Abstract
The importance of the HIV gp41 conserved disulfide loop to envelope function has been examined by mutational and functional analyses. Based on a luciferase-reporter entry assay, mutants gp41-CC/AA (C598A/C604A) and gp41-Δ (deletion of residues 596–606) result in a nonfunctional envelope protein. Western blot analysis shows both mutants to be properly expressed but not processed to form gp120 and gp41, which explains their nonfunctionality. The presence of mutant gp160 on the cell surface, as well as their ability to bind to sCD4, suggests that the mutations have disrupted processing at the furin recognition site encoded within the gp120 conserved domain 5, without resulting in an overall misfolding of the protein. With respect to the furin recognition site, the mutations are sequentially distant, which implies that the gp41 disulfide loop is interacting with gp120 C5 in gp160. In addition, we have modeled the gp120–gp41 interaction in unprocessed precursor gp160 using structural data available for gp120 and gp41 domains in isolation, supplemented by mutagenesis data. We suggest that the mutations have altered the interaction between gp120 C5 and the gp41 disulfide loop, resulting in decreased accessibility of the furin recognition site and implying that the interaction between the gp120 C5 and gp41 loop is a conformational requirement for gp160 processing. The sensitivity of this interaction could be exploited in future antivirals designed to disrupt HIV pathogenesis by disrupting gp160 processing.
Keywords: AIDS, HIV, gp41, gp160, gp120, SIV
Infection with the human immunodeficiency virus (HIV), and the analogous simian immunodeficiency virus (SIV), begins with the virus particle attaching itself to the target cell surface followed by fusion of the viral membrane with the target cell membrane, thereby allowing entry of the viral genetic material (Chan and Kim 1998; Gallo et al. 2003). The attachment and fusion steps are mediated by the viral glycoproteins gp120 and gp41, respectively (Freed and Martin 1995), which are produced by the proteolytic cleavage of the precursor gp160 by the cellular protease furin, or related proteases, in the Golgi compartment (Moulard and Decroly 2000). Processed envelope proteins are then presented on the surface of the infected cell as non-covalently associated complexes and, on subsequent budding, form the envelope of the new virions. Previous mutagenesis studies have shown that the furin recognition site consists of four residues (sequence, REKR) situated at the C terminus of conserved domain 5 (C5) of gp120 (Hallenberger et al. 1992; Doms et al. 1993). High-resolution structural information is available for the gp120 core; however, conserved domain 5, which contains the furin recognition site, is missing (Kwong et al. 1998). More recently, the structure of the gp120 C5 domain has been determined in isolation by solution NMR spectroscopy (Guilhaudis et al. 2002). In addition, there are numerous structures of the gp41 extracellular domain (Caffrey et al. 1997, 1998; Chan et al. 1997; Tan et al. 1997; Weissenhorn et al. 1997; Malashkevich et al. 1998; Yang et al. 1999; Caffrey 2001). In all cases, gp41 is a symmetric trimer composed of N and C helices that associate to form a “six helix bundle.” In the case of the NMR structure, gp41 contains the disulfide loop, which is conserved among retroviridae and connects the N and C helices (Caffrey et al. 1997). Based on molecular modeling and studies of synthetic peptides, Schulz et al. (1992) first suggested that the gp41 disulfide loop was the site of interaction with gp120. Subsequently, mutagenesis studies have clearly established that the gp41 disulfide loop is the site of non-covalent interactions with gp120 that are critical to HIV entry (Cao et al. 1993; Maerz et al. 2001; Binley et al. 2000; York and Nunberg 2004; Jacobs et al. 2005). During fusion, gp41 is thought to undergo a structural change from a “prefusogenic” state, in which the N and C helices are not associated, to a post-fusogenic state, in which the N and C helices are associated (i.e., the six helix bundle state). However, the gp41 loop, which connects the N and C helices, contains a conserved disulfide bond that may be expected to restrict conformational changes within the loop (Caffrey 2001; Caffrey et al. 1998). Indeed, at present there is no evidence for a structural change in the gp41 disulfide loop between the unprocessed gp160 and the gp41 prefusogenic and post-fusogenic states. In the present work, we have examined the role of the HIV gp41 disulfide loop in envelope processing and function by mutagenesis. In the first mutant, the disulfide bond of the loop was removed by simultaneous substitution of the cysteines by alanines (gp41-CC/AA; C598 and C604 replaced by alanines). We have previously shown that this mutation does not disrupt the structure of the post-fusogenic state of SIV gp41 (Caffrey et al. 1997). In the second mutant, the disulfide loop was largely removed by deletion of residues 596–606, which include the conserved cysteines (gp41-Δ; WGCSGKLICTT deleted).
Results and Discussion
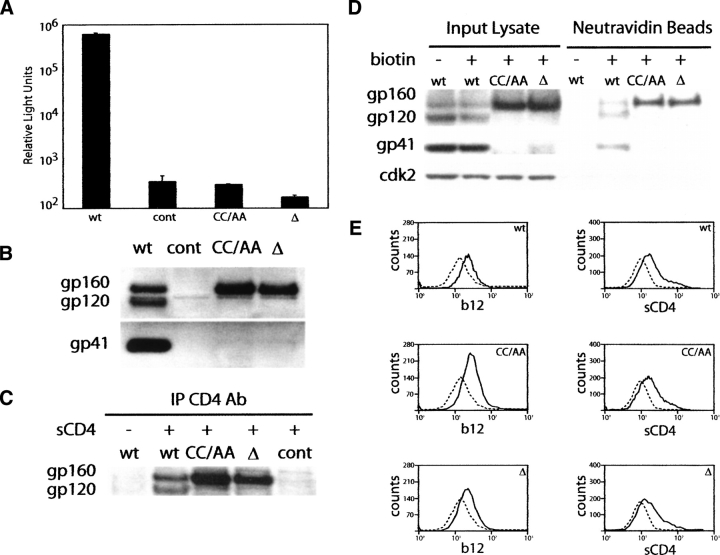
The functionality of gp41-CC/AA and gp41-Δ was first examined in a luciferase reporter assay, in which viral entry is proportional to the observed luciferase activity (Connor et al. 1995). As shown in Figure 1A, virus bearing gp41-CC/AA or gp41-Δ has negligible viral entry compared with that of the wild-type virus. Indeed, both mutants exhibited entry levels that were similar to virus lacking envelope, suggesting that they are not functional to support viral entry.
Figure 1.
(A) Luciferase reporter assay for viral entry of 293T cells transfected with plasmids expressing wild-type gp41 (WT), gp41-CC/AA (CCAA), or gp41-Δ (Δ). The control (cont) corresponds to mock transfection. The bar graph is presented on a log scale. Error bars, SD of three separate experiments. (B) Western blot analysis of envelope expression, processing, and gp120 association for HIV wild-type and mutants gp41-CC/AA and gp41-Δ in cell lysates. The control lane corresponds to mock transfection. (C) Immunoprecipitation assay using sCD4. (D) Western blot analysis of envelope surface exposure as assayed by biotinylation. (E) Immunofluorescence flow cytometry assay of b12 antibody and sCD4 binding to wild-type and mutant envelope. The dotted lines correspond to mock-transfected cells treated in the same manner as the cells expressing envelope.
The above observations lead to the question of the underlying factors involved in the effects of these mutations on gp41/envelope protein function, which could include reduced expression, stability, or processing of gp160; decreased strength of the non-covalent interaction between gp41 and gp120, thereby resulting in dissociation or “shedding” of gp120 into the media, and the formation of a gp41–gp120 complex that cannot support the attachment or fusion steps of viral entry. To distinguish which of the above scenarios is responsible for nonfunctionality of gp41-CC/AA and gp41-Δ, we probed for envelope proteins gp160, gp120, and gp41 present in 293T producer cell lysates by Western blotting. As shown in Figure 1B, the cells expressing wild-type gp41 exhibit the expected bands for gp160, gp120, and gp41, indicating that wild-type gp160 is well-expressed, stable, and processed to form gp120 and gp41 and that a stable gp41–gp120 complex has been formed. Interestingly, in contrast to wild type, gp120 and gp41 are clearly absent from the cells lysates of the mutants (Fig. 1B), suggesting that the mutations have disrupted envelope processing by furin. We note that the purified virus of the mutants exhibit small levels of gp160 and undetectable gp120 and gp41, consistent with disruption of furin processing.
It is next of interest to consider whether the mutations have resulted in large conformational changes. The conformational properties of the mutants were first probed by an immunoprecipitation assay using sCD4. Binding to sCD4 is indicative of the structural integrity of the gp120 envelope protein moiety (Schenten et al. 1999). In this assay, equivalent amounts of wild-type and mutant cell lysates were treated with sCD4 followed by incubation with the OKT4 antibody, which binds to CD4 (Reinherz et al. 1979). The envelope–sCD4–OKT4 complexes were immunoprecipitated with Protein G–Agarose beads and subjected to Western blotting analysis. As shown in Figure 1C, the expected bands are observed for gp160 and gp120 in the wild type and only gp160 in the mutant cell lysates, suggesting that the mutations have not disrupted the ability of the envelope to bind the CD4 receptor.
Cell surface expression was next tested by a surface biotinylation assay. In this assay, 293T cells expressing a wild-type or mutant envelope are exposed to a nonpermeable biotinylating reagent, and the resulting biotinylated proteins are isolated by affinity chromatography using neutravidin beads. The bound proteins are subsequently eluted and visualized by Western blot analysis. As shown in Figure 1D, in the wild-type gp160, gp120 and gp41 are biotinylated and thus taken to be exposed on the cell surface. In the case of gp41-CC/AA and gp41-Δ, the gp120 and gp41 bands are absent as expected, but gp160 is biotinylated and thus exposed on the cell surface. The presence of the cytosolic protein cdk2 in only the input fractions confirms a cell-surface–specific biotinylation reaction. We note that the surface exposure of unprocessed wild-type gp160 is in agreement with previous studies (Moulard et al. 1999). Moreover, surface expression of gp160 also suggests that the precursor protein is in a folded conformation that has passed the ER quality control. Thus, lack of processing in the mutants is not the result of a gross protein misfolding event.
We further probed the surface expression of the mutants using immunofluorescence flow cytometry assays employing the b12 antibody, a neutralizing antibody that binds to discontinuous epitopes within the CD4 binding site (Roben et al. 1994), and sCD4. In this assay, 293T cells expressing a wild-type or mutant envelope were treated or mock-treated with either IgG b12 antibody or sCD4. In the case of the sCD4 experiment, the cells were subsequently incubated with the anti-CD4 antibody OKT4. After incubation with anti-mouse IgG-FITC, the cells were analyzed by flow cytometry. FITC acquisition indicates binding of either b12 antibody or sCD4 to the envelope protein (gp120 and/or gp160) expressed on the cell surface. Figure 1E shows cell surface binding of the b12 antibody and sCD4, as indicated by the shift in mean fluorescence intensity compared with mock-transfected cells exposed to the fluorescent reporter. Clearly, gp41-CC/AA and gp41-Δ show a similar profile to the wild type, indicating that the mutant gp160 proteins are present on the cell surface and capable of binding b12 antibody and sCD4.
Finally, it is of interest to consider the underlying reasons for disruption of gp41-CC/AA and gp41-Δ processing. Interestingly, the gp41 disulfide loop, which encompasses the mutations, occurs ∼90 residues from the furin recognition site located in gp120 C5. The lack of furin processing suggests that in the wild-type gp160, the disulfide loop is interacting with the furin recognition site (or alternatively that the effects are propagated). A previous mutagenesis study has demonstrated an interaction between these two domains in processed envelope (Binley et al. 2000). The present data, as well as previous studies of similar mutations (Syu et al. 1991; Dedera et al. 1992), clearly suggest an interaction in the unprocessed state (i.e., gp160). Moreover, in a previous mutagenesis study of the gp41 disulfide loop, our group observed that two single-site mutants, L593A and T606A, which are proximal to the mutated sites of gp41-CC/AA and gp41-Δ, were also not processed (Jacobs et al. 2005). As discussed in the introduction, the gp41 disulfide loop has a well-established role in stabilizing the interaction between gp120 and gp41. The present work suggests that this interaction occurs in unprocessed gp160. Therefore, the gp41 disulfide loop clearly plays an important role in the orientation of the furin recognition site for efficient processing of gp160.
Two pertinent questions arise, accordingly. First, does the gp41 disulfide loop form part of the furin recognition site? According to this scenario, disruption of the gp41–gp120 interaction in the mutants would disrupt furin action. However, furin recognizes linear peptides containing the REKR sequence (c.f. Basak and Lotfipour 2005), suggesting that furin does not have a complex active site and that the gp41 loop would not provide part of the recognition site. Moreover, a large number of gp41 loop mutations have been shown to disrupt the gp41–gp120 interaction and lead to shedding of gp120, but do not affect gp160 processing (Cao et al. 1993; Merat et al. 1999; Maerz et al. 2001; York and Nunberg 2004; Jacobs et al. 2005). Consequently, a more likely scenario is that the mutants have altered the gp41–gp120 interaction to the point that the furin recognition site is no longer accessible to the enzyme. In this scenario, perturbation of the gp120–gp41 interaction results in a local conformational change and occlusion/burial of the processing site.
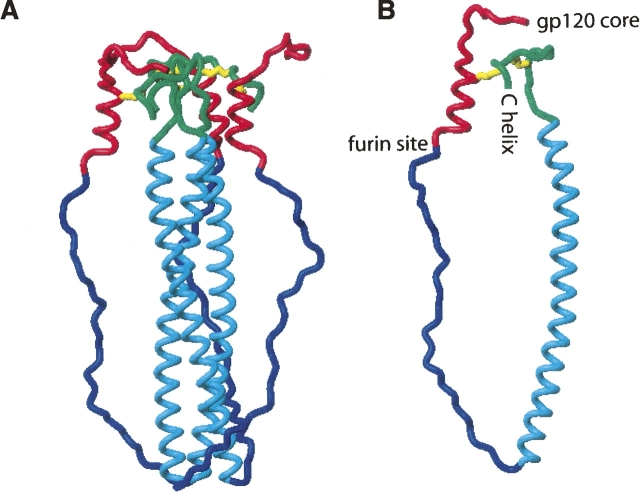
Given the structural knowledge of the gp120 C5 (Guilhaudis et al. 2002) and the gp41 disulfide loop (Caffrey 2001; Caffrey et al. 1998), can a direct interaction between the C5 and gp41 loop possibly exist in the unprocessed gp160? Accordingly, we have modeled this interaction in Figure 2 using the following assumptions: (1) The gp120 C5 and gp41 extracellular domains within gp160 are structurally similar to those previously characterized in isolation; (2) the N-terminal region of gp41, which consists of the fusion peptide and is missing from all available gp41 structures, is in an extended and flexible form; and (3) the gp41 disulfide loop is also in a flexible form. A torsion angle dynamics protocol was employed, which is similar to that previously described (Caffrey 2001). The model consists of residues 489–608 (HXB2 numbering; c.f. Binley et al. 2000), which comprise the gp120 C5 and gp41 fusion peptide, N-helix, and disulfide loop. Disulfide bonds between residue 501 of gp120 and residue 605 of gp41 (the nonnative disulfide bond characterized in Binley et al. 2000) and residues 598 and 604 of gp41 (the native disulfide) were maintained during the calculation. In Figure 2A, a representative low energy model of the envelope trimer is shown. In Figure 2B, the relative locations of the gp120 core, the furin recognition site, and the gp41 C-helix and transmembrane domain are noted. The model suggests that an interaction between gp120 C5 and the gp41 disulfide loop is indeed possible in unprocessed gp160. In this model, the hydrophobic side chains of the fusion peptide could be accommodated by hydrophobic side chains of the N-helix and/or gp120 domains not shown in the model.
Figure 2.
Model of the gp41–gp120 interaction of the trimer (A) and monomer (B). The gp120 conserved domain 5 is colored red, gp41 fusion peptide is colored blue, gp41 N-helix is colored cyan, gp41 disulfide loop is colored green, and the disulfide bonds are colored yellow.
In summary, the present observations demonstrate that the interaction between gp120 C5 and the conserved disulfide loop of gp41 plays a critical role in gp160 processing. The location of mutants with respect to the furin recognition site indicates that the two sites are interacting in unprocessed gp160. As suggested by previous mutational studies of gp41 (Cao et al. 1993; Merat et al. 1999; Maerz et al. 2001; York and Nunberg 2004; Jacobs et al. 2005), the gp120–gp41 interaction is apparently very sensitive to mutations in both unprocessed gp160 and processed gp120–gp41, and thus, the interaction must be relatively weak in nature. Taken together, the critical interactions of the gp41 disulfide loop suggest that it may be a novel site for therapeutic intervention.
Materials and Methods
Luciferase reporter assay for viral entry
Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene) with subsequent verification by DNA sequencing. Plasmids pHXB2 (Page et al. 1990) bearing wild-type or mutant gp41 and pNL4–3.Luc.R-E- (Connor et al. 1995) were cotransfected by calcium phosphate precipitation into 293T cells, which were maintained in Dulbecco's medium with 10% fetal bovine serum, 1 mM L-glutamine, 1% penicillin-streptomycin, and 0.5 mg/mL G418. Forty-eight hours post-transfection, the medium was harvested and filtered through a 0.45-μm filter to make the virus stock. U87.CD4.CXCR4 cells (Bjorndal et al. 1997), which were maintained in Dulbecco's medium with 15% fetal bovine serum supplemented with 1 μg/mL puromycin, 300 μg/mL G418, 1% L-glutamine, and 1% penicillin-streptomycin, were seeded to 1 × 105 cells/well of a 12-well cell culture plate in a volume of 1 mL. The following day, 400 μL of the virus stock was added to each of the wells of the U87 cells after removal of the medium. The plates were incubated overnight at 37°C in a CO2 incubator. After ∼16 h, the virus was aspirated and replaced with U87 medium, and the cells were allowed to grow for another 24 h. The luciferase activity was measured using the Luciferase Assay System from Promega and a Berthold FB12 luminometer running Sirius software.
Western blot analysis
Producer cell lysates were collected from 293T producer cells using lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 0.1% SDS). Cell lysates were normalized for total protein concentration using the Bradford Protein Assay Kit (Bio-Rad). After electrophoresis, transfer, washing (Tris-buffered saline plus 0.1% Tween-20 [TBST]), and blocking (TBST plus 5% dry milk), the blots were probed with either goat anti-HIV-1 gp120 polyclonal antibody (United States Biological) or with mouse anti-HIV-1 gp41 monoclonal antibody (Chessie 8, NIH AIDS Research and Reference Reagent Program). The secondary antibody used was peroxidase-conjugated affinipure donkey anti-goat IgG (H+L) (Jackson ImmunoResearch Labs, Inc.) or peroxidase-conjugated goat anti-mouse IgG (H+L) (Jackson ImmunoResearch Labs, Inc.). The blots were visualized by incubation with ECL detection reagents (Amersham Biosciences), exposed to Kodak X-Omat AR film, and developed using a Konika SRX-101A developer.
Immunoprecipitation assay
293T cells were incubated with sCD4 (5 μg/mL, Immunodiagnostics, Inc.) for 30 min on ice. Proteins bound to sCD4 were immunoprecipitated by treatment with OKT4 (1 ug/mL, eBiosciences) followed by Protein G beads (Invitrogen) and visualized by Western blotting.
Surface biotinylation
293T cells were incubated with Sulpho-NHS-SS-Biotin (Pierce) on ice. The reaction was quenched by washing the cells three times with CM-PBS/100 mM glycine. Cells were lysed in RIPA buffer, clarified by centrifugation, and immunoprecipitated overnight with Neutravidin beads. Samples were washed with RIPA buffer and bound proteins eluted by incubation with SDS-PAGE loading buffer containing 50 mM DTT for 1 h at 37°C. Biotinylated proteins were visualized by Western blotting.
Immunofluorescence flow cytometry
293T cells were treated with either IgG b12 antibody (2 μg/mL, NIH AIDS Research and Reference Reagent Program) or sCD4 (4 ug/mL) on ice and washed twice. In the case of the b12 antibody, the cells were exposed to anti-human IgG-FITC and analyzed by flow cytometry (DakoCytomation CyAn ADP). In the case of the sCD4 experiment, the cells were incubated with anti-CD4 antibody OKT4 on ice, washed, fixed with 1% paraformaldehyde on ice, washed again, stained with anti-mouse IgG-FITC (against OKT4), and analyzed by flow cytometry.
Molecular modeling
The model was determined by using a torsion angle dynamics protocol (c.f. Caffrey 2001). The experimental restraints for the gp41 moiety were taken from the HIV gp41 N-helix and disulfide loop Caffrey (2001), which is based on the NMR structure of the SIV gp41 ectdomain in the free state (Caffrey et al. 1998), and the experimental restraints of the HIV gp120 conserved domain 5 were taken from Guilhaudis et al. (2002).
Acknowledgment
This work was supported by NIH grant RO1 AI47674 to M.C.
Footnotes
Reprint requests to: Michael Caffrey, Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, IL 60607, USA; e-mail: caffrey@uic.edu; fax: (312) 413-0353.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072771407.
References
- Basak A. and Lotfipour, F. 2005. Modulating furin activity with designed mini-PDX peptides: Synthesis and in vitro kinetic evaluation. FEBS Lett. 579: 4813–4821. [DOI] [PubMed] [Google Scholar]
- Binley J., Sanders, R., Clas, B., Schuelke, N., Master, A., Guo, Y., Kajumo, F., Anselma, J., Maddon, P., Olson, W., et al. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74: 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndal A., Deng, H., Jansson, M., Fiore, J., Colognesi, C., Karlsson, A., Albert, J., Scarlatti, G., Littman, D., and Fenyo, E. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71: 7478–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. 2001. Model for the structure of the HIV gp41 ectodomain: Insight into the intermolecular interactions of the gp41 loop. Biochim. Biophys. Acta 1536: 116–122. [DOI] [PubMed] [Google Scholar]
- Caffrey M., Cai, M., Kaufman, J., Stahl, S., Wingfield, P., Gronenborn, A., and Clore, G. 1997. Determination of the secondary structure and global topology of the 44 kDa ectodomain of gp41 of the simian immunodeficiency virus by multidimensional nuclear magnetic resonance spectroscopy. J. Mol. Biol. 271: 819–826. [DOI] [PubMed] [Google Scholar]
- Caffrey M., Cai, M., Kaufman, J., Stahl, S., Wingfield, P., Covell, D., Gronenborn, A., and Clore, G. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17: 4572–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Bergeron, L., Helseth, E., Thali, M., Repke, H., and Sodroski, J. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67: 2747–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. and Kim, P. 1998. HIV entry and its inhibition. Cell 93: 681–684. [DOI] [PubMed] [Google Scholar]
- Chan D., Fass, D., Berger, J., and Kim, P. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89: 263–273. [DOI] [PubMed] [Google Scholar]
- Connor R., Chen, B., Choe, S., and Landau, N. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206: 935–944. [DOI] [PubMed] [Google Scholar]
- Dedera D., Gu, R., and Ratner, L. 1992. Conserved cysteine residues in the human immunodeficiency virus type 1 transmembrane envelope protein are essential for precursor envelope cleavage. J. Virol. 66: 1207–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R., Lamb, R., Rose, J., and Helenius, A. 1993. Folding and assembly of viral membrane proteins. Virology 193: 545–562. [DOI] [PubMed] [Google Scholar]
- Freed E. and Martin, M. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270: 23833–23836. [DOI] [PubMed] [Google Scholar]
- Gallo S., Finnegan, C., Viard, M., Raviv, Y., Dimitrov, A., Rawat, S., Puri, A., Durrell, S., and Blumenthal, R. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614: 36–50. [DOI] [PubMed] [Google Scholar]
- Guilhaudis L., Jacobs, A., and Caffrey, M. 2002. Solution structure of the HIV gp120 C5 domain. Eur. J. Biochem. 269: 4860–4867. [DOI] [PubMed] [Google Scholar]
- Hallenberger S., Bosch, V., Angliker, H., Shaw, E., Klenk, H.-D., and Garten, W. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360: 358–361. [DOI] [PubMed] [Google Scholar]
- Jacobs A., Sen, J., Rong, L., and Caffrey, M. 2005. Alanine scanning mutagenesis of the HIV gp41 loop. J. Biol. Chem. 280: 27284–27288. [DOI] [PubMed] [Google Scholar]
- Kwong P., Wyatt, R., Robinson, T., Sweet, R., Sodroski, J., and Hendrickson, W. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393: 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz A., Drummer, H., Wilson, K., and Poumbourios, P. 2001. Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J. Virol. 75: 6635–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich V., Chan, D., Chutkowski, C., and Kim, P. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: Conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. 95: 9134–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merat R., Raoul, H., Leste-Lasserre, T., Sonigo, P., and Pancino, G. 1999. Variable constraints on the principal immunodominant domain of the transmembrane glycoprotein of human immunodeficiency virus type 1. J. Virol. 73: 5698–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M. and Decroly, E. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469: 121–132. [DOI] [PubMed] [Google Scholar]
- Moulard M., Hallenberger, S., Garten, W., and Klenk, H. 1999. Processing and routage of HIV glycoproteins by furin to the cell surface. Virus Res. 60: 55–65. [DOI] [PubMed] [Google Scholar]
- Page K., Landau, N., and Littman, D. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64: 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinherz E., Kung, P., Goldstein, G., and Schlossman, S. 1979. Separation of functional subsets of human T cells by a monoclonal antibody. Proc. Natl. Acad. Sci. 76: 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roben P., Moore, J., Thali, M., Sodroski, J., Barbas, C., and Burton, D. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68: 4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D., Marcon, L., Karlsson, G., Parolin, C., Kodama, T., Gerard, N., and Sodroski, J. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73: 5373–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T., Jameson, B., Lopalco, L., Siccardi, A., Weiss, R., and Moore, J. 1992. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins? AIDS Res. Hum. Retroviruses 8: 1571–1580. [DOI] [PubMed] [Google Scholar]
- Syu W., Lee, W.-R., Du, B., Yu, Q.-C., Essex, M., and Lee, T.-H. 1991. Role of conserved gp41 cysteine residues in the processing of human immunodeficiency virus envelope precursor and viral infectivity. J. Virol. 65: 6349–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Liu, J.-H., Wang, J.-H., Shen, S., and Lu, M. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. 94: 12203–12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen, A., Harrison, S., Skehel, J., and Wiley, D. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387: 426–430. [DOI] [PubMed] [Google Scholar]
- Yang Z., Mueser, T., Kaufman, J., Stahl, S., Wingfield, P., and Hyde, C. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126: 131–144. [DOI] [PubMed] [Google Scholar]
- York J. and Nunberg, J. 2004. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J. Virol. 78: 4921–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]