Abstract
Malonyl-CoA: acyl carrier protein transacylase (MCAT) is a critical enzyme responsible for the transfer of the malonyl moiety to holo-acyl carrier protein (ACP) forming the malonyl-ACP intermediates in the initiation step of type II fatty acid synthesis (FAS II) in bacteria. MCAT has been considered as an attractive drug target in the discovery of antibacterial agents. In this study, the crystal structure of MCAT from Helicobacter pylori (Hp) at 2.5 Å resolution is reported, and the interaction of HpMCAT with HpACP is extensively investigated by using computational docking, GST-pull-down, and surface plasmon resonance (SPR) technology-based assays. The crystal structure results reveal that HpMCAT has a compact folding composed of a large subdomain with a similar core as in α/β hydrolases, and a similar ferredoxin-like small subdomain as in acylphosphatases. The docking result suggests two positively charged areas near the entrance of the active site of HpMCAT as the ACP-binding region. Binding assay research shows that HpMCAT demonstrates a moderately binding ability against HpACP. The solved 3D structure of HpMCAT is expected to supply useful information for the structure-based discovery of novel inhibitors against MCAT, and the quantitative study of HpMCAT interaction with HpACP is hoped to give helpful hints in the understanding of the detailed catalytic mechanisms for HpMCAT.
Keywords: Helicobacter pylori, Manoyl-CoA: acyl carrier protein transacylase (MCAT), acyl carrier protein (ACP), fatty acid biosynthesis, crystal structure
The biosynthesis of fatty acid (FAS) is an essential process for the survival of the organism (Magnuson et al. 1993; White et al. 2005). In bacteria, the process of FAS is completed by a series of individual enzymes, while in animals, only a single enzyme with several distinct domains is involved in all the reactions. Such a major difference between the animal and bacterial systems therefore makes the enzymes involved in the FAS process potential drug targets for the discovery of antibacterial agents (Campbell and Cronan Jr. 2001; Miesel et al. 2003; White et al. 2005).
Malonyl-CoA: acyl carrier protein transacylase (fabD; MCAT, EC2.3.1.39) is responsible for the transfer of malonyl moiety to holo-ACP forming malonyl-ACP intermediates to participate in fatty acid biosynthesis (Ruch and Vagelos 1973). It is reported that MCAT might also be involved in polyketide biosynthesis, producing one of the largest classes of secondary metabolites, such as the tetracyclines and erythromycins (Summers et al. 1995; Keatinge-Clay et al. 2003). Therefore, MCAT is considered to be a vital enzyme in bacterial metabolic activity.
To date, MCAT enzymes from several different species have been cloned and characterized, such as EcMCAT (Escherichia coli) (Serre et al. 1995), ScMCAT (Streptomyces coelicolor) (Keatinge-Clay et al. 2003), PfMCAT (Plasmodium falciparum) (Prigge et al. 2003), and HpMCAT (Helicobacter pylori) (Liu et al. 2006). The solved crystal structures of EcMCAT (PDB code: 1MLA) (Serre et al. 1995) and ScMCAT (PDB code: 1NM2) (Keatinge-Clay et al. 2003) demonstrated that both of these MCAT enzymes share similar structures and are composed of two subdomains. One is made up of a short four-stranded parallel β-sheet and 12 helices, and the other contains a four-stranded antiparallel β-sheet and two helices. It is found that MCAT could complete the malonyl transfer using the ping-pong mechanism with the His-Ser catalytic domain, which is commonly discovered in the serine-dependent acylhydrolases (Keatinge-Clay et al. 2003). It is generally regarded that MCAT has two motifs related to its biological function. One is the catalytic active site located in the deep gorge between two subdomains, and the other is the ACP-binding site present on the MCAT surface (Serre et al. 1994). According to the macromolecular docking result, the ACP binding site of MCAT is adjacent to the GQGXQ turn that is also found to be a highly conserved motif among different species (Keatinge-Clay et al. 2003; Liu et al. 2006).
H. pylori is a severe clinically pathogenic bacterium that is a great threat to public health and is related to the causes of peptic ulcer and gastric cancer (Schilling et al. 2002; Liu et al. 2005, 2006). In our previous work, we reported the characterization of MCAT from H. pylori strain SS1 (HpMCAT), and one natural inhibitor was first discovered (Liu et al. 2006). In this study, we performed the crystal structure analysis and ACP-binding investigation of HpMCAT. It is expected that the analyzed crystal structure might supply useful information for the structure-based discovery of novel inhibitors against MCAT, and the HpMCAT/HpACP interaction study might give helpful hints in the understanding of the detailed catalytic mechanisms for HpMCAT.
Results
HpMCAT structural analysis
The crystal structure of HpMCAT was solved with the molecular replacement (MR) method using the structure of ScMCAT (PDB code: 1NM2) as the MR model (Keatinge-Clay et al. 2003). Maximum-likelihood refinement was carried out with the CCP4 program REFMAC 5.0 (Murshudov et al. 1997) against 2.5 Å level data. Electron density interpretation and model building were performed by using the computer graphics program coot (Emsley and Cowtan 2004). The coordinates and structure factor of HpMCAT have been deposited in the Protein Data Bank (PDB code: 2H1Y).
The full-length HpMCAT has 309 residues, which can all be observed in the structure, suggesting a compact folding of HpMCAT. Two His residues from the His-tag and two residues from the vector before the N terminus were found and numbered “−3,” “−2,” “−1,” and “0.”
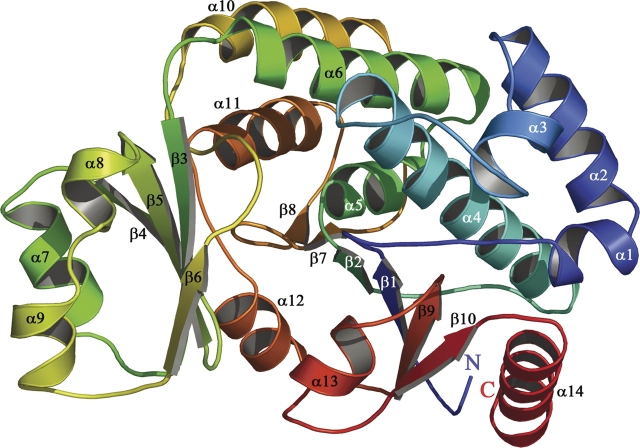
The current model contains two monomer molecules in the asymmetric unit. The HpMCAT monomer is composed of 14 α-helices (length ranging from three to 21 residues) and 10 β-sheets (length ranging from one to seven residues), and the total accessible surface is ∼13,569 Å2 as calculated with the program DSSP (Figs. 1 and 2A; Kabsch and Sander 1983). The structure could be divided into a large and a small subdomain. The large subdomain is made up of two noncontiguous segments (residues Met1–Asn127 and residues Val195–Val309), and its surface is ∼10,924 Å2. It contains a six-stranded (topology 10/9/1/2/7/8) β-sheet core, similar to the case in α/β hydrolases (Ollis et al. 1992), and an 11-α-helices cap, five of which form a helical flap (α1, α2, α3, α4, and α14) on the protein surface. The small subdomain (residues Lys128–Ser194) has a ferredoxin-like fold as observed in acylphosphatases (Keatinge-Clay et al. 2003). It consists of a four-antiparallel (topology 4/5/3/6) β-sheets core and a three-α-helices cap, with a surface of ∼4441 Å2.
Figure 1.
The overall structure of HpMCAT. The pictures are drawn by Pymol (DeLano 2004), and the secondary structures are assigned by the program DSSP (Kabsch and Sander 1983). The monomer of HpMCAT is colored blue to red from its N terminus to its C terminus; secondary structural elements are labeled.
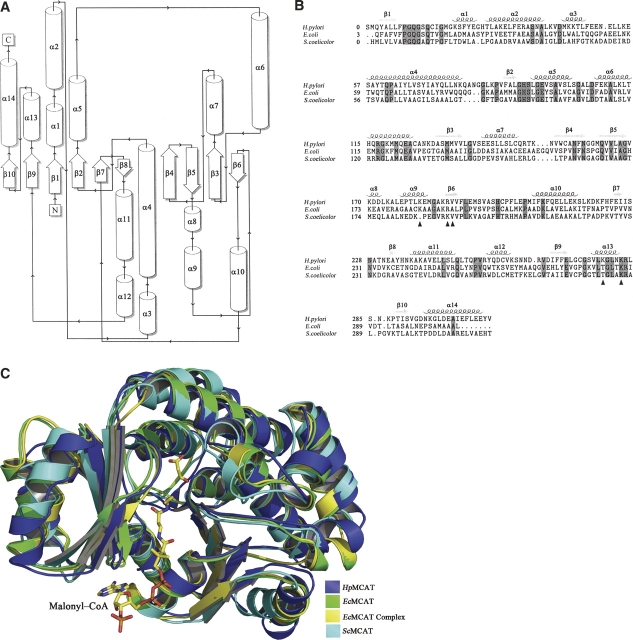
Figure 2.
(A) The topology diagram of HpMCAT. (Cylinders) Helices; (arrows) strands. The alignment was produced by the topology cartoon server from http://www.tops.leeds.ac.uk/. (B) The multiple structure-based sequence alignment of HpMCAT with EcMCAT and ScMCAT. Secondary structures of HpMCAT are labeled. Identical residues are shaded in cyan. (Blue triangles) These residues contribute to the interactions of MCAT with ACP or malonyl-CoA by H-bonds. The alignment was calculated by the CE algorithm (Shindyalov and Bourne 1998) and produced by the program STRAP (Gille et al. 2003) with slight modification. (C) The structure superposition diagram of HpMCAT, EcMCAT, ScMCAT, and EcMCAT complexed with malonyl-CoA. The structures of (blue) HpMCAT, (green) EcMCAT, (yellow) EcMCAT complex, and (cyan) ScMCAT. The malonyl-CoA (the malonyl moiety and the coenzyme A moiety) is labeled.
The amino acid sequence of HpMCAT has 32% identity with that of EcMCAT and 30% with that of ScMCAT (Liu et al. 2006). Through structural superposition (Fig. 2B,C), we found that these three enzymes share a similar overall structure. However, some differences could be observed: (1) In HpMCAT, residues Asn51–Leu53 form a loop between α3 and α4, while the corresponding residues in EcMCAT (Pro52–Glu55) and ScMCAT (Asp49–Glu52) form an α-helix. (2) α8 (residues Lys170–Asp172) and α9 (residues Glu177–Glu182) in HpMCAT correspond to a long helix in EcMCAT (residues Lys173–Lys184) and ScMCAT (residues Met174–Glu182). (3) β7 (residues Glu224–Ile225) in HpMCAT becomes a loop in EcMCAT (residues Pro227–Val228). (4) β8 (residue Ala233) in HpMCAT has no observable counterpart in either EcMCAT or ScMCAT, as is shown in Figure 2.
Structure-based HpMCAT catalysis assumption
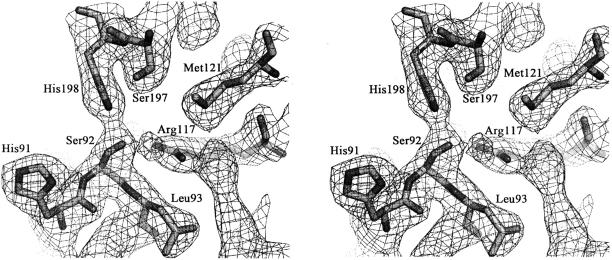
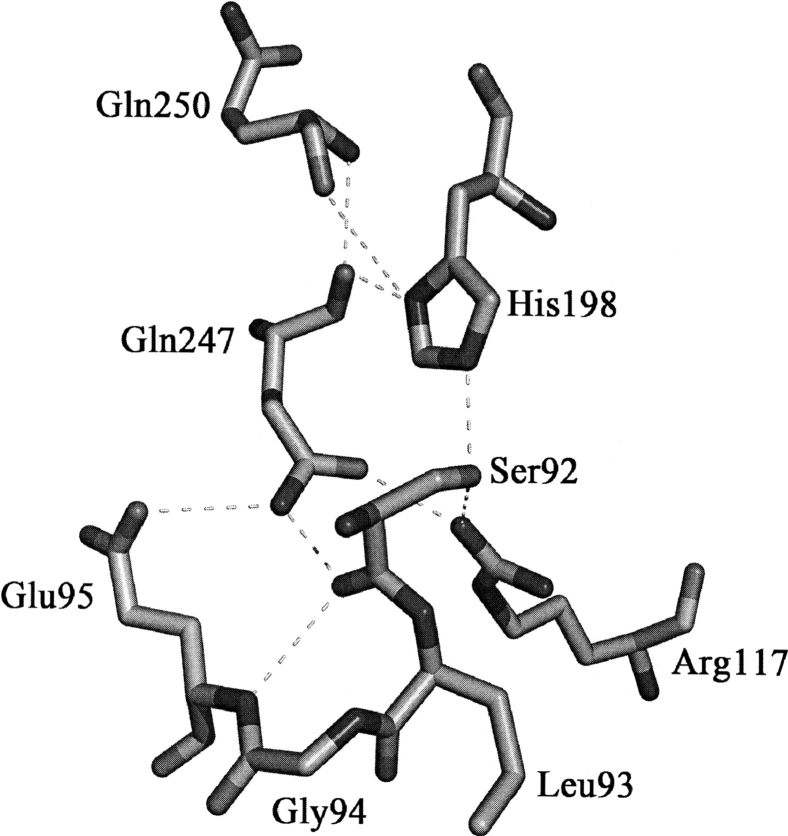
As previously reported, MCAT might adopt a ping-pong kinetic mechanism to transfer a malonyl from malonyl-CoA to ACP, and two key residues and a potential oxyanion hole mainly contribute to the catalysis reaction (Joshi and Wakil 1971; Keatinge-Clay et al. 2003). Based on the resolved HpMCAT structure (Figs. 1 and 3), residues Ser92 and His198 are in the deep gorge between the two subdomains. As a nucleophile, Ser92 lies in a highly conserved nucleophilic elbow Gly90-His91-Ser92-Leu93-Gly94 pentapeptide located in the sharp turn between β2 and α5, and its hydroxyl group interacts with Nɛ2 of His198 and Nη2 of Arg117 by forming H-bonds. His198, the proton acceptor, lies in the loop between β6 and α10, and its Nδ1 forms H-bonds with the backbone carbonyl oxygens of Gln247 and Gln250 (Fig. 4). As indicated in Figures 3 and 4, the plausible inactive oxyanion hole is formed by the backbone amide nitrogens of Gln10 and Leu93, while Gln10 is located in the other highly conserved peptide Pro8-Gly9-Gln10-Gly11-Ser12-Gln13 near the active-site Ser92.
Figure 3.
Stereoview of the final 2F o − F c electron density map contoured at 1.0 σ around the active site. Atoms are shown as sticks. (Bright orange) Carbon, (red) oxygen, (blue) nitrogen, and (orange) sulfur. The residues are labeled.
Figure 4.
H-bond network around the active site. Atoms are shown as sticks and H-bonds as yellow dashes. (Orange) Carbon, (red) oxygen, and (blue) nitrogen. The residues are labeled.
According to the published results, the overall catalytic reaction for MCAT is readily reversible and can be generally divided into two parts (Joshi and Wakil 1971). In the first part, the malonyl-CoA's binding to MCAT causes a subtle conformational change of the enzyme, and the oxyanion hole is activated. In the case of HpMCAT, it is assumed that in this part of the reaction, the malonyl-CoA binds to HpMCAT by H-bonds with residues Gln163, Arg187, and Arg283, and pushes residue Gln10 toward Leu93 (Oefner et al. 2006). Therefore, the distance between the amides of Gln10 and Leu93 becomes closer than the original 5.2 Å (5.5 Å in ScMCAT) (Keatinge-Clay et al. 2003), which causes the activation of the oxyanion hole. Subsequently, the thioester carbonyl of malonyl-CoA inserts into the hole and is held in the place by H-bond interaction with Gln10 and Arg117. At the same time, His198 extracts a proton from Ser92 and turns it into an active nucleophile. In the following, Ser92 exerts a nucleophilic attack on the thioester carbonyl and forms a tetrahedral intermediate. This intermediate is stabilized in the oxyanion hole through a charge–dipole interaction, which is an important feature to distinguish the catalytic mechanism of MCAT from those of other hydrolases (White et al. 2005). The presence of the active oxyanion hole may accelerate the nucleophilic attacks (White et al. 2005). After this nucleophilic attack, Arg117 moves toward α6; and the side chain of Ser92 flops ∼120° around the Cα–Cβ bond and points toward Asn157 (Oefner et al. 2006). In order to accommodate the malonyl group, His198 protonates CoA to release it from the protein, and a malonyl-MCAT complex is formed. Therefore, in the second part of HpMCAT's reaction, ACP binds to the surface of malonyl-MCAT, pushes Gln10 toward Leu93, and activates the oxyanion hole again. At this stage, the phosphopantetheinyl thiol of ACP enters the active site and donates a proton to His198. Consequently, this thiol of ACP becomes an active nucleophile and attacks the ester carbonyl of malonyl-Ser92 to form another intermediate with the malonyl. His198 then protonates the catalytic serine again and releases malonyl-ACP. After all these steps, the malonyl is successfully transferred from CoA to ACP.
Computational docking of HpMCAT binding to HpACP
In recent years, great attention has been paid to the interaction analysis between ACP and the individual enzymes in the FAS II system. However, it is hard to obtain the complex crystals (Zhang et al. 2001). In this study, the computational docking approach was carried out for investigating HpMCAT interaction with HpACP.
The optimal complex model was calculated as described in Materials and Methods. ZDOCK generated 54,000 conformations of the protein complex, the top 2000 of which were then refined and re-ranked by RDOCK. The top 298 poses ranked by RDOCK with energy scores <0 were clustered into 15 clusters according to the RMSD between any two ACP orientations. In the most reasonable cluster, the HpACP tends to bind to HpMCAT near the entrance to the active site. Poses in this cluster are energetically favorable and consistent with the putative binding site based on the surface electrostatic potential of the HpMCAT crystal structure.
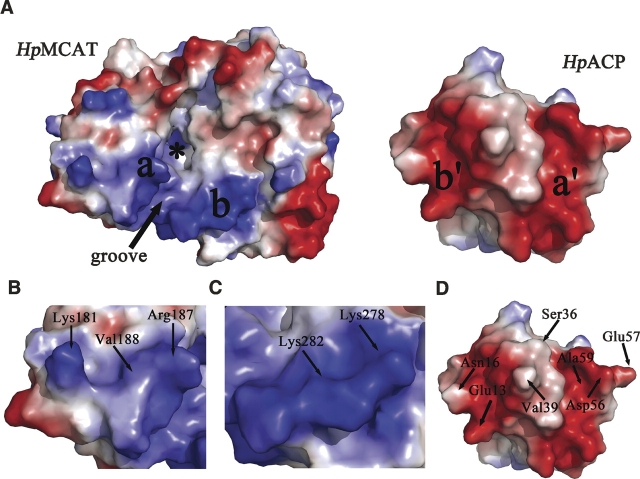
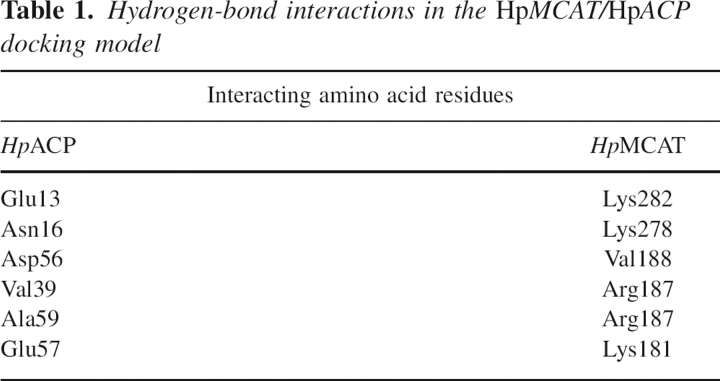
As indicated in the docking model of the HpMCAT/HpACP complex, the contacting surfaces are oppositely charged (Fig. 5). Two main positively charged areas on the surface of HpMCAT (here called “area a” and “area b”) are likely to bind to the two prominent negatively charged areas named a′ and b′ on HpACP. Area a is near the β6-strand and α9-helix of HpMCAT and interacts with a corresponding negatively charged area a′ on the surface of HpACP. Residues Lys181, Arg187, and Val188 from area a interact with Asp56, Glu57, and Ala59 from area a′ by H-bonds. Area b is the other positively charged area on the surface of HpMCAT near the α13-helix. Residues Lys278 and Lys282 from this area form H-bonds to residues Glu13 and Asn16 from area b′, which is the other negatively charged area of HpACP. Residues involved in H-bond interactions are shown in Table 1. Interestingly, such a contact through surface electrostatic interactions is a ubiquitous feature in the structures of type II fatty acid synthesis enzymes (Zhang et al. 2001, 2003a). For example, FabG and FabH both have predicted positively charged ACP-binding sites (Zhang et al. 2001, 2003b). The contact is also similar to that between ACP and the phosphopantetheinyl transferase ACPS (Zhang et al. 2003b), which is predominated by ionic interaction between positively charged residues on ACPS and negatively charged residues on ACP.
Figure 5.
HpACP docked to HpMCAT. (A) Electrostatic surfaces of HpMCAT and apo-HpACP. The extreme ranges of red (negative) and blue (positive) represent electrostatic potentials of −99 e/kT to +99 e/kT. Two positively charged areas on HpMCAT and two negatively charged areas on HpACP are labeled. If HpACP were rotated by 180° on top of HpMCAT, the same letter areas from each surface would match up. The active site of HpMCAT (*) and the critical groove are labeled. (B–D) Electrostatic surface of areas a, b, and HpACP. Residues contributing the H-bond interactions are labeled. The electrostatic surfaces are drawn by Pymol (DeLano 2004).
Table 1.
Hydrogen-bond interactions in the HpMCAT/HpACP docking model
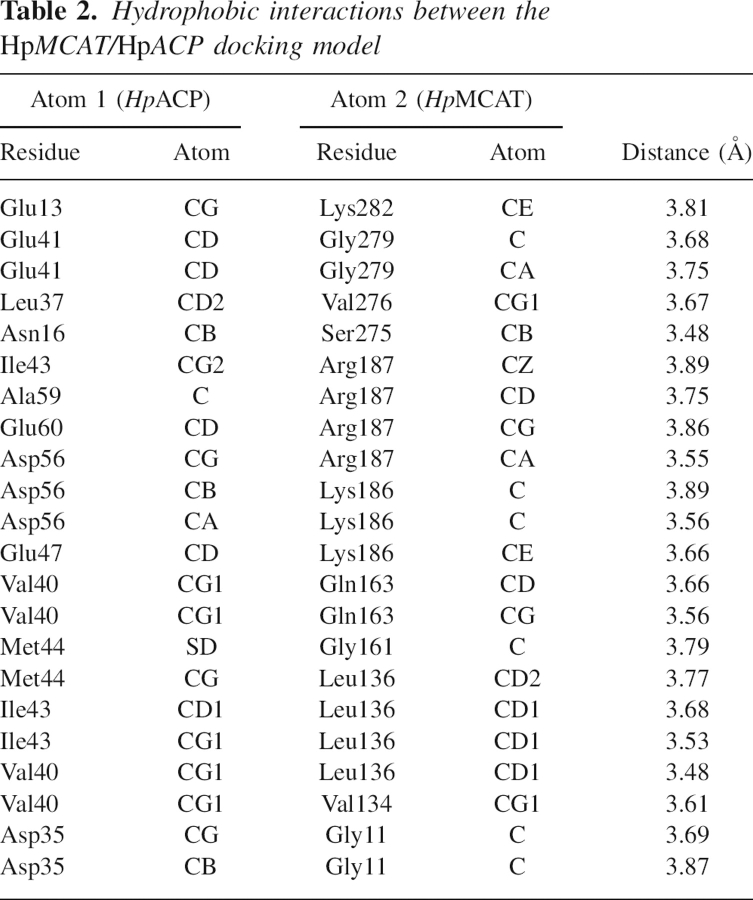
Apart from the electrostatic interactions between the two proteins, hydrophobic interactions were also significant in the HpMCAT/HpACP model. The highly conserved α2-helix of HpACP recognizes electropositive/hydrophobic areas adjacent to the active-site entrance of HpMCAT and stacks in the hydrophobic groove between the two positively charged areas on the surface of HpMCAT by hydrophobic interactions among numerous residues (Fig. 5). The residues involved in the hydrophobic interactions are shown in Table 2. Remarkably, as shown in Figure 2B, the interfacial residues of HpMCAT involved in the protein interaction are conserved. These characters are consistent with the hypothesis that the conserved α2-helix of ACP functions as a universal protein interaction domain (Zhang et al. 2003a,b) as verified in the FabH–ACP interaction analysis using computational docking, NMR, and site-directed mutation techniques (Zhang et al. 2001, 2003a,b). In addition, it is noticed that residue Ser36 on the end of HpACP's α2-helix, which is the attachment site for 4′-phosphopantetheine, is kept in the right place near the active-site entrance of HpMCAT and oriented appropriately for the insertion of the prosthetic group into the tunnel.
Table 2.
Hydrophobic interactions between the HpMCAT/HpACP docking model
In conclusion, in our HpACP and HpMCAT docking model, the ferredoxin-like subdomain of HpMCAT (β6-strand and α9-helix) contributes the main interactions, which was consistent with some published articles (Zhang et al. 2001; Lo Conte et al. 2002), but different from the docking results of ScMCAT and ScACP (Keatinge-Clay et al. 2003).
HpMCAT/HpACP interaction determined by GST-pull-down assay
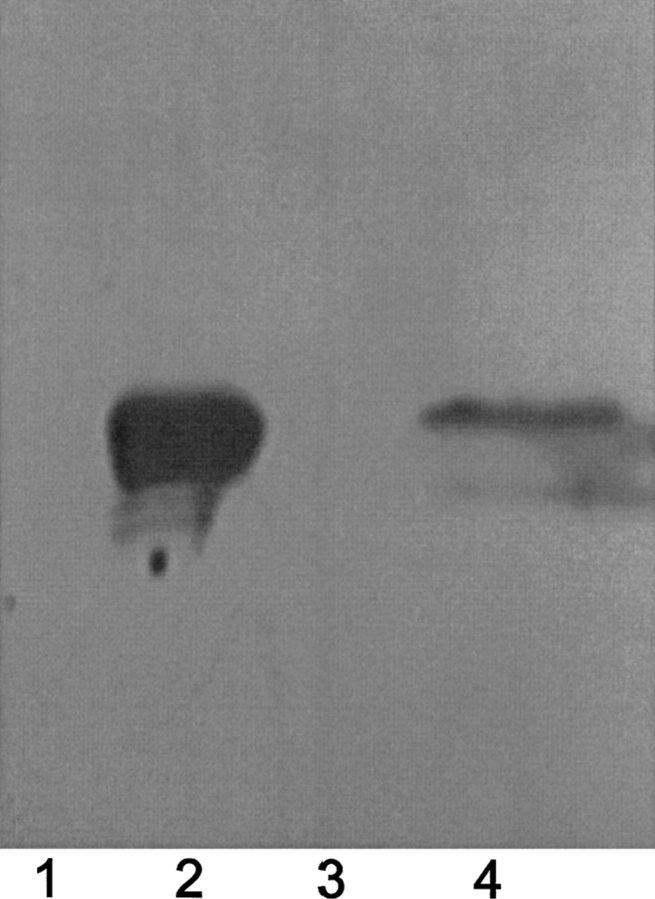
In the GST-pull-down assay, the purified GST-tagged HpACP combined with the glutathione-Sepharose beads as a bait protein according to the kit, while the His-tagged HpMCAT was obtained as the prey. The GST-HpACP and the possible partner were eluted by glutathione and analyzed by Western blot. As indicated in lane 4 of Figure 6, HpMCAT could be detected on the Western blot, suggesting that HpMCAT was eluted by glutathione together with protein GST-HpACP binding to the beads. These results thus indicated that GST-HpACP could specifically interact with HpMCAT in vitro.
Figure 6.
The direct interaction between HpMCAT and HpACP was determined by GST-pull-down. Samples were analyzed by Western blotting using anti-6×His antibody (Novagen). (Lane 1) The purified GST-tagged HpACP; (lane 2) the purified His-tagged HpMCAT; (lane 3) agarose gel control; (lane 4) GST-tagged HpACP and the pull-down HpMCAT.
SPR technology-based HpMCAT/HpACP interaction assay
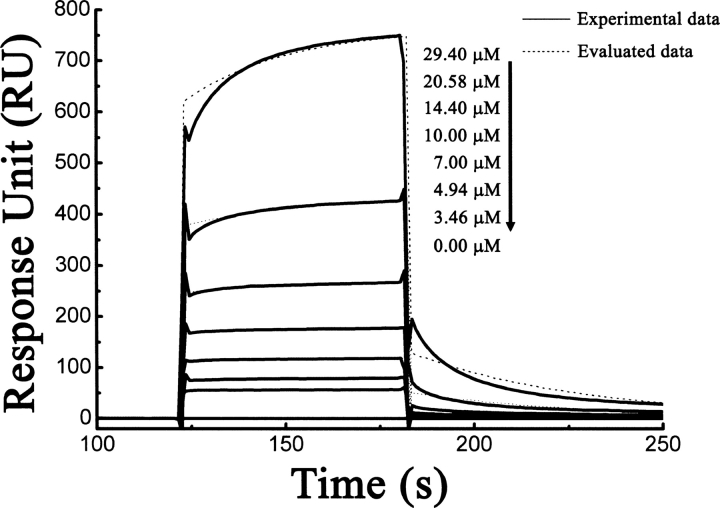
In order to further quantitatively characterize the binding of HpACP to HpMCAT in vitro, surface plasmon resonance (SPR) technology-based Biacore 3000 was applied. HpACP was immobilized on CM5 chip by 500RU, and a series of different concentrations of HpMCAT flowed through the reference and HpACP-immobilized cells in sequence. The kinetic parameters evaluating the HpACP/HpMCAT binding (k on and k off) were analyzed according to the 1:1 Langmuir binding model. As observed, the evaluated curves could overlay the experimental curves very well (Fig. 7). The SPR experimental data thus suggested that HpACP could bind to HpMCAT with the association rate constant of k on = 5.02 × 10−2 M−1 sec−1 and dissociation constant of k off = 2.17 × 10−2 sec−1, and the equilibrium dissociation constant K D (K D = k d/k a) was evaluated to be 4.31 × 10−5 M, which indicates that HpMCAT exhibited a moderately binding affinity against HpACP (Zhang et al. 2001).
Figure 7.
Sensorgrams of HpMCAT binding to the immobilized HpACP. The binding curves were fitted to a 1:1 Langmuir binding model. Superposition of fitting curves (…) to original curves (—).
Discussion
According to the superposition of HpMCAT, EcMCAT, and ScMCAT, the structure of HpMCAT is similar to the other two structures except for a few differences in secondary structure elements and the relatively more compact folding of MCAT (all residues could be observed in the crystal structure) (Fig. 2B,C). Most remarkable is an important G-H-S-L-G motif from HpMCAT. This pentapeptide belongs to the G-X-S-X-G sequence motif prevalent in α/β hydrolases (Keatinge-Clay et al. 2003), and is also observed in an extensively studied family of lipase from filamentous fungi as well as mammalian pancreatic lipases (Derewenda et al. 1994). The high conservation of this motif suggested that it might play an important role in the catalytic activity of the enzyme.
Recently, the crystal structure of EcMCAT complexed with malonyl-CoA was published, and several critical residues in the reaction were also found (Oefner et al. 2006). Comparing this structure with our docking model, it is interesting to find that the groove in HpMCAT that binds to the α2-helix of HpACP in our model corresponds to the same one as in EcMCAT that stacks the adenine of malonyl-CoA, and residues in HpMCAT that interact with HpACP by H-bonds are also contributed to the interaction with malonyl-CoA through structure-based alignment (residues are labeled by blue triangles in Fig. 2B). These “coincidences” suggest that malonyl-CoA and ACP might share similar binding areas on MCAT, and the 4′-phosphopantetheine arm of HpACP might stretch into the active site of MCAT in a similar way as malonyl-CoA. This conclusion was also supported by the docking results of ScACP and ScMCAT (Keatinge-Clay et al. 2003).
In terms of the quantitative HpMCAT interaction with HpACP using the SPR technique, a relative moderate binding affinity was detected (K D = 4.31 × 10−5 M), which has a large difference from the result for the interaction between HpFabZ (β-hydroxyacyl-acyl carrier protein dehydratase) and HpACP (K D = 1.2 × 10−8 M) (Liu et al. 2007) in the same conditions. This result therefore indicates that the mechanisms of the individual enzymes within the FASII pathway binding to ACP might exhibit subtle differences, which is also in accordance with the fact that a common ACP-binding motif is not found among the enzymes involved in the FASII pathway (Zhang et al. 2003b).
In conclusion, the crystal structure of HpMCAT was solved at 2.5 Å resolution. The highly similar structures among HpMCAT, EcMCAT, and ScMCAT prove that MCAT is a highly conserved enzyme, which makes it an excellent target for broad-spectrum antibacterial drug discovery. In addition, the interaction between HpMCAT and HpACP was simulated by the computational docking method and then investigated by using GST-pull-down and SPR techniques. Our present work is expected to supply useful information for the structure-based discovery of novel inhibitors against MCAT, and the quantitative study of the HpMCAT/HpACP interaction might give helpful hints in the understanding of the detailed catalytic mechanisms for HpMCAT.
Materials and Methods
Materials and strains
The H. pylori strain SS1 was obtained from our institute. The E. coli host strain M15 was purchased from QIAGEN. The E. coli host strain BL21 (DE3) was from Novagen. All other chemicals are of reagent grade or ultra-pure quality, and commercially available.
Expression and purification of the recombinant HpMCAT and HpACP
Expression and purification of His-tagged HpMCAT were performed as described previously (Liu et al. 2006). The homogeneity of HpMCAT was identified by SDS-PAGE, and the protein concentration was determined by measuring the absorbance at 280 nm with the extinction coefficient of 19,785 L/mol/cm.
The cloning, expression, and purification of His-tagged HpACP and GST-tagged HpACP were carried out according to the published method (Liu et al. 2007).
Crystallization and data collection
For crystallization, 1 μL of HpMCAT (∼35 mg/mL) in crystallization buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl) was mixed with an equal volume of reservoir solution containing 10% (v/v) PEG10000, 8% (v/v) MPD, and 0.1 M HEPES (pH 8.0). The mixture was equilibrated against 500 μL of the reservoir solution at 277 K by the hanging-drop vapor-diffusion method. Plate-like crystals of dimensions 0.7 × 0.5 × 0.1 mm3 grew after more than 13 mo, and one useable single crystal was then picked out from the cluster.
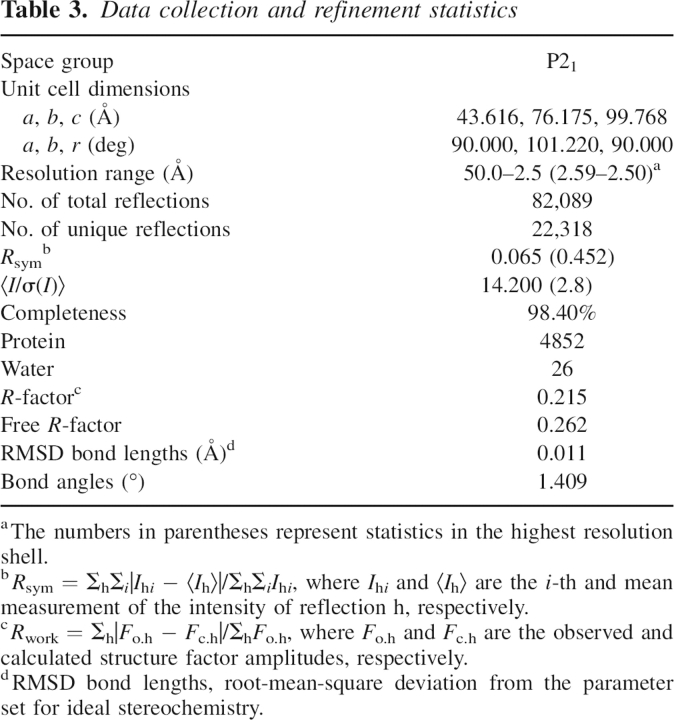
Diffraction data were collected in-house on a Rigaku rotating-anode X-ray generator operated at 100 kV and 100 mA (λ = 1.5418 Å). Diffraction images were recorded by a Rigaku R-AXIS IV++ imaging-plate detector with an oscillation step of 1°. The crystal was picked up with a nylon loop and flash-cooled in liquid nitrogen. Data collection was performed at 100 K using the original reservoir solution as cryoprotectant. The data were indexed, integrated, and scaled using the program suite HKL2000 (Otwinowski and Minor 1997). Analysis of the diffraction data indicated that the crystal belongs to space group P21. The related crystallographic statistics are summarized in Table 3.
Table 3.
Data collection and refinement statistics
Structural modeling of HpMCAT/HpACP complex
The 3D structure of HpACP was constructed by the homology modeling method based on the structure of EcACP (PDB code: 1T8K) as indicated in the Supplemental material. The complex model of HpMCAT/HpACP was constructed using ZDOCK 2.1 (Chen et al. 2003) and RDOCK (Li et al. 2003), which have proved to have an excellent predictive ability toward protein–protein complexes (Wiehe et al. 2005).
In the simulation, HpMCAT was kept fixed, whereas HpACP was allowed to rotate and translate around HpMCAT. Approximately 50,000 conformations were generated and ranked by pairwise shape complementarity (PSC) score when a complete sampling over the surface of HpMCAT was performed using ZDOCK. The rotational sampling interval was set at 6°, and all other default parameters were used. After that, the top-ranked poses were subjected to RDOCK to refine and re-rank with an energy minimization protocol. The top poses ranked by RDOCK were subjected to Gene Cluster3.0 (de Hoon et al. 2004) for k-means clustering. The top predictions from each cluster were then manually inspected and investigated by several criteria: RDOCK score, charge complementarity, hydrophobic interactions, and overall agreement with prior biological information. The interactions of the interface were plotted and analyzed using LIGPLOT (Wallace et al. 1995).
GST-pull-down assay
The HpMCAT/HpACP interaction was determined by GST-pull-down assay using the ProFound Pull-Down GST Protein: Protein Interaction Kit (Pierce) according to the published method (Luo et al. 2005). The purified GST-HpACP was used as the bait protein and loaded onto 60-μL glutathione-Sepharose beads, which was equilibrated with the pH 7.4 PBS buffer in the Handee Mini-spin column. The purified HpMCAT served as the prey protein to detect its interaction with the bait protein. The beads without the bait protein were used as a control to detect the nonspecific binding. In considering the fact that the molecular mass of GST-HpACP (∼35 kDa) is almost identical to that of HpMCAT (∼35 kDa), it is hard to discriminate between them in terms of the molecular weight by SDS-PAGE. Thereby the Western-blotting method was used to detect the existence of HpMCAT by using the anti-6×his tag antibody (Novagen).
Surface plasmon resonance (SPR) technology-based analysis
The binding of HpACP to HpMCAT was quantitatively assayed by using surface plasmon resonance (SPR) technology-based Biacore 3000 instrument (Biacore) (Rich and Myszka 2001; Luo et al. 2006). During the assay, the holo-HpACP was immobilized on the surface of a CM5 chip. The experiment was performed using pH 7.4 PBS buffer with 0.5% P20 at a speed of 20 μL/min, and the flow cells were regenerated by 1 mM NaOH after each injection. The reference cell was prepared similarly except that no HpACP was added. The association (k on) and dissociation rate (k off) constants as well as the equilibrium dissociation constant (K D) were achieved by fitting the data using 1:1 Langmuir binding model based on the BIAevaluation 3.1 software.
Electronic supplemental material
The Supplemental material contains three figures: Supplemental Figure A1, the homology model of HpACP; Supplemental Figure A2, the alignment sequence of HpACP with EcACP; and Supplemental Figure A3, Ramachandran plot of HpACP procheck. The homology modeling of HpACP can be found in the Supplemental material.
Acknowledgments
We are grateful to Eleanor Dodson and Kurt L. Krause for their generous help in the structure determinations. This work is supported by the State Key Program of Basic Research of China (grant 2004CB58905), the National Natural Science Foundation of China (grants 30525024, 20472095), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (grant 06JC14080), and a direction grant from CAS (grant KSCX2-YW-R-18).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Hualiang Jiang or Xu Shen, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China; e-mail: hljiang@mail.shcnc.ac.cn or xshen@mail.shcnc.ac.cn; fax: 86-21-50806918.
Abbreviations: FAS, fatty acid biosynthesis; ACP, acyl carrier protein; IPTG, isopropyl-β-D-thiogalasctopyranoside; SPR, surface plasmon resonance; FabZ, β-hydroxyacyl-ACP dehydratase; MCAT, manoyl-CoA: acyl carrier protein transacylase; FabG, β-ketoacyl-acyl carrier protein reductase; FabH, β-ketoacyl-acyl carrier protein synthase III; DSSP, definition of secondary structure of proteins given a set of 3D coordinates.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072757307.
References
- Campbell J.W. and Cronan Jr, J.E. 2001. Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu. Rev. Microbiol. 55: 305–332. [DOI] [PubMed] [Google Scholar]
- Chen R., Li, L., and Weng, Z. 2003. ZDOCK: An initial-stage protein-docking algorithm. Proteins 52: 80–87. [DOI] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto, S., Nolan, J., and Miyano, S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. 2004. Use of PyMOL as a communications tool for molecular science. Pap.-Am. Chem. Soc. 228: U313–U314. [Google Scholar]
- Derewenda U., Swenson, L., Green, R., Wei, Y., Dodson, G.G., Yamaguchi, S., Haas, M.J., and Derewenda, Z.S. 1994. An unusual buried polar cluster in a family of fungal lipases. Nat. Struct. Biol. 1: 36–47. [DOI] [PubMed] [Google Scholar]
- Emsley P. and Cowtan, K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Gille C., Lorenzen, S., Michalsky, E., and Frommel, C. 2003. KISS for STRAP: User extensions for a protein alignment editor. Bioinformatics 19: 2489–2491. [DOI] [PubMed] [Google Scholar]
- Joshi V.C. and Wakil, S.J. 1971. Studies on the mechanism of fatty acid synthesis. XXVI. Purification and properties of malonyl-coenzyme A–acyl carrier protein transacylase of Escherichia coli . Arch. Biochem. Biophys. 143: 493–505. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay A.T., Shelat, A.A., Savage, D.F., Tsai, S.C., Miercke, L.J., O'Connell III, J.D., Khosla, C., and Stroud, R.M. 2003. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure 11: 147–154. [DOI] [PubMed] [Google Scholar]
- Li L., Chen, R., and Weng, Z. 2003. RDOCK: Refinement of rigid-body protein docking predictions. Proteins 53: 693–707. [DOI] [PubMed] [Google Scholar]
- Liu W., Luo, C., Han, C., Peng, S., Yang, Y., Yue, J., Shen, X., and Jiang, H. 2005. A new β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Helicobacter pylori: Molecular cloning, enzymatic characterization, and structural modeling. Biochem. Biophys. Res. Commun. 333: 1078–1086. [DOI] [PubMed] [Google Scholar]
- Liu W., Han, C., Hu, L., Chen, K., Shen, X., and Jiang, H. 2006. Characterization and inhibitor discovery of one novel malonyl-CoA: acyl carrier protein transacylase (MCAT) from Helicobacter pylori . FEBS Lett. 580: 697–702. [DOI] [PubMed] [Google Scholar]
- Liu W., Du, L., Zhang, L., Chen, J., Shen, X., and Jiang, H. 2007. Helicobacter pylori acyl carrier protein: Expression, purification, and its interaction with β-hydroxyacyl-ACP dehydratase. Protein Expr. Purif. 52: 74–81. [DOI] [PubMed] [Google Scholar]
- Lo Conte L., Brenner, S.E., Hubbard, T.J., Chothia, C., and Murzin, A.G. 2002. SCOP database in 2002: Refinements accommodate structural genomics. Nucleic Acids Res. 30: 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Chen, Q., Chen, J., Chen, K., Shen, X., and Jiang, H. 2005. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 579: 2623–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Wu, D., Shen, C., Chen, K., Shen, X., and Jiang, H. 2006. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem. Cell Biol. 38: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson K., Jackowski, S., Rock, C.O., and Cronan Jr, J.E. 1993. Regulation of fatty acid biosynthesis in Escherichia coli . Microbiol. Rev. 57: 522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel L., Greene, J., and Black, T.A. 2003. Genetic strategies for antibacterial drug discovery. Nat. Rev. Genet. 4: 442–456. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Oefner C., Schulz, H., D'Arcy, A., and Dale, G.E. 2006. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 62: 613–618. [DOI] [PubMed] [Google Scholar]
- Ollis D.L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S.M., Harel, M., Remington, S.J., Silman, I., Schrag, J., et al. 1992. The α/β hydrolase fold. Protein Eng. 5: 197–211. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Prigge S.T., He, X., Gerena, L., Waters, N.C., and Reynolds, K.A. 2003. The initiating steps of a type II fatty acid synthase in Plasmodium falciparum are catalyzed by pfACP, pfMCAT, and pfKASIII. Biochemistry 42: 1160–1169. [DOI] [PubMed] [Google Scholar]
- Rich R.L. and Myszka, D.G. 2001. BIACORE J: A new platform for routine biomolecular interaction analysis. J. Mol. Recognit. 14: 223–228. [DOI] [PubMed] [Google Scholar]
- Ruch F.E. and Vagelos, P.R. 1973. The isolation and general properties of Escherichia coli malonyl coenzyme A-acyl carrier protein transacylase. J. Biol. Chem. 248: 8086–8094. [PubMed] [Google Scholar]
- Schilling C.H., Covert, M.W., Famili, I., Church, G.M., Edwards, J.S., and Palsson, B.O. 2002. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol. 184: 4582–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre L., Swenson, L., Green, R., Wei, Y., Verwoert, I.I., Verbree, E.C., Stuitje, A.R., and Derewenda, Z.S. 1994. Crystallization of the malonyl coenzyme A-acyl carrier protein transacylase from Escherichia coli . J. Mol. Biol. 242: 99–102. [DOI] [PubMed] [Google Scholar]
- Serre L., Verbree, E.C., Dauter, Z., Stuitje, A.R., and Derewenda, Z.S. 1995. The Escherichia coli malonyl-CoA: acyl carrier protein transacylase at 1.5-Å resolution. Crystal structure of a fatty acid synthase component. J. Biol. Chem. 270: 12961–12964. [DOI] [PubMed] [Google Scholar]
- Shindyalov I.N. and Bourne, P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11: 739–747. [DOI] [PubMed] [Google Scholar]
- Summers R.G., Ali, A., Shen, B., Wessel, W.A., and Hutchinson, C.R. 1995. Malonyl-coenzyme A: acyl carrier protein acyltransferase of Streptomyces glaucescens: A possible link between fatty acid and polyketide biosynthesis. Biochemistry 34: 9389–9402. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski, R.A., and Thornton, J.M. 1995. LIGPLOT: A program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 8: 127–134. [DOI] [PubMed] [Google Scholar]
- White S.W., Zheng, J., Zhang, Y.M., and Rock, C.O. 2005. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74: 791–831. [DOI] [PubMed] [Google Scholar]
- Wiehe K., Pierce, B., Mintseris, J ., Tong, W.W., Anderson, R., Chen, R., and Weng, Z. 2005. ZDOCK and RDOCK performance in CAPRI rounds 3, 4, and 5. Proteins 60: 207–213. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Rao, M.S., Heath, R.J., Price, A.C., Olson, A.J., Rock, C.O., and White, S.W. 2001. Identification and analysis of the acyl carrier protein (ACP) docking site on β-ketoacyl-ACP synthase III. J. Biol. Chem. 276: 8231–8238. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Marrakchi, H., White, S.W., and Rock, C.O. 2003a. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J. Lipid Res. 44: 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Wu, B., Zheng, J., and Rock, C.O. 2003b. Key residues responsible for acyl carrier protein and β-ketoacyl-acyl carrier protein reductase (FabG) interaction. J. Biol. Chem. 278: 52935–52943. [DOI] [PubMed] [Google Scholar]