Abstract
Prp8 is a critical pre-mRNA splicing factor. Prp8 is proposed to help form and stabilize the spliceosome catalytic core and to be an important regulator of spliceosome activation. Mutations in human Prp8 (hPrp8) cause a severe form of the genetic disorder retinitis pigmentosa, RP13. Understanding the molecular mechanism of Prp8’s function in pre-mRNA splicing and RP13 has been hindered by its large size (over 2000 amino acids) and remarkably low-sequence similarity with other proteins. Here we present the crystal structure of the C-terminal domain (the last 273 residues) of Caenorhabditis elegans Prp8 (cPrp8). The core of the C-terminal domain is an α/β structure that forms the MPN (Mpr1, Pad1 N-terminal) fold but without Zn2+ coordination. We propose that the C-terminal domain is a protein interaction domain instead of a Zn2+-dependent metalloenzyme as proposed for some MPN proteins. Mapping of RP13 mutants on the Prp8 structure suggests that these residues constitute a binding surface between Prp8 and other partner(s), and the disruption of this interaction provides a plausible molecular mechanism for RP13.
Keywords: Prp8, retinitis pigmentosa, MPN domain, crystal structure
Pre-mRNA splicing is carried out by the spliceosome, a huge RNA/protein complex made of five snRNAs and over 100 protein factors. Prp8 is a key factor for pre-mRNA splicing. Prp8 was proposed to help form/stabilize the catalytic core and to be an important regulator in spliceosome activation (Collins and Guthrie 2000; Grainger and Beggs 2005). Prp8 is the only spliceosome protein that extensively cross-links with the 5′ splice site, the 3′ splice site, the branch point sequence, and the U5 and U6 snRNAs, all of which are considered components of the catalytic core (Grainger and Beggs 2005 and references therein). In addition, Prp8 interacts with a number of protein partners required for spliceosome activation (Grainger and Beggs 2005 and references therein; Brenner and Guthrie 2006; Liu et al. 2006; Small et al. 2006). Many Prp8 mutations suppress or enhance splicing defects caused by splice-site mutations and other mutations affecting spliceosome activation or the second step of splicing (Grainger and Beggs 2005 and references therein). Several mutations in the C terminus of Prp8 are associated with RP13, a severe form of human genetic disorder retinitis pigmentosa, which leads to progressive photoreceptor degeneration in the retina and eventual blindness (McKie et al. 2001; van Lith-Verhoeven et al. 2002; Kondo et al. 2003; Martinez-Gimeno et al. 2003).
Understanding the molecular mechanism of Prp8’s function in pre-mRNA splicing and RP13 has been difficult due to its large size and low-sequence similarity with other proteins. Prp8 is one of the largest (over 2000 amino acids in length) proteins in the nucleus. Prp8 has remarkably low-sequence similarity with other proteins, making it difficult to deduce function from sequence analyses. However, Prp8 is highly conserved across species, highlighting its essential structural/functional role for pre-mRNA splicing. To better understand the structure and function of Prp8, we determined the crystal structure of the last 273 residues of Caenorhabditis elegans Prp8 (residues 2057–2329) to 2.3 Å resolution. We designate this fragment cC273 and the corresponding fragments in human and yeast hC273 and yC273. The high-sequence identity between C. elegans and human and yeast Prp8 (86% and 59%, respectively) makes cC273 a good homology model for the corresponding human and yeast Prp8s (yPrp8).
Results and Discussion
Overall structure
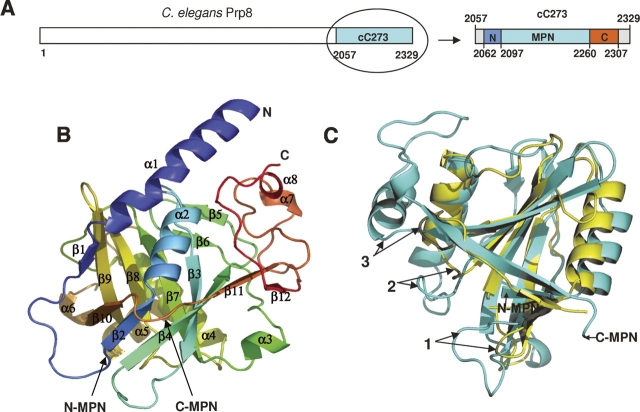
cC273 forms a compact single-domain structure that can be further characterized as an MPN domain core (174 residues) tightly wrapped by a small N-terminal (35 residues) and C-terminal (47 residues) extension (Fig. 1A,B). The extreme N-terminal five residues and C-terminal 22 residues of cC273 are disordered and cannot be seen in the crystal structure. The MPN core contains a β-barrel flanked by helices on opposite sides (Fig. 1C). The overall fold is similar to the MPN domain structure of Af2198 (a hypothetical protein from Archaeoglobolus fulgidus) (Tran et al. 2003; Ambroggio et al. 2004), although cC273 has much longer insertions between secondary structures compared with Af2198 (Figs. 1C, 2). The dominant feature of the N-terminal extension is a long helix, and the C-terminal extension contains a significant portion of loops. Residues in these loops interact extensively with the rest of the protein and every loop in the C-terminal extension has a well-defined conformation, with similar B factors and electron density quality as the rest of the protein.
Figure 1.
Structure of cC273. (A) Schematic view of the cC273 construct and its MPN core as well as the N- and C-terminal extensions in the context of the full-length C. elegans Prp8 (not drawn to proportion). (B) Overall structure of cC273 with secondary structures labeled. The structure is colored from blue to red in a rainbow spectrum from the N terminus to the C terminus. (C) Superimposition of the MPN domains of cC273 (blue) and Af2198 (yellow). The MPN domain is rotated 90° compared with B to best show the comparison. Labels 1–3 are examples of insertions that are much longer in cC273 than Af2198.
Figure 2.
Multiple sequence alignment among cC273, hC273, yC273, and Af2198. The three Prp8 sequences are aligned using MultAlin (Corpet 1988). Af2198 is aligned with cC273 based on structural superimposition. Secondary structures as they are observed in the crystal structures of cC273 and Af2198 are labeled on top of the cC273 sequence and the bottom of the Af2198 sequence, respectively. 310 helices are not differentiated from α helices and are both labeled as helices with prefix α for simplicity. Gray represents residues that are disordered in the crystal structure. Gray bars highlight residues in the JAMM motif. Boxed and underlined residues are missense and frameshift RP13 mutants, respectively.
MPN domains are present in proteins of several multisubunit complexes (Aravind and Ponting 1998; Glickman et al. 1998; Hofmann and Bucher 1998) and a subset of MPN domains were proposed to be Zn2+-dependent isopeptidases, which remove ubiquitin or ubiquitin-like molecules from target proteins (Maytal-Kivity et al. 2002; Verma et al. 2002; Yao and Cohen 2002). Bioinformatics analyses predicted the presence of the MPN domain, one of the only two small putative domains predicted for Prp8 (Maytal-Kivity et al. 2002; Grainger and Beggs 2005). However, the MPN domain in cC273 is 43 residues longer at the C terminus than the domain boundary predicted by bioinformatics analyses (Maytal-Kivity et al. 2002; Grainger and Beggs 2005). Expression of the MPN domain predicted by bioinformatics analyses resulted in completely insoluble proteins (data not shown) (Bellare et al. 2006), likely due to improper folding resulting from the incorrect C-terminal domain boundary.
We found that the MPN domain, even with the correct domain boundaries, does not exist stably on its own: It is an integral component of the larger cC273 structure, even though previous literature has widely regarded the MPN domain as an independent domain (Aravind and Ponting 1998; Glickman et al. 1998; Hofmann and Bucher 1998; Maytal-Kivity et al. 2002). When we removed the N-terminal or C-terminal extension or both, the truncated cC273 expressed at similar levels as the entire cC273, yet the majority of the protein was insoluble (Supplemental Fig. 1). We suggest that the entire cC273 is a single domain structure whose central MPN core is a “subdomain” instead of an independent “domain.”
C273 is likely a protein-interaction domain
There has long been speculation whether the presence of the MPN domain in Prp8 represents a unique connection between Prp8 and de-ubiquitination activity as proposed for some MPN proteins (Maytal-Kivity et al. 2002; Tran et al. 2003). This subset of the MPN proteins contains the JAMM motif (E-x[n]-H-x-H-x[7]-S-x[2]-D, where x denotes any amino acid). These proteins were thought to coordinate a Zn2+ ion (through the conserved His and Asp residues) and function as Zn2+-dependent isopeptidases to remove ubiquitin or ubiquitin-like molecules from target proteins (Maytal-Kivity et al. 2002; Verma et al. 2002; Yao and Cohen 2002). The MPN domain of Prp8 contains a partial JAMM motif, where the first Glu and one of the His in the canonical JAMM motif are changed to Gln (Fig. 2). The change from His to Gln does not necessarily exclude Zn2+ binding, since Gln can still coordinate Zn2+ (Lesburg et al. 1997). Using anomalous data collected at the wavelength of zinc's absorption edge, we demonstrated that cC273 does not bind Zn2+ even when soaked with 1mM ZnCl2 (data not shown). There is no water or other cations occupying the equivalent Zn2+ position. Consistent with this observation, mutation of all four non-Ala residues in the JAMM motif to Ala in yPrp8 did not produce any phenotype at a permissive temperature, suggesting that the JAMM motif is not important for Prp8’s function (Bellare et al. 2006). Taken together, Prp8 does not bind Zn2+ and is unlikely a Zn2+-dependent isopeptidase like some MPN proteins.
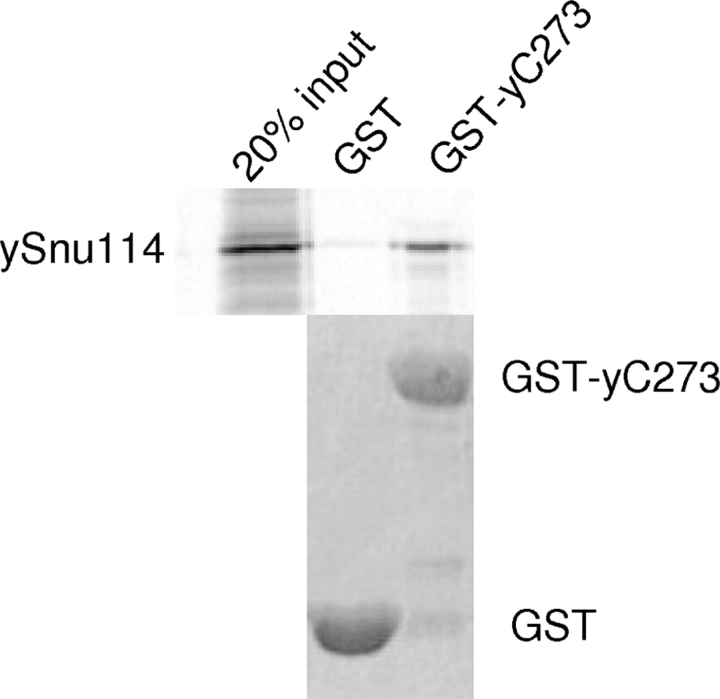
Instead, available evidence suggests that C273 is a domain specialized for protein–protein interaction in Prp8. yC273 was found to bind ubiquitin, even though it does not have deubiquitination activity (Bellare et al. 2006). The last 94 residues in hPrp8 (which mostly fall in the C-terminal extension of C273) were reported to bind a multifunctional protein PAI-2 (Fan et al. 2004). Yeast two-hybrid analyses showed that the C-terminal region of yPrp8 interacts with Brr2 (van Nues and Beggs 2001) and hPrp8 (1986–2335) interacts with Brr2, Snu114, and the N-terminal domain of Prp8 (Liu et al. 2006). To examine whether C273 directly interacts with some of the protein factors listed in the above yeast two-hybrid analyses, we performed GST-pull down assay using yeast yC273. We demonstrated that GST-yC273 specifically pulled down in vitro-translated yeast Snu114 (ySnu114) (Fig. 3), confirming the direct physical interactions between C273 and Snu114. Previous data also demonstrated the interaction between the N-terminal region of Prp8 and many other proteins (Boon et al. 2005; Grainger and Beggs 2005 and references therein; Liu et al. 2006). Taken together, the large Prp8 may segregate its functions into different regions of the protein: Regions in the middle of Prp8 contact RNA (Reyes et al. 1996; Turner et al. 2006), whereas the N- and C-terminal regions are likely the protein interaction domains that associate with other spliceosome proteins. Prp8 may use these interactions to regulate the assembly or activation of the spliceosome (Kuhn et al. 1999, 2002; Kuhn and Brow 2000; Bellare et al. 2006; Small et al. 2006).
Figure 3.
GST-yC273 can pull down in vitro-translated ySnu114 (residues 618–1008). GST alone and GST-yC273 were expressed in E. coli, purified with glutathione resin, incubated with in vitro-translated ySnu114 (35S-labeled), washed, and separated by SDS-PAGE. The polyacrylamide gel was stained with Coomassie Blue to show equal loading of GST and GST-yC273 (bottom). The gel was dried, and bound ySnu114 was visualized by autoradiography (top).
Retinitis pigmentosa mutants likely disrupt protein interactions
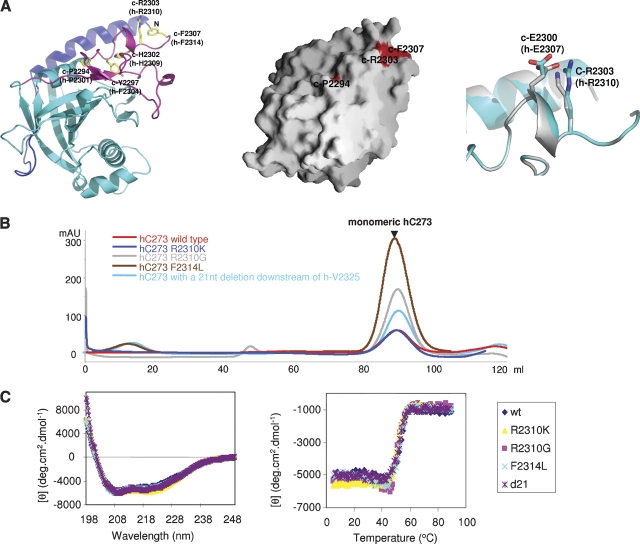
C273 contains all known RP13 mutants. To gain insight into the RP13 disorder, we mapped equivalent RP13 mutants onto the cC273 structure (the equivalent C. elegans residues are designated by prefix c-, and their corresponding human residues are in parentheses). The structure shows that all 12 RP13 mutations are located in the C-terminal extension (Figs. 2, 4A). Of these, the eight residues located at and downstream from c-R2303 (h-R2310) are particularly revealing for understanding the molecular mechanism of RP13. c-R2303 and c-F2307 (h-R2310 and h-F2314) reside near the end of the C-terminal extension and face the solvent (Fig. 4A). The remaining five frameshift and one missense RP13 mutations are downstream from c-F2307 (h-R2314) and fall within the terminal peptide that is disordered in the cC273 structure. We reason that because these residues either face the solvent or reside on disordered terminal peptides, mutations at these sites are unlikely to affect the overall structure of cC273.
Figure 4.
RP13 mutations. (A, left) Residues corresponding to the human RP13 mutations (ball and stick models) are located on the C-terminal extension of the cC273 structure (ribbon diagram). Dark blue, cyan, and purple backbones represent the N-terminal extension, MPN core, and C-terminal extension, respectively. (Middle) RP13 mutants on a surface representation (Nicholls et al. 1991) of the cC273 structure. Note that c-P2294 is partially exposed to the protein surface and c-Y2297 and c-H2302 are completely buried and invisible from the protein surface. (Right) Structural comparison of the cC273 R2303K mutant (silver backbone and silver residues) and wild-type cC273 (blue backbone and blue residues). (B) S200 gel filtration profiles and (C) CD spectra of four RP13 mutants indicate that these mutations do not affect the overall folding of the protein. All proteins were expressed and purified under identical conditions.
To test this hypothesis, we generated hC273 carrying four RP13 mutants at or downstream from h-R2310 (h-R2310K, h-R2310G, h-F2314L, and a 21-nt deletion immediately downstream from h-V2325). All mutants were expressed in Escherichia coli at similar levels as the wild type and led to production of well-behaved soluble proteins. Gel-filtration chromatography showed that all four mutants eluted identically to the wild type as monomers (Fig. 4B). Additionally, circular dichroism (CD) spectroscopy indicated that all four proteins had essentially identical secondary structure content and melting temperatures (Fig. 4C).
We further determined the crystal structure of cC273 R2303K mutant (equivalent to h-R2310K) to 2.1 Å resolution. The cC273 R2303K mutant had identical solution behavior as the wild type and crystallized under similar conditions. The crystal structure of this mutant is identical to the wild type, with the only exception being that c-R2303 was replaced with Lys and the side chain of c-E2300 moves slightly (OE2 moves 2.5Å) to form a hydrogen bond with c-K2303 in the mutant structure (Fig. 4A). Since the c-R2303K mutant causes zero disturbance to the overall structure of cC273 and both c-E2300 and c-R2303 are completely exposed to solvent, the most logical explanation for the biological effect of this mutation is that it disrupts the interaction(s) between Prp8 and other partner(s).
The above data support the hypothesis that the extreme C-terminal peptide of Prp8 (c2303-c2329, corresponding to h2310-h2335) constitutes a direct binding interface between Prp8 and other partner(s). Mutations on this surface, therefore, could lead to the disruption of the critical protein–protein interactions necessary for function, providing a possible molecular mechanism that underlies the RP13 disorder. Although the peptide downstream from c-F2307 (h-F2314) is disordered in our crystal structure, it may take on a defined conformation upon interaction with its partner. The three residues in this region, where missense mutations occur (c-R2303, c-F2307, and c-F2328, corresponding to h-R2310, h-F2314, and h-Y2334), are likely key residues on this interface that interact with Prp8’s binding partner. Since the side-chain conformation of c-E2300 (h-E2307) is altered in the c-R2303K mutant, c-E2300 (h-E2307) may contribute to this interaction as well.
Inspection of the remaining RP13 mutants on the four residues upstream of c-R2303 (h-R2310) on the cC273 structure further supports the role of the lost protein/protein interaction in the RP13 disorder. Three of these residues (c-P2294, c-Y2297, and c-H2302, equivalent to h-P2301, h-F2304, and h-H2309) are located immediately upstream of c-R2303 (h-R2310) pointing inward toward the protein and interacting with other residues on the protein (Fig. 4A). Mutations of these residues are very likely to disrupt these interactions, consequently altering the conformation of the downstream C-terminal peptide. These mutations may also destabilize the protein to a certain extent, since the hydrophobic protein surface that is normally covered by these residues will now be exposed. The frameshift mutation located at c-L2291 (h-L2298) will have a similar effect as the three missense mutations, altering the conformation of the downstream C-terminal peptide and indirectly disrupting the interaction between Prp8 and other partners.
RP13 mutants possibly disrupt the binding between Prp8 and one of its spliceosome partners (such as Brr2, Snu114, or the N-terminal region of Prp8) or ubiquitinated splicing factor(s). A retina-specific splicing or alternative splicing product may be highly sensitive to these disruptions. Alternatively, the RP13 mutations may disrupt the interaction between Prp8 and a yet to be defined partner, which reflects an unknown function of Prp8 specific to the retina.
Implications for the structure and function of other MPN proteins
A subset of the MPN proteins containing the JAMM motif was proposed to be metalloenzymes, but the general function of many “plain” MPN proteins remains unknown. cC273 is the first MPN protein structure determined outside of the archaea MPN protein realm, providing a unique opportunity to evaluate the structure and function of eukaryotic MPN proteins in general. Contrary to the popular notion that the MPN domain is an independent structure and function unit, we demonstrated that the MPN domain in Prp8 is an integral component of the larger single-domain structure of cC273. Tran et al. (2003) also noted that with the exception of archaea Af2198, none of the typical MPN domains can be expressed as soluble proteins. Multiple sequence alignment of cC273 and six representative MPN proteins show that a small N-terminal extension and a much longer C-terminal extension are always present on each side of the MPN domain (Supplemental Fig. 2). An attractive possibility is that, like cC273, the MPN domain in other proteins is also an integral component of the structure and function of the entire protein and should be more accurately regarded as “subdomain.” The MPN domain, in general, may serve as a common scaffold that accommodates different N- or C-terminal extensions to create unique protein-interaction surfaces (together with the MPN domain core) in different MPN proteins.
Supporting the above idea, Rpn8 and Rpn11 (two MPN proteins in the proteasome lid complex) are involved in extensive interactions with other subunits (Fu et al. 2001; Sharon et al. 2006) and the MPN domain and residues in the C-terminal extension are both required for these interactions (Fu et al. 2001). Although the JAMM-containing MPN proteins were thought to act as metalloenzymes, none of these proteins have been demonstrated to have enzymatic activity on their own (Maytal-Kivity et al. 2002; Verma et al. 2002; Yao and Cohen 2002; Tran et al. 2003; Ambroggio et al. 2004). It is worth considering whether these MPN proteins also bind another protein(s) that together fulfill substrate binding and/or enzymatic activity. The possible function of the MPN domain as a common scaffold for creating unique protein-binding surfaces in each MPN protein may also be the underlying reason why MPN domains appear in several multiprotein complexes with drastically diverse functions—from protein degradation (proteasome and signalosome) to splicing (spliceosome) to translation (eIF3).
Materials and Methods
Protein expression and purification
C. elegans Prp8 residues 2057–2329 (cC273) were subcloned into a pGEX-6P-1 vector (GE Healthcare) and expressed in E. coli strain XA90 as a GST-fusion protein. The fusion protein was first purified using glutathione sepharose resin and cleaved using PreScission protease (GE Healthcare). The resultant cC273 was further purified on a Superdex-200 (S200) gel-filtration column and concentrated to 12 mg/mL for crystallization trials.
Crystallization and data collection
cC273 was crystallized by the hanging-drop vapor diffusion method using a well solution containing 0.1 M Hepes (pH 7.0), 14% PEG4000, and 0.2 M MgCl2. The cC273 R2303K mutant was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and was expressed and purified using a protocol similar to that used for the native protein. cC273 R2303K was crystallized using a well solution containing 0.1 M Hepes (pH 7.0), 14% PEG4000, and 0.22 M MgCl2. All crystallographic data were collected using the mail-in data collection program at the National Synchrotron Light Source (NSLS). Data were processed using the HKL2000 package (Otwinowski and Minor 1997) and data statistics are shown in Supplemental Table 1.
Structural determination
The structure of cC273 was determined using the Se-Met SAD method and programs Solve and Resolve (Terwilliger and Berendzen 1999) with Se-Met positions identified by program HKL2MAP (Page and Schneider 2004). The initial electron density map was of excellent quality and most of the main chain and side chains were traced without any ambiguity. The structure of the R2303K mutant was solved with the Molecular Replacement method using the native cC273 as a model (with R2303 replaced with Ala). Refinement of both structures was performed using CNS (Brunger et al. 1998) and refinement statistics are shown in Supplemental Table 1. Coordinates of both structures have been deposited in the Protein Data Bank with ID code 2P87 (cC273) and 2P8R (R2303K).
CD spectroscopy
CD spectroscopy was performed on a Jasco-810 spectrometer using 0.2 mg/mL protein in 25 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Each spectrum was an average of eight wavelength scans collected at 0.2-nm intervals from 198 to 250 nm. Temperature-induced denaturation was monitored by CD at a wavelength of 222 nm using 0.6 mg/mL protein from 5°C to 90°C. CD data points were taken at 1°C intervals at a scan rate of 60°C/hour.
GST pull down
ySnu114 (residues 618–1008) was subcloned into a pTNT vector (Promega) and in vitro translated using 35S-Met (GE Healthcare) and the TnT Quick Coupled Transcription/Translation System (Promega) following the manufacturer's instructions. In vitro-translated ySnu114 was incubated with glutathione resin containing GST-yC273 in binding buffer (50 mM Tris-HCl at pH7.5, 120 mM NaCl, 2 mM EDTA, 0.2% NP40, 1 mg/mL BSA) at 4°C for 1 h. Resin was washed four times with binding buffer minus BSA, added with 2X SDS sample buffer and boiled for 5 min. The sample was analyzed on a 12% SDS–polyacrylamide gel, stained with Coomassie Blue, dried, and visualized by autoradiograph.
Note added in proof
While this manuscript was in preparation, the structure of the C-terminal domain of yPrp8 was published (scPrp8p2147–2397) (Pena et al. 2007). We and Pena et al. both reached similar conclusions independently. The two structures are highly similar (overall RMSD 1.5Å for Cα atoms) with differences concentrated on the loop regions (Supplemental Fig. 3). cC273 should be a better homologous model for hPrp8, considering hPrp8 is 74% identical to cPrp8 and 43% identical to yPrp8 in this region. The last nine residues in scPrp8p2147–2397 protrude from the compact portion of the structure and engage in crystal packing contacts. Most of the equivalent residues and the additional 16 C-terminal residues in our structure are disordered. The apparent flexibility of this C-terminal peptide supports the hypothesis that this peptide may become ordered and adopt different conformations upon interacting with other protein(s). We showed that yC273 pulled down in vitro-translated ySnu114, which complements the yeast two-hybrid analyses performed by Pena et al. (2007) with hPrp8. The similar conclusions independently arrived from structural studies of Prp8 from two species strongly support the general function of the C-terminal domain as a protein-interaction domain and the role of disrupted protein/protein interaction in RP13.
Acknowledgments
We thank the University of Colorado at Denver and Health Sciences Center's (UCDHSC) X-ray core facility, which is supported by the University of Colorado Cancer Center. Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and from the Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources of the National Institutes of Health (NIH). We thank Drs. X. Chen, E. Eisenmesser, D. Brow, D. Bentley, M. Churchill, J. Kieft, and G. Zhang for their helpful discussion or critical reading of the manuscript. ySnu114 clone was a generous gift of Dr. J. Staley, University of Chicago. This work is supported in part by an American Heart Association Scientist Development Grant and an American Cancer Society Research Scholar Grant to R.Z. R.Z. is also a V Scholar and a Kimmel Scholar.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Rui Zhao, M.S. 8101, P.O. Box 6511, Aurora, CO 80045, USA; e-mail: rui.zhao@uchsc.edu; fax: (303) 724-3215.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072872007.
References
- Ambroggio X.I., Rees, D.C., and Deshaies, R.J. 2004. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. and Ponting, C.P. 1998. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 7: 1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P., Kutach, A.K., Rines, A.K., Guthrie, C., and Sontheimer, E.J. 2006. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA 12: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K.L., Norman, C.M., Grainger, R.J., Newman, A.J., and Beggs, J.D. 2005. Prp8p dissection reveals domain structure and protein interaction sites. RNA 12: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner T.J. and Guthrie, C. 2006. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA 12: 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Collins C.A. and Guthrie, C. 2000. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 7: 850–854. [DOI] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhang, Y.Q., Li, P., Tong, C., Tan, L., and Zhu, Y.S. 2004. Interaction between plasminogen activator inhibitor type-2 and pre-mRNA processing factor 8. Acta Biochim. Biophys. Sin (Shanghai) 36: 623–628. [DOI] [PubMed] [Google Scholar]
- Fu H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. 2001. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20: 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623. [DOI] [PubMed] [Google Scholar]
- Grainger R.J. and Beggs, J.D. 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. and Bucher, P. 1998. The PCI domain: A common theme in three multiprotein complexes. Trends Biochem. Sci. 23: 204–205. [DOI] [PubMed] [Google Scholar]
- Kondo H., Tahira, T., Mizota, A., Adachi-Usami, E., Oshima, K., and Hayashi, K. 2003. Diagnosis of autosomal dominant retinitis pigmentosa by linkage-based exclusion screening with multiple locus-specific microsatellite markers. Invest. Ophthalmol. Vis. Sci. 44: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Kuhn A.N. and Brow, D.A. 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A.N., Li, Z., and Brow, D.A. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3: 65–75. [DOI] [PubMed] [Google Scholar]
- Kuhn A.N., Reichl, E.M., and Brow, D.A. 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 99: 9145–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburg C.A., Huang, C., Christianson, D.W., and Fierke, C.A. 1997. Histidine → carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity. Biochemistry 36: 15780–15791. [DOI] [PubMed] [Google Scholar]
- Liu S., Rauhut, R., Vornlocher, H.P., and Luhrmann, R. 2006. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 12: 1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gimeno M., Gamundi, M.J., Hernan, I., Maseras, M., Milla, E., Ayuso, C., Garcia-Sandoval, B., Beneyto, M., Vilela, C., Baiget, M., et al. 2003. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 44: 2171–2177. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis, N., Hofmann, K., and Glickman, M.H. 2002. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie A.B., McHale, J.C., Keen, T.J., Tarttelin, E.E., Goliath, R., van Lith-Verhoeven, J.J., Greenberg, J., Ramesar, R.S., Hoyng, C.B., Cremers, F.P., et al. 2001. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum. Mol. Genet. 10: 1555–1562. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology (eds. C.W. Carter and R.M. Sweet), pp. 307–326. Academic Press, New York. [DOI] [PubMed]
- Page T. and Schneider, T.R. 2004. HKL2MAP: A graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37: 843–844. [Google Scholar]
- Pena V., Liu, S., Bujnicki, J.M., Luhrmann, R., and Wahl, M.C. 2007. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell 25: 615–624. [DOI] [PubMed] [Google Scholar]
- Reyes J.L., Kois, P., Konforti, B.B., and Konarska, M.M. 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2: 213–225. [PMC free article] [PubMed] [Google Scholar]
- Sharon M., Taverner, T., Ambroggio, X.I., Deshaies, R.J., and Robinson, C.V. 2006. Structural organization of the 19S oroteasome lid: Insights from MS of intact complexes. PLoS Biol. 4: e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E.C., Leggett, S.R., Winans, A.A., and Staley, J.P. 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell 23: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.J., Allen, M.D., Lowe, J., and Bycroft, M. 2003. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42: 11460–11465. [DOI] [PubMed] [Google Scholar]
- Turner I.A., Norman, C.M., Churcher, M.J., and Newman, A.J. 2006. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA 12: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lith-Verhoeven J.J., van der Velde-Visser, S.D., Sohocki, M.M., Deutman, A.F., Brink, H.M., Cremers, F.P., and Hoyng, C.B. 2002. Clinical characterization, linkage analysis, and PRPC8 mutation analysis of a family with autosomal dominant retinitis pigmentosa type 13 (RP13). Ophthalmic Genet. 23: 1–12. [DOI] [PubMed] [Google Scholar]
- van Nues R.W. and Beggs, J.D. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae . Genetics 157: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind, L., Oania, R., McDonald, W.H., Yates 3rd, J.R., Koonin, E.V., and Deshaies, R.J. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615. [DOI] [PubMed] [Google Scholar]
- Yao T. and Cohen, R.E. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407. [DOI] [PubMed] [Google Scholar]