Abstract
The sorting nexins (SNXs) constitute a large group of PX domain-containing proteins that play critical roles in protein trafficking. We report here the solution structure of human sorting nexin 22 (SNX22). Although SNX22 has <30% sequence identity with any PX domain protein of known structure, it was found to contain the α/β fold and compact structural core characteristic of PX domains. Analysis of the backbone dynamics of SNX22 by NMR relaxation measurements revealed that the two walls of the ligand binding cleft undergo internal motions: on the picosecond timescale for the β1/β2 loop and on the micro- to millisecond timescale for the loop between the polyproline motif and helix α2. Regions of the SNX22 structure that differ from those of other PX domains include the loop connecting strands β1 and β2 and the loop connecting helices α1 and α2, which appear to be more mobile than corresponding loops in other known structures. The interaction of dibutanoyl-phosphatidylinositol-3-phosphate (dibutanoyl-PtdIns(3)P) with SNX22 was investigated by an NMR titration experiment, which identified the binding site in a basic cleft and indicated that ligand binding leads only to a local structural rearrangement as has been found with other PX domains. Because motions in the loops are damped out when dibutanoyl-PtdIns(3)P binds, entropic effects could contribute to the lower affinity of SNX22 for this ligand compared to other PX domains.
Keywords: domains and motifs, protein trafficking/sorting, NMR spectroscopy
The sorting nexin (SNX) family consists of a diverse array of proteins that are involved in endocytosis and cellular protein trafficking (Worby and Dixon 2002). The molecular functions of SNX members in these processes mainly rely on their phosphatidylinositol (PI) binding motif, the phox homology (PX) domain (Xu et al. 2001b; Ellson et al. 2002). The PX domain, a 100–130 residue module, was originally identified in the p40phox and p47phox subunits of NADPH oxidases (Ponting 1996). With its appearance in >100 eukaryotic proteins (Sato et al. 2001; Xu et al. 2001b; Ellson et al. 2002), the PX domain has emerged as a major player in PI signaling. SNX is one of the largest protein subgroups that contain PX domains. The human SNX family consists of 29 members (Xu et al. 2001b), many of which have been identified through database searches. On the basis of their divergent domain architecture, SNXs are divided into three groups: Seven SNXs consist of little more than a PX domain; 13 SNXs contain C-terminal coiled-coil regions that may mediate oligomerization of these proteins; the remaining identified SNXs contain protein domains in addition to PX.
Recent structural and biochemical studies (Bravo et al. 2001; Cheever et al. 2001; Hiroaki et al. 2001; Xing et al. 2004) have determined that the general biological functions of PX domains are to regulate protein sorting and membrane trafficking. The common structural features of a PX domain are an α/β fold containing a basic PI binding cleft formed by the residues from the β3/α1 junction, the N-terminal ends of strand β2 and helix α2, and loop α1/α2. In addition, PX domains also contain a characteristic polyproline motif at loop α1/α2, which is the primary interaction site between the PX domain and SH3 domain-containing proteins (Hiroaki et al. 2001). Thus, the functions of PX domains involve both protein–protein and protein–PI interactions. Different PX domains exhibit a wide range of PI binding specificities and affinities. The PX domains from Saccharomyces cerevisiae preferentially bind phosphatidylinositol-3-phosphate (PtdIns[3]P) over other PIs (Yu and Lemmon 2001), and they can be divided further into two groups on the basis of their PtdIns(3)P binding affinities (Yu and Lemmon 2001): those with high affinities (dissociation constant Kd = 2–3 μM), and those with low affinities (Kd ∼ 100 μM). In contrast to the yeast PX domains, the PX domains of higher eukaryotes show specificities to more divergent substrates (Kanai et al. 2001; Song et al. 2001; Xu et al. 2001a; Zhong et al. 2002; Du et al. 2003). The structural origins of this diverse PI binding activity is unclear at present, because of the limited number of available structures of PX domains and the generally low sequence identity between PX domains.
Human SNX22 is a single PX domain protein and a putative member of the SNX family (Worby and Dixon 2002). The sequence identity between SNX22 and other structurally characterized SNXs, such as SNX1 (Zhong et al. 2002) and SNX3 (Xu et al. 2001a), is lower than 30%. We report here the NMR solution structure of SNX22 and an investigation of its interaction with dibutanoyl-PtdIns(3)P (a soluble form of PtdIns[3]P). The results confirmed that SNX22 contains a characteristic PX domain and identified the dibutanoyl-PtdIns3P binding site. The high mobility observed for loop β1/β2 and loop α1/α2 of free SNX22 suggests a possible explanation for the protein's weak binding affinity for PtdIns(3)P.
Results
Solution conditions and NMR data collection
SNX22 has a strong tendency to precipitate in 10 mM Bis-Tris, 100 mM NaCl at pH 7.0. We found it possible to stabilize the protein by adding 50 mM arginine and 50 mM glutamate to the NMR buffer (Golovanov et al. 2004). Under these conditions 0.5 mM SNX22 was stable for one month, and it was possible to collect a full suite of multidimensional NMR spectra for determining the structure and dynamics of SNX22 and its interaction with PtdIns(3)P.
Three-dimensional structure
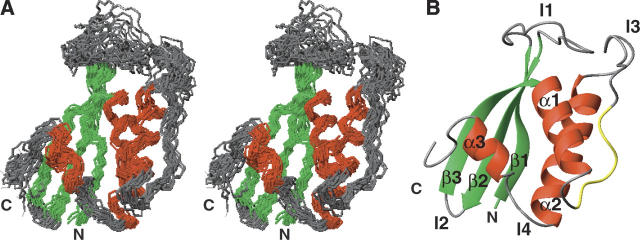
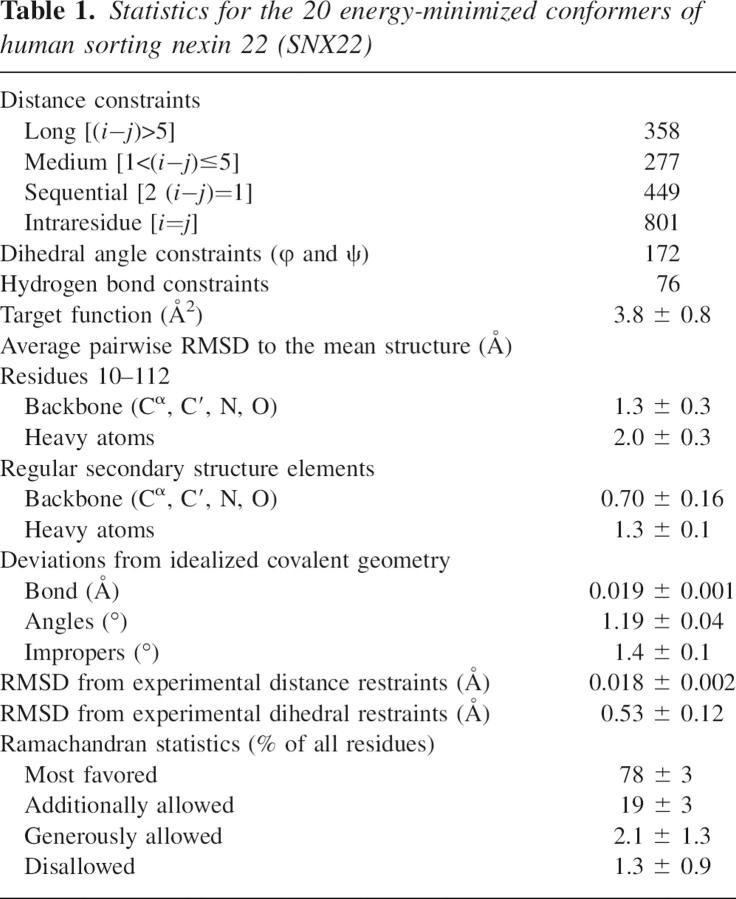
The 20 conformers of SNX22 that represent its solution structure superimposed by the backbone atoms of their structured region are shown in Figure 1A. The structural statistics are summarized in Table 1. All the residues that fall into the disfavored region in the Ramachandran plot are located in the poorly defined loops. The overall fold of SNX22 consists of an N-terminal antiparallel β-sheet and a C-terminal helical subdomain, packed against each other (Fig. 1B). The β-sheet is formed by three β-strands (β1: 3–10, β2: 25–33, and β3: 36–43), while the helical subdomain consists of two long α-helices (α1: 52–61 and α2: 84–101) and a short helix (α3: 99–104) at the C terminus. Other notable structural features include two long loops, held by β1 and β2 (β1/β2: 11–25), and α1 and α2 (α1/α2: 62–83), respectively. The polyproline motif contained in loop α1/α2, V60-P61-D62-F63-P64-S65-K66, adopts a left-handed polyproline II helix conformation (PPII) and is exposed to the solvent, making it accessible to SH3 domain-containing proteins. As in other PX domains, loop β1/β2 and loop PPII/α2 line up to form a solvent accessible cleft at the interface of the two subdomains. The N-terminal Ext-G(His6)-LE tag and the C-terminal fragment (113–120) are both structurally disordered, as evidenced by their chemical shifts and NOE values.
Figure 1.
Solution structure and backbone mobility of human sorting nexin 22 (SNX22). (A) Stereoview of the ensemble of 20 conformers representing the structure colored to distinguish residues in helices (red), β-strands (green), and loops (gray). (B) Ribbon representation of a representative conformer (that with the lowest energy), with individual elements of secondary structure labeled and colored as in (A) and the proline-rich sequence (V60–K66) in yellow. For clarity, the disordered eight residues from the N terminus and eight residues from the C terminus are not shown.
Table 1.
Statistics for the 20 energy-minimized conformers of human sorting nexin 22 (SNX22)
Backbone dynamics
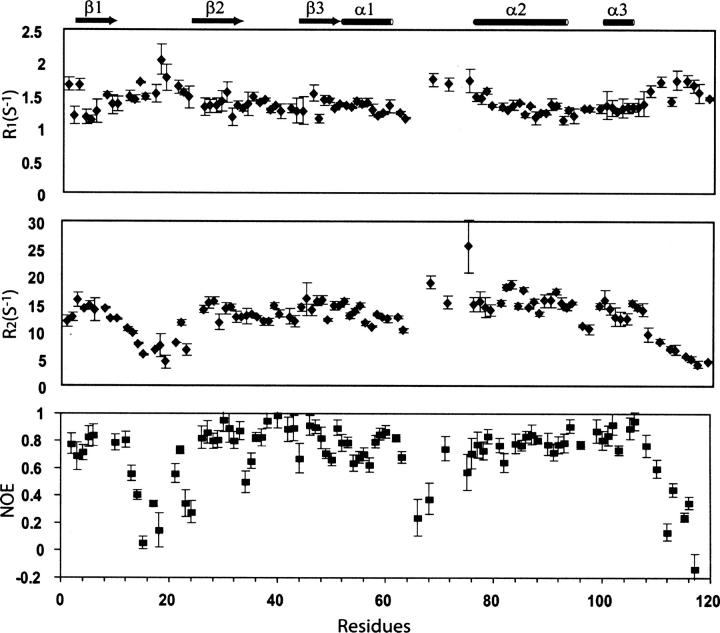
The backbone dynamics of SNX22 were determined from measurements of 15N R1, R2 relaxation rates and 15N-NOE values (Kay et al. 1989). Because the limited sets of NMR data did not support accurate analysis by the model-free approach (Lipari and Szabo 1982a,b), we chose to infer the internal motion of SNX22 from the parameters directly. These parameters are sensitive to the molecular tumbling time as well as to local motions on the pico- to nanosecond timescale. R2 relaxation has additional contributions from slow chemical exchange on the micro- to millisecond timescale. The 15N-NOE values (Fig. 2) observed for residues participating in secondary structural elements are 0.6 or higher, consistent with their being highly ordered. By contrast, the NOE values observed for the C terminus, loop β1/β2, PPII, and loop PPII/α2 are significantly lower than those of the rest of the sequence, indicating high mobility of the peptide backbone. Interestingly, analysis of the R1 and R2 values from these flexible regions further revealed that their respective motions occur on different timescales: Loop β1/β2 and the C terminus show R1 values higher than average but R2 values lower than average, as is characteristic for internal dynamics on the pico- to nanosecond timescale (Kay et al. 1989); by contrast, the R1 and R2 values of PPII and loop PPII/α2 are both significantly higher than average, as is characteristic for internal dynamics on the micro- to millisecond timescale (Kay et al. 1989). Millisecond scale motions for the PPII and loop PPII/α2 regions are also supported by the observation of severe signal broadenings (data not shown).
Figure 2.
15N R2/R1 ratios and 15N-NOEs for SNX22 plotted as a function of residue number.
Sequence and structure comparisons with other PX domains
The scaffold of SNX22 is analogous to those observed for other PX domains (Fig. 3). Despite their low sequence identities (Fig. 3A), the structures of SNX22, p40phox (Bravo et al. 2001) and the yeast SNX3 ortholog, Grd19p (Zhou et al. 2003) align well with each other (Fig. 3A). The superposition of Cα atoms, obtained using the DALI server (Holm and Sander 1995), yielded an RMS deviation of 2.8 Å for 101 aligned residues between SNX22 and Grdp19, 3.7 Å for 96 aligned residues between SNX22 and p40phox, and 3.0 Å for 113 aligned residues between Grd19p and p40phox. The most similar structural elements are the antiparallel β-sheet and the α2-helix (Fig. 3A). The α1-helices, whereas well superimposed at their N termini, have slightly different orientations: The α1-helices from SNX22 and p40phox deviate from that of Grd19p by about 20°. The observed close alignment of the structural cores of all three proteins (SNX22, p40phox, and Grdp19) is consistent with the relatively high sequence homology in these regions (Fig. 3B). In addition, key ligand-coordinating residues identified in other PX domains, including yeast SNARE protein Vam7p (Cheever et al. 2001), p40phox (Bravo et al. 2001), and Grd19p (Zhou et al. 2003), are also conserved in SNX22 as residues R43, Y44, S45, K66, and R79 (Fig. 3B).
Figure 3.
(A) Superimposed structures of: (left) SNX22 (red; this work) and Grd19p (blue; Zhou et al. 2003); (middle) SNX22 (red) and P40phox (green; Bravo et al. 2001); (right) Grd19p (blue) and P40phox (green). (B) Alignment of PX domain sequences with residues colored to indicate those that are strictly conserved (red highlighted in yellow), identical in two of the three sequences (green highlight), and similar (cyan highlight).
By contrast, the largest differences in sequence and structure among the three PX domains are found in loops β1/β2 and α1/α2. The 20-residue β1/β2 of SNX22 is significantly longer than its counterparts in Grd19p and p40phox (10 residues and five residues, respectively), and contains three proline residues (P11, P16, and P20) not present in the sequences of the other two proteins (Fig. 3B). The large structural variation observed in loop α1/α2 arises mainly from the deletion of 10–11 residues in SNX22 that are present in Grd19p and p40phox (Fig. 3B). Variations in the loops of different PX domains may be expected to lead to differences in internal dynamics. For example, whereas the β1/β2 loop of free SNX22 was found to be highly dynamic from NMR relaxation data, the equivalent loop in free Grd19p appeared to be well ordered on the basis of its well-defined electron density (Zhou et al. 2003). Although the orientation of the α3-helix of SNX22 differs from its counterparts in the other two structures, this difference may simply be the result of the scarcity of long-range NOEs rather than a true divergence between SNX22 and the other two PX domains.
Dibutanoyl-PtdIns(3)P titration
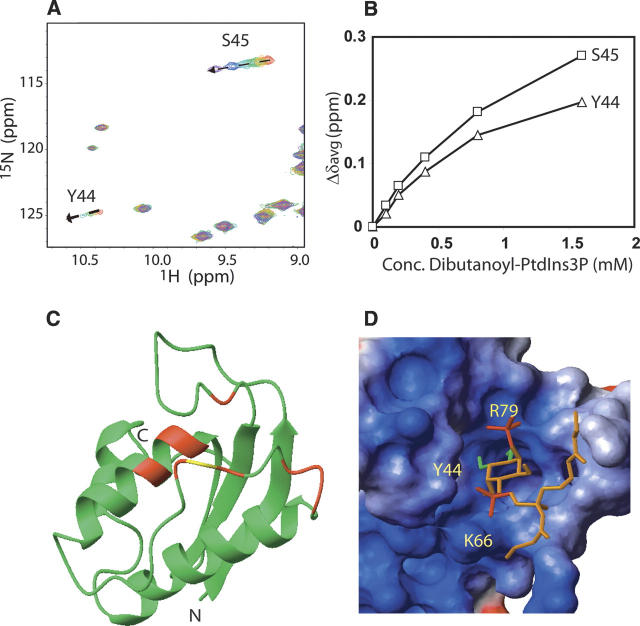
We chose PtdIns(3)P, the most common ligand for PX domains (Ellson et al. 2002), to investigate the ligand binding activity of SNX22. Unlabeled dibutanoyl-PtdIns(3)P, a more soluble form of PtdIns(3)P, was titrated into [U-13C,15N]-SNX22. A series of [1H,15N]-HSQC spectra were collected to follow changes in chemical shifts upon the progressive addition of dibutanoyl-PtdIns(3)P (0 mM, 0.1 mM, 0.2 mM, 0.4 mM, 0.8 mM, and 1.6 mM). Changes in 1HN and 15N chemical shifts between the free state of SNX22 and the ligand-bound state were calculated according to the formula Δδave = [(ΔδHN2 + (ΔδN/5)2)/2]1/2, where ΔδHN and ΔδN correspond, respectively, to the chemical shift differences in 1HN and 15N. The analysis showed that the NMR peaks of a number of residues are increasingly perturbed by adding dibutanoyl-PtdIns(3)P (Fig. 4A,B). Those residues displaying the largest chemical shift changes (Δδave > 0.05 ppm) included S23, Y44, S45, H48, K66, L68, W71, R72, and T73; residues showing smaller but significant chemical shifts (0.05 > Δδave > 0.035ppm) included G10, G75, Q78, and R79. When mapped onto the structure of the SNX22, these residues are located in loop β1/β2, loop PPII/α2, and the N-terminal region of α1 (Fig. 4C); they are spatially proximate to each other in the structure of SNX22 and define the dibutanoyl-PtdIns(3)P binding site as being centered on the basic cleft (Fig. 4C), as found for PtdIns(3)P binding to other PX domains. In particular, key ligand-coordinating residues observed in complexes of Vam7p (Cheever et al. 2001), P40phox (Bravo et al. 2001), and Grd19p (Zhou et al. 2003) are also associated with ligand binding in SNX22 (residues Y44, S45, K66, and R79). The chemical shift perturbations observed as a function of the dibutanoyl-PtdIns(3)P concentration (Fig. 4B) were consistent with a 1:1 stoichiometry, and were analyzed to yield a dissociation constant of 1.1 ± 0.3 mM.
Figure 4.
(A) Overlaid 2D [1H,15N]-HSQC spectra highlight the progressive chemical shift changes of selected residues (dashed lines). The spectra were collected for 0.2 mM [U-15N,13C]-SNX22 in the presence of various concentrations of dibutanoyl-PtdIns(3)P: (red) 0 mM, (orange) 0.1 mM, (green) 0.2 mM, (cyan) 0 4 mM, (blue) 0.8 mM, (purple) 1.6 mM. (B) Plot of Δδave for S43 and Y44 as a function of the concentration of the dibutanoyl-PtdIns(3)P. (C) Ribbon representation of the solution structure of SNX22 showing the residues (red) that display the largest chemical shift changes (Δδave > 0.05 ppm) between samples containing 0 mM and 1.6 mM dibutanoyl-PtdIns(3)P. Signal broadening prevented observation of the NMR peak from R67 (yellow). (D) Surface representation of the structure SNX22:dibutyl-PtdIns(3)P complex generated from the experimental NMR data by the program HADDOCK (Dominguez et al. 2003). The 1- and 3-phosphate groups, and the 4- and 5-OH groups of dibutanoyl-PtdIns(3)P are colored red and green, respectively.
We used the HADDOCK program (Dominguez et al. 2003) to construct a structural model of the complex of SNX22 with dibutanoyl-PtdIns(3)P. On the basis of data on solvent accessibility, chemical shift perturbations, and sequence conservation, we identified R43, Y44, K66, and R79 from SNX22 and the inositol ring, 1-phosphate and 3-phosphate groups from dibutanoyl-PtdIns(3)P as the “active” set to model the interactions. From 100 structures calculated after water refinement, we selected the 20 with the lowest energies for further analysis. In all the analyzed structural models, the 3-phosphate group, 4- and 5-OH groups of dibutanoyl-PtdIns(3)P are inserted into the basic pocket formed in SNX22 (Fig. 4D). At the edge of the pocket, the positively charged side chains of K66 and R79 interact, respectively, with the 1-phosphate and 3-phosphate groups of dibutanoyl-PtdIns(3)P, while the aromatic ring of Y44 stacks with the inositol ring and the dibutanoyl tail protrudes into the solvent.
Discussion
SNX22, together with SNX13, SNX14, SNX24, and SNX25, belongs to a small subgroup of SNXs that contain only a PX domain and very little else (Worby and Dixon 2002). The physiological functions of members of this group of SNXs have not been determined. However, it is generally accepted that they function to recruit other proteins to the membrane via protein–protein and protein– ligand interactions. The three-dimensional structure of SNX22 presented here represents the first for a member of this subclass. Despite its low sequence identity with other proteins of known structure, SNX22 contains an α/β fold typical of PX domains. The structure of SNX22 most closely resembles those of PX domain proteins p40phox and Grd19p. Superposition of the three structures showed close agreement in the positions of all helices and strands. The largest variations were in loop β1/β2 and loop α1/α2.
Backbone dynamics determined from 15N R1, R2 relaxation rates, and 15N-NOEs revealed that residues of SNX22 involved in secondary structure are relatively rigid, whereas those in loop β1/β2, PPII, loop PPII/α2, and the C terminus are mobile. Although loops β1/β2 and PPII/α2 are located next to one other, they exhibit motions on different timescales: pico- to nanosecond for loop β1/β2 but micro- to millisecond for loop PPII/α2. Although the dynamic properties of other PX domains have not been studied in solution, the crystallographic B-factors of loop β1/β2, PPII, and loop PPII/α2 in the X-ray structures of all ligand-free PX domains determined so far, such as Grd19p (Zhou et al. 2003), p47phox (Hiroaki et al. 2001), and the cytokine-independent survival kinase (CISK) (Xing et al. 2004), are relatively higher than those of the rest of the structure, implying certain conformational plasticity. Because the PPII domain is involved in interactions with SH3 domains, its internal motions may facilitate conformational reorientation required for protein–protein interaction. On the other hand, the lengths of these loops vary between different proteins. For example, in the solution structure of free Vam7p, the PPII/α2 loop is highly disordered and characterized by sharp NMR signals (Lu et al. 2002) that suggest motions on the picosecond timescale (Kay et al. 1989). The differential selectivity of PI binding of various PX domains likely is a function of the combined differences in the structure and dynamics of loops β1/β2 and PPII/α.
The PI ligand binding mechanism determined for SNX22 is similar to those for other PX domains (Bravo et al. 2001; Zhou et al. 2003). The ligand binding site is defined by a basic cleft rimmed by loop β1/β2, loop PPII/α2, and the N terminus of α1. The extent and magnitude of chemical shift perturbations and the solution structure of the ligand-free SNX22 suggests that ligand binding induces only local conformational changes, consistent with previous observations for other PX domains (Bravo et al. 2001; Cheever et al. 2001; Zhou et al. 2003; Xing et al. 2004). However, the binding affinity between PtdIns(3)P and SNX22 is relatively low (Kd ∼ 1 mM), about 200 times weaker than that of p40phox (Bravo et al. 2001) and 2000 times than that of the Grd19p (Zhou et al. 2003). Because the residues important to binding of PtdIns(3)P (R43, Y44, S45, K66, and R79 of SNX22) are conserved in all three PX domains, the large differences in affinity probably involve secondary elements. For example, the loops surrounding the ligand binding pocket, with high sequence variation (Fig. 3B), may encode differential dynamics in these PX domains. Binding of PtdIns(3)P is expected to damp out loop motions present in the free protein, as shown by analysis of differences in the normalized crystallographic B factors for unligated (PDB 1ocs) and ligated (PDB 1ocu) Grd19p (data not shown). Thus, we suggest that high local mobility in loops β1/β2 and loop PPII/α2 of SNX22 may explain the low affinity of SNX22 for PtdIns(3)P.
We attempted to determine the interaction between PtdIns(3)P-bound SNX22 and membrane, by using dodecylphosphocholine (DPC) as a membrane mimetic. The addition of 1.2 mM DPC resulted in no appreciable change in the [1H,15N]-HSQC spectrum of SNX22 (Supplementary Fig. S1). However, large spectral changes occurred when the concentration of added DPC was raised above its normal critical micelle concentration (le Maire et al. 2000) to 2.4 mM (Supplementary Fig. S1). Whereas a small fraction of peaks remained unchanged, the majority of the peaks exhibited large shifts or were missing in the NMR spectrum. This spectral change was reversed when the DPC concentration was diluted back to 1 mM. These large, reversible spectral changes indicate that SNX22 undergoes a major conformational rearrangement upon binding to DPC micelles. The physiological relevance of this observation remains to be determined. This result with SNX22 differs from that observed for Vam7p (Cheever et al. 2001), which exhibited only a local conformational change in the presence of micelles.
Materials and Methods
Informatics
We used the Sesame Laboratory Information Management System (Zolnai et al. 2003) developed at the Center for Eukaryotic Structural Genomics to select this target and to track all subsequent steps in this study.
Cloning
The cDNA encoding human SNX22 (GenBank Accession: BC019655.1) was subcloned into the peU–(N)–His6 vector (a gift from Professor Endo at Ehime University, Matsuyama, Japan.), used in the cell-free translation reaction, by PCR. The peU–(N)–His6–SNX22 construct was then sequenced to confirm the absence of any spurious mutations in the coding region.
Protein production
We used a wheat germ cell-free protein expression protocol to prepare the labeled protein samples used in this NMR study: [15N]-Next-SNX22 and [13C,15N]-Next-SNX22. The wheat germ extract used for in vitro translation reactions was purchased from CellFree Sciences, Ltd. The protocols used for in vitro transcription and translation were based on published procedures (Madin et al. 2000; Sawasaki et al. 2002; Vinarov et al. 2004). The protein sample used for the NMR measurements corresponded to the full-length SNX22 gene product plus an N-terminal extension, Next− ≡ G(H)6LE-, added in the cloning process (128 amino acid residues in total). The numbering system used here is according to the native sequence of SNX22. SDS-PAGE was used to determine the purity of the protein, and the BCA method (Smith et al. 1985) was used to determine the protein concentration. The NMR sample for the structure determination contained 0.5 mM [U-13C,U-15N]-labeled SNX22, 10 mM Bis-Tris, 100 mM NaCl, 50 mM sodium glutamate, 50 mM arginine chloride, 10 mM DTT, and 90%H2O/10% D2O at pH 7.0.
NMR spectroscopy
All NMR spectra were recorded at the National Magnetic Resonance Facility at Madison (NMRFAM) on a Varian INOVA 600 spectrometer equipped with a cryogenic triple-resonance probe. The sample temperature was controlled at 25°C. We collected a suite of pulse field gradient-enhanced NMR experiments, including HNCACB, CBCA(CO)NH, HNCO, HBHA(CO)NH, C(CO)NH, and HCCH-TOCSY, as input for sequential backbone and nonaromatic side chain assignments. We collected a 3D 15N-edited NOESY-HSQC (τmix = 100 msec) and a 3D 13C-edited NOESY-HSQC (τmix = 100 msec) data set to measure NOEs used for additional assignments (side chain amide groups and aromatic groups) and distance constraints. The spectra were processed and analyzed, respectively, with the NMRPipe (Delaglio et al. 1995) and Sparky (http://www.cgl.ucsf.edu/home/sparky) software packages. The raw, time-domain NMR data sets and chemical shift assignments have been deposited in the BioMagResBank (BMRB) database under accession number 6396. A sample of 0.5 mM [U-15N]-SNX22 was used for the collection of steady state 15N-NOE and 15N relaxation (R1, R2) data by standard pulse sequences (Farrow et al. 1994). To determine the 15N R1 values, NMR spectra were recorded with delays of 110, 250, 530, 710, and 950 msec. To determine 15N R2 values, NMR spectra were recorded with delays of 10, 30, 50, 70, and 90 msec. The relaxation rates were calculated by least-squares fitting of peak heights versus relaxation delay to a single exponential decay. The reported error estimates are standard deviations derived from fitting the data. Steady-state 15N-NOE values were calculated from the ratio of peak heights in a pair of NMR spectra acquired with and without proton saturation. The signal-to-noise ratio in each spectrum was used to estimate the experimental uncertainty. [1H,15N]-HSQC spectra were collected to detect the binding of unlabeled dibutanoyl-PtdIns(3)P to [U-15N,U-13C]-labeled SNX22. Throughout the titration, the concentration of SNX22 was maintained at 0.2 mM, and the concentration of PtdIns(3)P was varied to gives a series of different SNX22:PidIns(3)P molar ratios (1:0, 1:0.5, 1:1, 1:4, and 1:8). The sample buffer used for PtdIns(3)P titration is the same as that used for structure determination.
Structure determination
Interproton distance constraints were derived from a 3D 15N-edited NOESY-HSQC spectrum and a 3D 13C-edited NOESY-HSQC spectrum. Backbone ϕ and ψ angles were derived from TALOS-based analysis of backbone chemical shifts (Cornilescu et al. 1999). The automated iterative refinement module (Güntert et al. 1997) in the CYANA software package (Herrmann et al. 2002) was used to generate the initial NOE assignments and the initial set of structural models. These NOE assignments were subsequently subject to correction through iterative analysis of NMR spectra and structural calculation. In the final round of CYANA refinement, 1885 NOE restraints and 172 dihedral angle restraints were used. In addition, 76 hydrogen bond restraints, generated from analysis of the secondary chemical shifts and from observed NOEs characteristic for α-helices and β-sheets, were also added to the set of restraints. Out of 100 final structures calculated by CYANA, the 20 structures with the lowest target functions were chosen for further refinement by X-PLOR, with use of a physical force field and explicit water solvent terms. The final 20 conformers were validated by Procheck-NMR (Laskowski et al. 1996), and the statistics for these are listed in Table 1.
PDB Deposition
Structure determined under the National Institutes of Health, NIGMS Protein Structure Initiative; coordinates and related data have been deposited at PDB (2ETT) and NMR data at BMRB (bmr 6866).
Acknowledgments
This work was supported by the National Institutes of Health, Protein Structure Initiative through grants P50 GM64598 and U54 GM074901. NMR data were collected at the National Magnetic Resonance Facility at Madison, which is supported in part by NIH Grants P41 RR02301 and P41 GM66326. We thank the many team members from the Center for Eukaryotic Structural Genomics who provided the infrastructure for this work.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: John L. Markley, Center for Eukaryotic Structural Genomics, Department of Biochemistry, 433 Babcock Drive, University of Wisconsin-Madison, Madison, WI 53706-1544, USA; e-mail: markley@nmrfam.wisc.edu; fax: (608) 262-3759.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072752407.
References
- Bravo J., Karathanassis, D., Pacold, C.M., Pacold, M.E., Ellson, C.D., Anderson, K.E., Butler, P.J., Lavenir, I., Perisic, O., Hawkins, P.T., et al. 2001. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell 8: 829–839. [DOI] [PubMed] [Google Scholar]
- Cheever M.L., Sato, T.K., de Beer, T., Kutateladze, T.G., Emr, S.D., and Overduin, M. 2001. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3: 613–618. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Dominguez C., Boelens, R., and Bonvin, A.M. 2003. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125: 1731–1737. [DOI] [PubMed] [Google Scholar]
- Du G., Altshuller, Y.M., Vitale, N., Huang, P., Chasserot-Golaz, S., Morris, A.J., Bader, M.F., and Frohman, M.A. 2003. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson C.D., Andrews, S., Stephens, L.R., and Hawkins, P.T. 2002. The PX domain: A new phosphoinositide-binding module. J. Cell Sci. 115: 1099–1105. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Golovanov A.P., Hautbergue, G.M., Wilson, S.A., and Lian, L.Y. 2004. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 126: 8933–8939. [DOI] [PubMed] [Google Scholar]
- Güntert P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert, P., and Wüthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319: 209–227. [DOI] [PubMed] [Google Scholar]
- Hiroaki H., Ago, T., Ito, T., Sumimoto, H., and Kohda, D. 2001. Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 8: 526–530. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20: 478–480. [DOI] [PubMed] [Google Scholar]
- Kanai F., Liu, H., Field, S.J., Akbary, H., Matsuo, T., Brown, G.E., Cantley, L.C., and Yaffe, M.B. 2001. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3: 675–678. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Torchia, D.A., and Bax, A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 28: 8972–8979. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8: 477–486. [DOI] [PubMed] [Google Scholar]
- le Maire M., Champeil, P., and Moller, J.V. 2000. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508: 86–111. [DOI] [PubMed] [Google Scholar]
- Lipari G. and Szabo, A. 1982a. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules.1. Theory and range of validity. J. Am. Chem. Soc. 104: 4546–4559. [Google Scholar]
- Lipari G. and Szabo, A. 1982b. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104: 4559–4570. [Google Scholar]
- Lu J., Garcia, J., Dulubova, I., Sudhof, T.C., and Rizo, J. 2002. Solution structure of the Vam7p PX domain. Biochemistry 41: 5956–5962. [DOI] [PubMed] [Google Scholar]
- Madin K., Sawasaki, T., Ogasawara, T., and Endo, Y. 2000. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. 97: 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. 1996. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: Binding partners of SH3 domains? Protein Sci. 5: 2353–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T.K., Overduin, M., and Emr, S.D. 2001. Location, location, location: Membrane targeting directed by PX domains. Science 294: 1881–1885. [DOI] [PubMed] [Google Scholar]
- Sawasaki T., Hasegawa, Y., Tsuchimochi, M., Kamura, N., Ogasawara, T., Kuroita, T., and Endo, Y. 2002. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 514: 102–105. [DOI] [PubMed] [Google Scholar]
- Smith P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85. [DOI] [PubMed] [Google Scholar]
- Song X., Xu, W., Zhang, A., Huang, G., Liang, X., Virbasius, J.V., Czech, M.P., and Zhou, G.W. 2001. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 40: 8940–8944. [DOI] [PubMed] [Google Scholar]
- Vinarov D.A., Lytle, B.L., Peterson, F.C., Tyler, E.M., Volkman, B.F., and Markley, J.L. 2004. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat. Methods 1: 149–153. [DOI] [PubMed] [Google Scholar]
- Worby C.A. and Dixon, J.E. 2002. Sorting out the cellular functions of sorting nexins. Nat. Rev. Mol. Cell Biol. 3: 919–931. [DOI] [PubMed] [Google Scholar]
- Xing Y., Liu, D., Zhang, R., Joachimiak, A., Songyang, Z., and Xu, W. 2004. Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX). J. Biol. Chem. 279: 30662–30669. [DOI] [PubMed] [Google Scholar]
- Xu Y., Hortsman, H., Seet, L., Wong, S.H., and Hong, W. 2001a. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3: 658–666. [DOI] [PubMed] [Google Scholar]
- Xu Y., Seet, L.F., Hanson, B., and Hong, W. 2001b. The Phox homology (PX) domain, a new player in phosphoinositide signalling. Biochem. J. 360: 513–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W. and Lemmon, M.A. 2001. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276: 44179–44184. [DOI] [PubMed] [Google Scholar]
- Zhong Q., Lazar, C.S., Tronchere, H., Sato, T., Meerloo, T., Yeo, M., Songyang, Z., Emr, S.D., and Gill, G.N. 2002. Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. 99: 6767–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.-Z., de La Sierra-Gallay, I.L., Quevillon-Cheruel, S., Collinet, B., Minard, P., Blondeau, K., Henckes, G., Aufrère, R., Leulliot, N., Graille, M., et al. 2003. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278: 50371–50376. [DOI] [PubMed] [Google Scholar]
- Zolnai Z., Lee, P.T., Li, J., Chapman, M.R., Newman, C.S., Phillips Jr, G.N., Rayment, I., Ulrich, E.L., Volkman, B.F., and Markley, J.L. 2003. Project management system for structural and functional proteomics: Sesame. J. Struct. Funct. Genomics 4: 11–23. [DOI] [PubMed] [Google Scholar]