulin is the ruler (Labeit et al. 1991). In these cases, the molecular ruler is a protein in an elongated state that determines the number of subunits of another protein that assembles into a filamentous structure, hence regulating its length.
Another example is the needle or injectisome of Yersinia. The length of the Yersinia injectisome is 58 ± 10 nm, and is controlled by yscP, a homolog of fliK (Journet et al. 2003). See Figure 3 for a comparison of FliK and YscP sequences. yscP null mutants have a polyneedle phenotype in which the needle of the injectisome is of indeterminate length much like the polyhooks seen in fliK mutants (Magdalena et al. 2002; Tamano et al. 2002; Journet et al. 2003). Insights into the needle-length control mechanism were first glimpsed from comparison of the YscP proteins from Yersinia enterocolitica E40 and Yersinia pestis KIM5 (Journet et al. 2003). YscP from Y. pestis KIM5 is 90% identical in sequence to YscP from Y. enterocolitica but is shorter because it lacks a duplication of 60 central residues (Payne and Straley 1999; Stainier et al. 2000). A Y. enterocolitica yscP mutant can be complemented by yscP from Y. pestis, yet the needles are shorter (Journet et al. 2003). When the repeated sequence in YscP from Y. enterocolitica was deleted, the needles were again shorter (Journet et al. 2003). It was found that any YscP protein variant without the first 35 or the last 130 residues was unable to control needle length, while YscP protein variants with deletions up to 126 amino acids between residues 36 and 360 were still able to control needle length, though the needle would be correspondingly shorter (Journet et al. 2003). Furthermore, when the 60-residue repeat sequence from yscPentero was inserted multiple times, the needles were correspondingly longer. This strict linear relationship between the number of amino acids of YscP and the needle length strongly suggests that YscP acts as a molecular ruler. Although YscP and FliK are similar (∼36% amino acid similarity; Fig. 3), the differences present are apparently too great to allow sufficient interactions with export apparatus proteins (Agrain et al. 2005). It has been suggested that YscP, InvJ, Spa32, and FliK, all proteins involved in length control, diverged more during evolution than other proteins from type-III secretion systems (Journet et al. 2003).
A protein secondary structure prediction analysis of various FliK proteins carried out by us suggests the protein is unlikely to regulate hook length by acting as a molecular ruler. Other biological rulers function because the ruler consists predominantly of stretches of polypeptide in an α-helical state (Katsura and Hendrix 1984; Katsura 1987; Labeit et al. 1991). From the amino acid sequence and secondary structure predictions of FliK, it is highly unlikely that any long α-helical stretches exist, particularly in the N-terminal region and almost definitely not in the proline-rich central region. Furthermore, the asymmetric nature of FliK, with the N and C termini being quite different from each other, is not characteristic of a molecular ruler (Kawagishi et al. 1996). Finally, FliK would have to be anchored on the outside of the cell (if it were always measuring the hook length, substrates would then not be able to pass through the narrow inner channel), yet it has never been found there. Moreover, as mentioned above, true molecular rulers will yield shorter structures when they themselves are shortened, as long as the termini are left intact (Katsura and Hendrix 1984; Katsura 1987; Labeit et al. 1991), but there are many examples in which internal deletions of fliK still result in the polyhook phenotype, casting doubt on its potential as a molecular ruler (Williams et al. 1996).
In favor of the molecular ruler hypothesis, it does seem that bacteria with longer hook lengths also tend to have longer FliK proteins (Table 1). For example the C. coli hook is approximately twice the length of that from S. enterica, and its FliK protein is much longer (628 amino acids vs. 409).
With doubts about the molecular ruler mechanism, it was postulated that FliK's role in measuring hook length could be related to a temporal change, particularly as there is a link between flagellar morphogenesis and the cell cycle (Nishimura and Hirota 1989). Experiments were performed using plasmid-based expression of fliK that was independent from chromosomal control, and no effect in hook length was noted (Minamino et al. 1999). Thus, a temporal change in fliK expression is most likely not the cause of the substrate specificity switch (Minamino et al. 1999).
The measuring cup theory
S. enterica mutants that produced shorter hooks were found for the first time by Makishima et al. (2001). These mutations mapped to either fliG, fliM, or fliN (Makishima et al. 2001). The mutations resulted in either filament-less (Fla−) mutants (due to major defects) or motility-less (Mot−, which had filaments) or chemotaxis-less (Che−) mutants from minor defects, and the mutants had shorter hooks of either ∼25 nm or 45 nm (Makishima et al. 2001). FliG forms the rotor, and a complex of FliM and FliN works in switching the rotational direction (Macnab 2003). Without the switch proteins, the flagellum is not formed beyond the MS ring complex (Kubori et al. 1992). These switch proteins form a hollow-cup structure called the C-ring beneath the flagellar basal body (Fig. 1; Khan et al. 1992; Berg 2003; Thomas et al. 2006).
Based on this evidence, a “quantized measuring cup” model was proposed for hook-length regulation (Makishima et al. 2001). The length of the hook would be determined by the number of FlgE subunits that fit within the C ring. Mutations in the switch proteins might decrease the holding capacity of the C ring. Once all the FlgE proteins were exported, FliK could access FlhB and cause the substrate specificity switch (Makishima et al. 2001). This theory was quickly dismissed by others (Ferris et al. 2005; Moriya et al. 2006). According to the measuring cup model of regulating hook length, even in flgD mutants, which can export FlgE but cannot form hooks in the absence of the hook-cap protein, the switch in substrate specificity should occur (Moriya et al. 2006). It was noted that flgD mutants, however, can never switch substrate specificity, and the cell simply continues to export FlgE (Ohnishi et al. 1994; Minamino and Macnab 1999). The substrate specificity switch occurs regardless of how many FlgE molecules are exported. In another counterexample, flgEΔ(9–20) mutants produce FlgE variants that are efficiently exported but cannot polymerize effectively (Moriya et al. 2006). The substrate specificity switch never occurs in these mutants either. The model also fails to explain the lack of a difference in hook length when FlgE is overproduced (Muramoto et al. 1999; Moriya et al. 2006). An increase in the number of FlgE molecules would be expected to result in an increase in hook length as FliK would be prevented from stopping hook elongation as long as the “measuring cup” remained full of FlgE protein. However, it is worth noting that although the average hook length in cells overproducing FlgE was unchanged, the frequency of polyhooks was increased (Moriya et al. 2006).
Requirement of FliK N terminus in hook-length control
In a study by Hirano et al. (2005), the authors made various N-terminal deletions of FliK, deleting from the first amino acid through residue 13, 19, 39, 59, 79, 99, 119, 139, 159, 179, or 199. They found that FliKΔ39 and FliKΔ99 showed small swarm rings when not overproduced, and swarm sizes of 30% and 60% of wild-type swarming, respectively, when overproduced, indicating that these fragments of the protein could impart limited motility (Hirano et al. 2005). FliKΔ13 and FliKΔ59 failed to swim without IPTG induction, and showed tiny rings with IPTG (Hirano et al. 2005). FliK with an N-terminal truncation >100 amino acids did not swarm at all. It was suggested that the region 119–199 may be part of the C-terminal domain (Hirano et al. 2005), although an analysis of the domain structure of FliK suggest this is highly unlikely. The N-terminally truncated FliK variants were not secreted into the media via the type-III secretion system, as expected (Hirano et al. 2005). Also constructed were N- and C-terminal fusions of FliK with cyan fluorescent protein (CFP) (Hirano et al. 2005). With CFP at the C terminus, FliK–CFP was nonfunctional. With CFP at the N terminus, CFP–FliK was not exported but could still restore partial motility to fliK mutants when the fusion protein was overproduced.
From the above evidence, the authors concluded that the N terminus is dispensable for hook-length control (Hirano et al. 2005). However, as FliKΔ19 and FliKΔ79 produced swarm rings which are virtually indistinguishable from the vector control, more investigations should be conducted before the N terminus is viewed as dispensable. It is also curious that a large truncation, FliKΔ99, complemented fliK mutants better than smaller truncations. The stability of each truncated FliK variant was not discussed, which is important in order to correctly interpret any negative results. Nevertheless, all truncated proteins, even when overproduced, showed lesser motility compared to the wild-type. The role of the FliK N terminus is still quite unclear, but although the protein can withstand greater disruption in this domain it would be premature at this stage to suggest that this part of the protein is entirely dispensable for function.
FliK as an infrequent molecular ruler
Moriya et al. (2006) described five distinct flgE mutant strains that displayed a weakly motile phenotype and produced shorter hooks with broader distributions than those of wild-type cells. All flgE mutants described prior to this study had been nonmotile and did not secrete flagellin protein. These flgE mutants had missense mutations in their sequences, had the same concentration of mutant FlgE in the cell, and a higher concentration in the supernatant compared to wild-type FlgE concentrations. Furthermore, all were flagellated, showing that substrate specificity could be switched during flagellum assembly. It was concluded that these flgE mutants were less motile probably due to a decrease in polymerization efficiency that results in shorter hooks; accordingly, overproduction of these FlgE mutant proteins improved motility (Moriya et al. 2006).
Based on this and previously discussed evidence, a model whereby FliK controls hook length by an infrequent molecular ruler mechanism was proposed (Moriya et al. 2006). In this model, the size of the hook structure depends on the rate of hook polymerization (governed by FlgE concentration) and the frequency of FliK export (in turn, governed by the interaction rate of FliK with FlgE and FlgD). The diameter of the central channel of the flagellum is only 2 nm (Yonekura et al. 2003) so according to this model FliK must be in an extended conformation in the central channel. FliK is likely to operate as a hook-length controller only during its infrequent export process, so this may be why the hook length is not strictly controlled but has a relatively broad distribution. It was proposed that the N-terminal domain of FliK interacts strongly with FlgD and weakly with FlgE during its export. Thus, when the hook is within a certain range of an appropriate length (a “window of opportunity,” between 45 nm and 65 nm in S. enterica as an arbitrary example), the binding of FliKN to FlgD and FlgE stabilizes FliKC in the proper conformation or position to promote the interaction of FliKC with FlhBC. This interaction results in the termination of hook elongation and the switch in export apparatus substrate specificity. Previous evidence can be explained by this mechanism. Overproduction of FliK slightly shortens hooks compared to the wild-type (Muramoto et al. 1998), probably because it increases the chances of FliK being exported, thus increasing the number of opportunities for FliKC to interact with FlhBC earlier. When cellular levels of FliK are reduced, the probability of FliK measuring the growing hook length during export is decreased (since it will rarely be exported itself), so the FliKC–FlhBC interaction may never occur, resulting in polyhooks (Muramoto et al. 1998). The fact that there exist polyhook-filament phenotypes implies that the FliK–FlhB interaction can indeed take place even when FliKN can no longer interact with FlgD. The weak interaction with FlgE may allow for this interaction, though it may be much less frequent, resulting in the wide distribution of lengths in polyhook-filament cells (Moriya et al. 2006). Longer hooks are observed when wild-type FlgE is overproduced because it decreases the chances of FliK's export, and thus of the FliKC–FlhBC interaction, delaying the timing of the switch (Moriya et al. 2006). The shorter hooks in the weakly motile flgE mutants are due to more FliK molecules being exported per polymerized FlgE, allowing more opportunities for the FliKC–FlhBC interaction earlier in the window of opportunity. Overproduction of the FlgE mutant protein restores the normal hook length because the export ratio of FlgE over FliK increases the frequency of the FliK–FlhB interaction back to normal levels (Moriya et al. 2006).
While the infrequent ruler mechanism can explain many observations, several matters remain to be investigated. First, while the binding pattern of FliK to FlgD and FlgE does lend support to the theory that FliK may measure the hook by binding to FlgD, no control was performed to establish the binding pattern of FlgE. It is easy to imagine that in order to quickly construct the hook apparatus, hook subunits may also bind strongly to FlgD and weakly to polymerized FlgE to be “pulled along” the central channel to the distal tip. Hence, it is possible that FliK's binding pattern to FlgD and FlgE may simply be a result of its status as an exported hook-type protein.
If the infrequent ruler mechanism is correct, then control of hook length is based on a fine balance of the relative concentrations of FlgE and FliK. It could be assumed that FliK has a window of opportunity within which it can efficiently transmit the signal that the hook is of appropriate length to FlhB. FliK molecules exported before the hook reaches 45 nm, for example, might not cause the substrate specificity switch, while FliK exported after the hook passes 65 nm might not bind FlgD and so will not effectively transmit the signal. When wild-type FliK is overproduced, estimated to be at a concentration of 2000–6000 molecules per cell as opposed to the estimated wild-type concentration of 40–80 molecules per cell, slightly shorter hooks averaging 46 ± 7 nm are observed (Muramoto et al. 1998). While an increase in the relative concentration of FliK to FlgE would be expected to produce shorter hooks, as observed, it would also be expected to produce a narrower distribution. This expectation is because, with a greater frequency of FliK export, the switch will occur much sooner after the hook enters the “window of opportunity,” producing shorter hooks, and will definitely happen within a smaller time period, producing a narrower distribution. However, the observation that overproduced FliK results in a hook distribution of ±7 nm, similar to the distribution of ±6 nm seen for wild-type levels (Hirano et al. 1994), cannot be explained by the infrequent ruler mechanism alone.
The “molecular clock”
An overlapping hook-length control mechanism that is dependent on the rate of hook elongation has also been proposed (Moriya et al. 2006). This model argues in favor of an intrinsic timing device that programs the export machinery to switch its substrate specificity, or at least to slow down or stop rod/hook-type export, regardless of hook length, but only once hook elongation has initiated. The complete switch to initiate filament-type export would still be dependent on FliK. Observations to support this theory are as follows. (1) The hooks of the weakly motile flgE mutants are shorter than wild-type hooks, as the polymerization rate is slower. If the timing device stops hook growth at a shorter length, this can be overcome by overexpression of mutant FlgE proteins. The infrequent ruler mechanism, however, can already explain this phenomenon. As the FlgE proteins inefficiently polymerize, the ratio of exported FliK to polymerized FlgE is increased in these mutants compared to the wild-type, ensuring that FliK will initiate the substrate specificity switch sooner once the hook enters the window of opportunity. Overexpression of FlgE simply decreases the exported FliK/polymerized FlgE ratio, increasing hooks to wild-type lengths. (2) Another observation to support the timing device theory is the fact that overexpression of wild-type FlgE increases hook length, presumably due to an increase in the rate of polymerization. Yet this observation is also already explained by the infrequent ruler mechanism. The overexpression of FlgE simply decreases the exported FliK/polymerized FlgE ratio, ensuring that on average, FliK will initiate the substrate specificity switch later in its window of opportunity. (3) Finally, polyhook mutants show a long monotonic tailing toward much longer lengths than wild-type, but the peak in the length distribution is still 55 nm (Koroyasu et al. 1998). It is argued that the timing device slows hook export after 55 nm. The peak at 55 nm has been observed in a number of mutants (Hirano et al. 1994; Williams et al. 1996; Muramoto et al. 1998, 1999). Another simpler explanation of this phenomenon may be that diffusion of proteins through the nascent flagellar structure slows hook growth after 55 nm. Before 55 nm, interaction of the exported hook subunits with the FlgD cap may speed up the polymerization rate; after 55 nm, contact with FlgD may not occur until a slower process of diffusion takes place, effectively slowing the growth rate of the hook. This slower process would be expected to continue at a constant rate, which has been observed (Koroyasu et al. 1998). Therefore, in a population of growing hooks, lengths around 55 nm are reached quickly, after which the slower yet constant growth occurs. So hook lengths would always peak near to 55 nm.
If the timing device does exist, what is it? The cleavage of FlhBC was postulated to be a likely candidate (Moriya et al. 2006). Uncleaved FlhBC might efficiently export rod/hook-type proteins, and the autocleavage event, occurring after a certain period of time, may slow this export process (Moriya et al. 2006). Supporting evidence for this lies in the observations that cleavage is an autocatalytic event that is independent of FliK (Ferris et al. 2005); coproduced FlhBΔCC and FlhBCC cannot restore motility of an flhB mutant to wild-type levels (Minamino and Macnab 2000a); and if cleavage is prevented, the export apparatus remains locked in the rod/hook-type specificity state (Fraser et al. 2003). This model does not suggest a role for FliK or explain how it determines hook length and signals this to FlhB. If cleavage is indeed a molecular clock, then when FlhBΔCC and FlhBCC are coproduced in an flhB–fliK double null mutant, the peak hook length in the distribution of polyhooks should be shorter. If cleavage is prevented, by a N269A mutation for instance, then the resulting population of polyhooks should show no bias to a length of 55 nm (unless the diffusion process discussed above does indeed have the predicted effect). Data from these experiments would help to elucidate the existence or absence of the timing device.
Conclusions and future perspectives
As hook length directly affects the performance of the flagella in producing translational motion, it is crucial that bacteria tightly regulate the process of hook assembly. As discussed above, even single amino acid changes in FliK can have severe effects on motility. From the published evidence concerning how FliK actually regulates flagellar hook length, the most strongly supported hypothesis at present would seem to be the FliK as an infrequent molecular ruler model. This mechanism posits that hook length is regulated neither by an intrinsic property of the hook proteins themselves, nor by the intrinsic protein holding capacity of a preexisting substructure (i.e., the C-ring), but rather that FliK determines the hook length during its own export and, should the hook be of a suitable length, transmits a signal to FlhB. However, we are not at a stage where the biochemical basis of how this signal is transmitted can be accurately hypothesized. Whatever the signal constitutes, it results in the termination of hook assembly and the subsequent initiation of filament protein synthesis and export.
In the absence of hook-length control, a cell will continue to transcribe class II genes and produce an overabundance of these proteins. Whole genome array profiling of flagellar mutants has successfully identified additional genes involved in flagellar biosynthetic pathways (Niehus et al. 2004; Wang et al. 2006) and a similar approach could be used with a FliK mutant to further expand the class II and III regulons. There are almost certainly more general, as well as species-specific proteins involved in flagella biogenesis that have not yet been identified, a point illustrated by the recent identification of HP1575 as a so-called “spare part,” which can partially replace FlhB in H. pylori (Wand et al. 2006). Moreover the FlhD2C2 master regulon continues to grow as novel promoter binding and gene array experiments have identified other genes (some flagellar some metabolic) under the control of this regulator (Pruss et al. 2003; Kapatral et al. 2004; Stafford et al. 2005). FliW, a new flagellin assembly protein in T. pallidum which has orthologs in many related species, has also recently been identified (Titz et al. 2006).
Despite the many ingenious experiments which have helped us to understand how hook-length control is regulated, it is likely that the next advance in our understanding of how this mechanism works will come when a FliK protein crystal structure is finally solved.
Acknowledgments
This work was supported by grants from the Irish Research Council for Science Engineering and Technology (to K.A.R.), Science Foundation Ireland (to P.W.O.T.), and a scholarship from the George Mitchell Foundation (to R.C.W.).
Footnotes
Reprint requests to: Kieran A. Ryan, Department of Microbiology, University College Cork, Cork, Ireland; e-mail: kieran.ryan@ucc.ie; fax: 353 21 490 3101.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072785407.
References
- Abram D., Mitchen, J.R., Koffler, H., and Vatter, A.E. 1970. Differentiation within the bacterial flagellum and isolation of the proximal hook. J. Bacteriol. 101: 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrain C., Callebaut, I., Journet, L., Sorg, I., Paroz, C., Mota, L.J., and Cornelis, G.R. 2005. Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56: 54–67. [DOI] [PubMed] [Google Scholar]
- Amsler C.D., Cho, M., and Matsumura, P. 1993. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175: 6238–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvray F., Ozin, A.J., Claret, L., and Hughes, C. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: Interaction with acidic phospholipids and FliH. J. Mol. Biol. 318: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H.C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72: 19–54. [DOI] [PubMed] [Google Scholar]
- Caspar D.L. 1963. Assembly and stability of the tobacco mosaic virus particle. Adv. Protein Chem. 18: 37–121. [DOI] [PubMed] [Google Scholar]
- DePamphilis M.L. and Adler, J. 1971. Fine structure and isolation of the hook–basal body complex of flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenhofer E., Rachel, R., Haslbeck, M., and Scharf, B. 2006. MotD of Sinorhizobium meliloti and related {α}-proteobacteria is the flagellar-hook-length regulator and therefore reassigned as FliK. J. Bacteriol. 188: 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G. and Soll, D. 1988. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 52: 354–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F. and Macnab, R.M. 1996. Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271: 31981–31988. [DOI] [PubMed] [Google Scholar]
- Fan F., Ohnishi, K., Francis, N.R., and Macnab, R.M. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Ferris H.U. and Minamino, T. 2006. Flipping the switch: Bringing order to flagellar assembly. Trends Microbiol. 14: 519–526. [DOI] [PubMed] [Google Scholar]
- Ferris H.U., Furukawa, Y., Minamino, T., Kroetz, M.B., Kihara, M., Namba, K., and Macnab, R.M. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 280: 41236–41242. [DOI] [PubMed] [Google Scholar]
- Francis N.R., Sosinsky, G.E., Thomas, D., and DeRosier, D.J. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235: 1261–1270. [DOI] [PubMed] [Google Scholar]
- Fraser G.M., Hirano, T., Ferris, H.U., Devgan, L.L., Kihara, M., and Macnab, R.M. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48: 1043–1057. [DOI] [PubMed] [Google Scholar]
- Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68: 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L. and Boller, T. 2002. Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 7: 251–256. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pedrajo B., Ballado, T., Campos, A., Sockett, R.E., Camarena, L., and Dreyfus, G. 1997. Structural and genetic analysis of a mutant of Rhodobacter sphaeroides WS8 deficient in hook-length control. J. Bacteriol. 179: 6581–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Yamaguchi, S., Oosawa, K., and Aizawa, S. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176: 5439–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Shibata, S., Ohnishi, K., Tani, T., and Aizawa, S. 2005. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol. Microbiol. 56: 346–360. [DOI] [PubMed] [Google Scholar]
- Hirsh D. and Gold, L. 1971. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J. Mol. Biol. 58: 459–468. [DOI] [PubMed] [Google Scholar]
- Homma M., Iino, T., and Macnab, R.M. 1988. Identification and characterization of the products of six region III flagellar genes (flaAII.3 through flaQII) of Salmonella typhimurium. J. Bacteriol. 170: 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis T.J., Robertson, T., Woods, D.E., Dutton, N., and Chang, B.J. 2003. Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect. Immun. 71: 2280–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda S. and Kutsukake, K. 1995. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol. Gen. Genet. 249: 417–424. [DOI] [PubMed] [Google Scholar]
- Journet L., Agrain, C., Broz, P., and Cornelis, G.R. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Kagawa H., Owaribe, K., Asakura, S., and Takahashi, N. 1976. Flagellar hook protein from Salmonella SJ25. J. Bacteriol. 125: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H., Aizawa, S.I., Yamaguchi, S., and Ishizu, J.I. 1979. Isolation and characterization of bacterial flagellar hook proteins from salmonellae and Escherichia coli. J. Bacteriol. 138: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V., Campbell, J.W., Minnich, S.A., Thomson, N.R., Matsumura, P., and Pruss, B.M. 2004. Gene array analysis of Yersinia enterocolitica FlhD and FlhC: Regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiology 150: 2289–2300. [DOI] [PubMed] [Google Scholar]
- Kato S., Aizawa, S., and Asakura, S. 1982. Reconstruction in vitro of the flagellar polyhook from Salmonella. J. Mol. Biol. 161: 551–560. [DOI] [PubMed] [Google Scholar]
- Katsura I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327: 73–75. [DOI] [PubMed] [Google Scholar]
- Katsura I. and Hendrix, R.W. 1984. Length determination in bacteriophage lambda tails. Cell 39: 691–698. [DOI] [PubMed] [Google Scholar]
- Kawagishi I., Homma, M., Williams, A.W., and Macnab, R.M. 1996. Characterization of the flagellar hook-length control protein fliK of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 178: 2954–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I.H., Reese, T.S., and Khan, S. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. 89: 5956–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Minamino, T., Yamaguchi, S., and Macnab, R.M. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov S.M., Castrisios, M., and Shaw, J.G. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72: 1939–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y., Silverman, M., and Simon, M. 1978. Identification of the structural gene for the hook subunit protein of Escherichia coli flagella. J. Bacteriol. 133: 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya K., Shibano, N., Higano, T., Azuma, N., Yamaguchi, S., and Aizawa, S.I. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34: 767–779. [DOI] [PubMed] [Google Scholar]
- Kornacker M.G. and Newton, A. 1994. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol. Microbiol. 14: 73–85. [DOI] [PubMed] [Google Scholar]
- Koroyasu S., Yamazato, M., Hirano, T., and Aizawa, S.I. 1998. Kinetic analysis of the growth rate of the flagellar hook in Salmonella typhimurium by the population balance method. Biophys. J. 74: 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Shimamoto, N., Yamaguchi, S., Namba, K., and Aizawa, S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226: 433–446. [DOI] [PubMed] [Google Scholar]
- Kubori T., Okumura, M., Kobayashi, N., Nakamura, D., Iwakura, M., and Aizawa, S.I. 1997. Purification and characterization of the flagellar hook–basal body complex of Bacillus subtilis. Mol. Microbiol. 24: 399–410. [DOI] [PubMed] [Google Scholar]
- Kutsukake K. 1997. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J. Bacteriol. 179: 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Suzuki, T., Yamaguchi, S., and Iino, T. 1979. Role of gene flaFV on flagellar hook formation in Salmonella typhimurium. J. Bacteriol. 140: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Ohya, Y., and Iino, T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Minamino, T., and Yokoseki, T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176: 7625–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima G., Kawagishi, I., Homma, M., Asaka, J., Kondo, E., and Macnab, R.M. 1989. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc. Natl. Acad. Sci. 86: 4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S., Gibson, T., Lakey, A., Leonard, K., Zeviani, M., Knight, P., Wardale, J., and Trinick, J. 1991. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett. 282: 313–316. [DOI] [PubMed] [Google Scholar]
- Limberger R.J., Slivienski, L.L., and Samsonoff, W.A. 1994. Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis. J. Bacteriol. 176: 3631–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R.M. 1977. Bacterial flagella rotating in bundles: A study in helical geometry. Proc. Natl. Acad. Sci. 74: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R.M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26: 131–158. [DOI] [PubMed] [Google Scholar]
- Macnab R.M. 1999. The bacterial flagellum: Reversible rotary propellor and type III export apparatus. J. Bacteriol. 181: 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R.M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57: 77–100. [DOI] [PubMed] [Google Scholar]
- Magdalena J., Hachani, A., Chamekh, M., Jouihri, N., Gounon, P., Blocker, A., and Allaoui, A. 2002. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184: 3433–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima S., Komoriya, K., Yamaguchi, S., and Aizawa, S.I. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291: 2411–2413. [DOI] [PubMed] [Google Scholar]
- Minamino T. and Macnab, R.M. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181: 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. and Macnab, R.M. 2000a. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182: 4906–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T. and MacNab, R.M. 2000b. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37: 1494–1503. [DOI] [PubMed] [Google Scholar]
- Minamino T. and MacNab, R.M. 2000c. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35: 1052–1064. [DOI] [PubMed] [Google Scholar]
- Minamino T., Gonzalez-Pedrajo, B., Yamaguchi, K., Aizawa, S.I., and Macnab, R.M. 1999. FliK, the protein responsible for flagellar hook-length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34: 295–304. [DOI] [PubMed] [Google Scholar]
- Minamino T., Chu, R., Yamaguchi, S., and Macnab, R.M. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182: 4207–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Gonzalez-Pedrajo, B., Kihara, M., Namba, K., and Macnab, R.M. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Saijo-Hamano, Y., Furukawa, Y., Gonzalez-Pedrajo, B., Macnab, R.M., and Namba, K. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 341: 491–502. [DOI] [PubMed] [Google Scholar]
- Minamino T., Ferris, H.U., Moriya, N., Kihara, M., and Namba, K. 2006. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar Type III export apparatus. J. Mol. Biol. 362: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Moriya N., Minamino, T., Hughes, K.T., Macnab, R.M., and Namba, K. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 359: 466–477. [DOI] [PubMed] [Google Scholar]
- Muramoto K., Makishima, S., Aizawa, S.I., and Macnab, R.M. 1998. Effect of cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J. Mol. Biol. 277: 871–882. [DOI] [PubMed] [Google Scholar]
- Muramoto K., Makishima, S., Aizawa, S., and Macnab, R.M. 1999. Effect of hook subunit concentration on assembly and control of length of the flagellar hook of Salmonella. J. Bacteriol. 181: 5808–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E., Gressmann, H., Ye, F., Schlapbach, R., Dehio, M., Dehio, C., Stack, A., Meyer, T.F., Suerbaum, S., and Josenhans, C. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52: 947–961. [DOI] [PubMed] [Google Scholar]
- Nishimura A. and Hirota, Y. 1989. A cell division regulatory mechanism controls the flagellar regulon in Escherichia coli. Mol. Gen. Genet. 216: 340–346. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Ohto, Y., Aizawa, S., Macnab, R.M., and Iino, T. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176: 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole P.W., Kostrzynska, M., and Trust, T.J. 1994. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol. Microbiol. 14: 691–703. [DOI] [PubMed] [Google Scholar]
- Pallen M.J., Penn, C.W., and Chaudhuri, R.R. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13: 143–149. [DOI] [PubMed] [Google Scholar]
- Patterson-Delafield J., Martinez, R.J., Stocker, B.A., and Yamaguchi, S. 1973. A new fla gene in Salmonella typhimurium–flaR–and its mutant phenotype-superhooks. Arch. Mikrobiol. 90: 107–120. [DOI] [PubMed] [Google Scholar]
- Payne P.L. and Straley, S.C. 1999. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J. Bacteriol. 181: 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power M.E., Alm, R.A., and Trust, T.J. 1992. Biochemical and antigenic properties of the Campylobacter flagellar hook protein. J. Bacteriol. 174: 3874–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss B.M., Campbell, J.W., Van Dyk, T.K., Zhu, C., Kogan, Y., and Matsumura, P. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner–Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185: 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.A., Karim, N., Worku, M., Penn, C.W., and O'Toole, P.W. 2005. Helicobacter pylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J. Bacteriol. 187: 5742–5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M. and Simon, M. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249: 73–74. [DOI] [PubMed] [Google Scholar]
- Smith K.D. and Ozinsky, A. 2002. Toll-like receptor-5 and the innate immune response to bacterial flagellin. Curr. Top. Microbiol. Immunol. 270: 93–108. [DOI] [PubMed] [Google Scholar]
- Stafford G.P., Ogi, T., and Hughes, C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiol. 151: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier I., Bleves, S., Josenhans, C., Karmani, L., Kerbourch, C., Lambermont, I., Totemeyer, S., Boyd, A., and Cornelis, G.R. 2000. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 37: 1005–1018. [DOI] [PubMed] [Google Scholar]
- Suzuki T. and Iino, T. 1981. Role of the flaR gene in flagellar hook formation in Salmonella spp. J. Bacteriol. 148: 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T. and Komeda, Y. 1981. Incomplete flagellar structures in Escherichia coli mutants. J. Bacteriol. 145: 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino, T., Horiguchi, T., and Yamaguchi, S. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J. Bacteriol. 133: 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Yonekura, K., and Namba, K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337: 105–113. [DOI] [PubMed] [Google Scholar]
- Tamano K., Katayama, E., Toyotome, T., and Sasakawa, C. 2002. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J. Bacteriol. 184: 1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampakaki A.P., Fadouloglou, V.E., Gazi, A.D., Panopoulos, N.J., and Kokkinidis, M. 2004. Conserved features of type III secretion. Cell. Microbiol. 6: 805–816. [DOI] [PubMed] [Google Scholar]
- Thomas D.R., Francis, N.R., Xu, C., and DeRosier, D.J. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188: 7039–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B., Rajagopala, S.V., Ester, C., Hauser, R., and Uetz, P. 2006. Novel conserved assembly factor of the bacterial flagellum. J. Bacteriol. 188: 7700–7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand M.E., Sockett, R.E., Evans, K.J., Doherty, N., Sharp, P.M., Hardie, K.R., and Winzer, K. 2006. Helicobacter pylori FlhB function: The FlhB C-terminal homologue HP1575 acts as a “spare part” to permit flagellar export when the HP0770 FlhBCC domain is deleted. J. Bacteriol. 188: 7531–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Mariconda, S., Suzuki, A., McClelland, M., and Harshey, R.M. 2006. Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188: 7981–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerlund-Wikstrom B. 2000. Peptide display on bacterial flagella: Principles and applications. Int. J. Med. Microbiol. 290: 223–230. [DOI] [PubMed] [Google Scholar]
- Williams A.W., Yamaguchi, S., Togashi, F., Aizawa, S.I., Kawagishi, I., and Macnab, R.M. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178: 2960–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K., Maki-Yonekura, S., and Namba, K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424: 643–650. [DOI] [PubMed] [Google Scholar]
- Zhu K., Gonzalez-Pedrajo, B., and Macnab, R.M. 2002. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41: 9516–9524. [DOI] [PubMed] [Google Scholar]