Abstract
The cysteine-rich somatomedin B domain (SMB) of the matrix protein vitronectin is involved in several important biological processes. First, it stabilizes the active conformation of the plasminogen activator inhibitor (PAI-1); second, it provides the recognition motif for cell adhesion via the cognate integrins (αvβ3, αvβ5, and αIIbβ3); and third, it binds the complex between urokinase-type plasminogen activator (uPA) and its glycolipid-anchored receptor (uPAR). Previous structural studies on SMB have used recombinant protein expressed in Escherichia coli or SMB released from plasma-derived vitronectin by CNBr cleavage. However, different disulfide patterns and three-dimensional structures for SMB were reported. In the present study, we have expressed recombinant human SMB by two different eukaryotic expression systems, Pichia pastoris and Drosophila melanogaster S2-cells, both yielding structurally and functionally homogeneous protein preparations. Importantly, the entire population of our purified, recombinant SMB has a solvent exposure, both as a free domain and in complex with PAI-1, which is indistinguishable from that of plasma-derived SMB as assessed by amide hydrogen (1H/2H) exchange. This solvent exposure was only reproduced by one of three synthetic SMB products with predefined disulfide connectivities corresponding to those published previously. Furthermore, this connectivity was also the only one to yield a folded and functional domain. The NMR structure was determined for free SMB produced by Pichia and is largely consistent with that solved by X-ray crystallography for SMB in complex with PAI-1.
Keywords: vitronectin, PAI-1, NMR, amide hydrogen (1H/2H) exchange, mass spectrometry, disulfide-knot, somatomedin B domain, protein structure
Vitronectin is an abundant glycoprotein primarily found circulating in blood (400 mg/L or 5 μM), where it is believed to be involved in regulation of hemostasis and vascular remodeling. Comparative studies using vitronectin-deficient mice highlight a role of vitronectin in stabilizing formed thrombi. Genetic ablation of vitronectin consequently leads to an increased frequency of embolization due to the instability of the generated thrombi (Reheman et al. 2005). Elevated concentrations of plasma vitronectin correlate to the severity of coronary atherosclerosis (Ekmekci et al. 2002). A matrix-deposited form of vitronectin plays a role in integrin-mediated cellular adhesion and migration through its RGD-motif (residues 45–47). Furthermore, vitronectin is required for urokinase-type plasminogen activator receptor (uPAR)-dependent activation of the Rac-mediated signaling pathway leading to increased cell motility (Kjøller and Hall 2001).
Vitronectin is composed of several functional domains, which bind a plethora of ligands. Its hemopexin-like domains are, for example, reported to bind plasminogen, urokinase-type plasminogen activator (uPA), terminal complement components, collagen, and heparin (Preissner and Seiffert 1998). Of particular interest is the cysteine-rich, N-terminal somatomedin B (SMB) domain, which contains the binding determinants for the plasminogen activator inhibitor-1 (PAI-1) (Seiffert and Loskutoff 1991; Deng et al. 1996b), the urokinase-type plasminogen activator receptor (uPAR) (Wei et al. 1994; Deng et al. 1996a; Gårdsvoll and Ploug 2007), and the integrins αvβ3, αvβ5, and αIIbβ3 (Cheresh et al. 1989; Wayner et al. 1991).
The interaction of this small SMB domain with PAI-1 stabilizes the serpin in its active form by attenuating the latency transition to the inactive form (Seiffert et al. 1994). The RGD sequence found adjacent to the SMB domain binds to several members of the integrin family (αvβ3, αvβ5, and αIIbβ3), which causes integrin-expressing cells to adhere to and migrate on vitronectin substrates (Cheresh et al. 1989; Wayner et al. 1991). The RGD-motif is inaccessible in plasma vitronectin, suggesting that vitronectin exists in multiple conformations (Seiffert and Smith 1997). The SMB domain also binds the glycolipid-anchored uPAR (Deng et al. 1996a; Gårdsvoll and Ploug 2007). This interaction promotes integrin-independent cell adhesion in a process that is facilitated by receptor occupancy by the primary ligand, the serine protease uPA (Wei et al. 1994). The binding sites for PAI-1 and uPAR on the SMB domain are overlapping, and these ligands compete mutually for this binding site (Deng et al. 1996a). Binding of PAI-1 to vitronectin also competes with integrin binding to the RGD-motif, causing detachment of cells from the vitronectin matrix (Stefanson and Lawrence 1996). Cell adhesion and migration are important processes for normal tissue remodeling as typified by wound healing and for pathological conditions such as cancer invasion and metastasis.
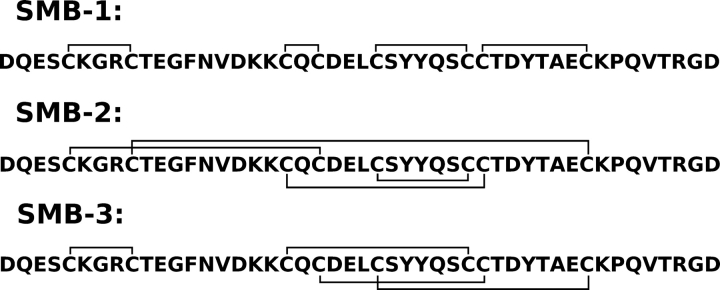
No experimental high-resolution structure of intact vitronectin has been presented so far, but studies on monomeric, plasma-derived vitronectin by small-angle X-ray scattering reveal a roughly peanut-shaped molecule with a maximum length of 110 Å (Lynn et al. 2005). In contrast, several structures have been reported for its cysteine-rich SMB domain (Zhou et al. 2003; Kamikubo et al. 2004; Mayasundari et al. 2004). Nonetheless, considerable controversy exists in the literature regarding the disulfide connectivity of this domain. Three different disulfide connectivities (▶) have thus been assigned by either X-ray crystallography (Zhou et al. 2003) or by biochemical methods (Kamikubo et al. 2002; Horn et al. 2004). The SMB preparations used were produced either recombinantly in Escherichia coli (Kamikubo et al. 2002; Zhou et al. 2003) or derived from plasma vitronectin by CNBr cleavage (Horn et al. 2004). Adding to this controversy, different three-dimensional structures have been solved for these preparations. Several explanations have been put forward to explain these inconsistencies. It has been proposed that bacterial expression and refolding introduce erroneous disulfide bridges (Horn et al. 2004), that multiple different disulfide connectivities can yield biologically active SMB (Kamikubo et al. 2004; Cheek et al. 2006), that the plasma-derived SMB represents a biologically inactive form (Kamikubo et al. 2006), or that significant thiol–disulfide interchange has occurred during biochemical assignment of the disulfide arrangements (Li et al. 2007).
Figure 1.
Disulfide connectivities reported for the SMB domain of human vitronectin. Three different disulfide connectivities have been assigned experimentally for the SMB domain—SMB-1: connectivity determined biochemically after partial reduction of refolded SMB produced in E. coli (Kamikubo et al. 2002); SMB-2: connectivity determined by X-ray crystallography of refolded SMB produced in E. coli (Zhou et al. 2003); and SMB-3: connectivity determined biochemically after partial reduction of SMB derived from plasma vitronectin by CNBr cleavage (Horn et al. 2004).
To circumvent the possible pitfalls of recombinant expression in prokaryotes, we have in the present study chosen to express the SMB domain in two different eukaryotic expression systems: Pichia pastoris and S2-cells from Drosophila melanogaster. To verify that our recombinant products are similar to plasma-derived SMB, we made a direct comparison of the global amide hydrogen (1H/2H) exchange kinetics of these domains, and investigated their functional status by comparing the protection conferred by binding to PAI-1. Both recombinant products were shown to behave similarly to plasma SMB. The recombinant SMB produced in P. pastoris was used for three-dimensional structure determination by nuclear magnetic resonance (NMR) spectroscopy utilizing the possibility for stable isotope labeling offered by this expression host.
Results
Expression and labeling of SMB produced in P. pastoris
In this study, we chose to express the SMB domain of vitronectin in two different eukaryotic expression systems (P. pastoris and D. melanogaster S2 cells). This strategy is expected to minimize the potential risk of generating an ensemble of misfolded proteins with nonnative disulfide pairing similar to that reported previously for the expression of recombinant SMB in E. coli, which yielded only ∼10% binding-active SMB (Kamikubo et al. 2002).
It is well established that Pichia is able to produce high amounts of disulfide-rich proteins that often fail to express properly in E. coli (White et al. 1994). Furthermore, Pichia can grow on minimal medium, thus enabling stable isotope labeling for NMR studies (Laroche et al. 1994). The vector for expression in Pichia was constructed to ensure that the protein retained its native N terminus as only one Kex2 site was maintained for cleavage of the α-factor signal (see Materials and Methods), and the vector also contained a C-terminal histidine tag for purification. Expression of SMB (residues 1–47) in labeled minimal medium yields 3–4 mg of purified SMB/L medium with a purity of >95% as assessed by SDS-PAGE and MALDI-TOF MS (data not shown).
With a view to the existing controversy in the literature regarding the structure and disulfide configurations of the SMB domain, it is extremely important to ensure that our recombinant SMB domains are identical to the physiologically relevant SMB domain. Thus for comparison, we also used S2 cells from D. melanogaster as an expression host for the production of recombinant SMB (Gårdsvoll et al. 2007), utilizing the full protein-processing machinery of a higher eukaryote to ensure proper folding. Accordingly, we have previously used S2 cells to express highly disulfide-bonded proteins in their active, native conformation (Gårdsvoll et al. 2004, 2006, 2007), and importantly, the two possible N-linked glycosylation sites of the chosen purification tag (i.e., uPAR domain III) provide a retention signal for the small non-glycosylated SMB domain, enabling its processing by the calrecticulin/calnexin complex and the associated protein-disulfide isomerase (Molinari 2007).
Comparison of recombinant SMB with SMBplasma derived from purified native vitronectin
To determine if our recombinant SMB domains produced in the two different eukaryotic hosts have the same overall structure as native SMBplasma that was derived from purified plasma vitronectin by CNBr cleavage, we compared their global amide hydrogen (1H/2H) exchange kinetics, to assess their overall solvent exposure. Secondary and tertiary structural elements in proteins render some amides inaccessible for interaction with the surrounding solvent, which is revealed as slowly exchanging amides in isotope exchange experiments. The hydrogen exchange profile of a given protein thus constitutes a sensitive and powerful tool to uncover subtle changes in solvent exposure or stability among various protein preparations. Hydrogen (1H/2H) exchange kinetics is sensitive to differences in both structure and dynamics of the molecules and probes the structure of any subpopulation in the entire protein preparation.
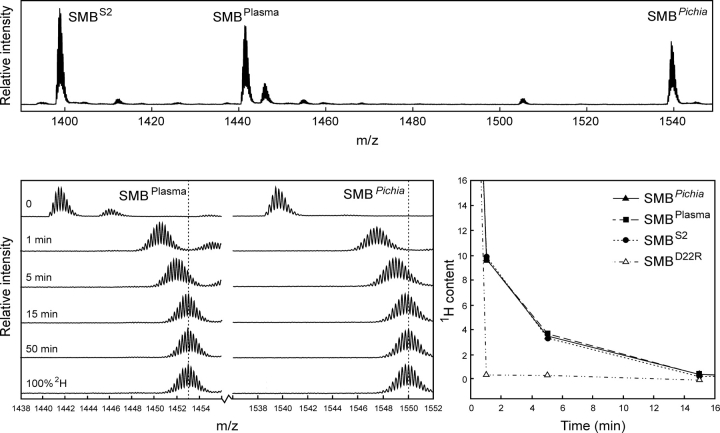
The three different preparations of SMB used in this study have different molecular masses and can therefore be resolved in a single mass spectrum (▶, upper panel). This facilitates a direct comparison and greatly improves the data quality, because it allows the hydrogen exchange to be performed simultaneously for a mixture of the three SMB preparations. Importantly, this experimental setup enables a meticulous comparison of the global exchange kinetics for all three SMB variants, as they encounter exactly identical exchange times during sampling as well as back-exchange during desalting. The progress of the hydrogen exchange is revealed by a gradual shift in the mass envelope for SMB domains (▶, lower left panel). These mass envelopes are subsequently processed to calculate the average number of unexchanged amides remaining in the respective SMB preparation as a function of exchange time. This serves to eliminate the differences in the number of incorporated deuterons caused by varying chain lengths of the presumed unstructured C-termini and allows a direct comparison of the number of protected amides in the three different SMB variants. Such thorough examinations reveal indistinguishable exchange kinetics, implicating the same number of protected amides for all three variants of SMB (▶, lower right panel). This clearly indicates that they have comparable overall structures and conformational stability within the timescale addressed by the isotope exchange experiment. To verify that the exchange kinetics is, indeed, a sensitive probe of structural perturbations, we furthermore tested a single-site mutant of SMBD22R. As shown in ▶ (lower right panel), there is a dramatic increase in the rate of isotopic exchange in SMBD22R compared to the wild-type SMB preparations, which highlights the sensitivity of amide hydrogen (1H/2H) exchange to probe structural perturbations.
Figure 2.
Comparison of global hydrogen (1H/2H) exchange properties of different preparations of SMB. (Upper panel) A full scan mass spectrum of the quadruple charge state of the three forms of the SMB domain that we tested simultaneously in this study. The monoisotopic masses are shown, which translate into the following neutral molecular masses—SMBS2: 5588.33 Da (Δm = 0.17 Da); SMBplasma: 5758.51 Da (Δm = 0.18 Da); and SMBPichia: 6150.67 Da (Δm = 0.21 Da). Deviations from the theoretical masses are shown in brackets. SMBplasma contains an additional molecular species (5776.50 Da) due to an incomplete conversion between homoserine and homoserine lactone after CNBr cleavage. (Lower left panel) The mass spectra obtained for SMBplasma and SMBPichia are shown as a function of exchange time after dilution into deuterium oxide at 25°C. Notably, only one isotope envelope is produced by this exchange experiment for each SMB variant. (Lower right panel) The residual 1H content as a function of exchange time is illustrated for the three different wild-type SMB preparations, as well as a single-site mutant (SMBD22R) with impaired binding to PAI-1 and uPAR.
Functional status of recombinant and native SMB
The differences between the three-dimensional structures reported for SMB could possibly be explained if only one of the forms retained biological activity and the other represented a stable but misfolded state without biological activity. Both SMB variants produced in E. coli bind PAI-1, which demonstrates that they have a biologically active conformation (Kamikubo et al. 2002; Zhou et al. 2003). This led to the proposition that the native SMB domain derived from purified plasma vitronectin used by Peterson and coworkers is not biologically active (Kamikubo et al. 2006). As a consequence, we tested the functional status of our recombinant SMB and plasma-derived SMB by comparing the hydrogen (1H/2H) exchange profile for these SMB preparations in the presence and absence of the cognate high-affinity ligand PAI-1. As shown in ▶ (lower panel), the complex formation between PAI-1 and SMB caused a dramatic retardation in the exchange rates for approximately nine amides in all three SMB domains compared in this study. The slow dissociation of the SMB·PAI-1 complex and the stability of its corresponding binding interface is illustrated by the longevity of the protection observed for amide hydrogens in the complex, where an average of more than five amide hydrogen atoms were retained in the complex after 50 min of isotope exchange at 25°C (▶, lower panel). The three SMB preparations tested were thus still indistinguishable when the exchange was performed in the presence of PAI-1. This finding further substantiates that both our recombinant SMB domains are structurally and functionally identical to the plasma-derived SMB. In conclusion, we find that both the plasma-derived and our recombinant forms of SMB are in a stable and biologically active conformation.
Figure 3.
PAI-1 induces similar protection of amide hydrogens in recombinant and plasma-derived SMB as monitored by global hydrogen (1H/2H) exchange. The global exchange profiles recorded by mass spectrometry for SMBPichia alone (upper left panel) and in complex with PAI-1 (upper right panel) are shown as time-course experiments. In the SMB·PAI-1 complex experiment, 7 μM SMB was preincubated with 60 μM PAI-1 before the exchange was initiated by a 25-fold dilution in deuterium oxide. (Lower panel) A summary of the calculated residual 1H content in SMBPichia, SMBS2, and SMBplasma as a function of exchange time and occupancy with PAI-1.
Importantly, this hydrogen (1H/2H) exchange experiment also revealed structural and functional homogeneity of the entire population of our His-tag purified SMB produced in Pichia. The isotope mass distributions in all of our exchange experiments move as single envelopes (▶ and ▶), which unambiguously demonstrates that our SMB preparations are 100% homogeneous with respect to their global stability as well as their PAI-1-binding properties. This is in sharp contrast to expression in E. coli, where only ∼10% of the His-tag purified recombinant SMB was reported to be biologically active (Kamikubo et al. 2002).
Impact of disulfide connectivity on stability and function of SMB
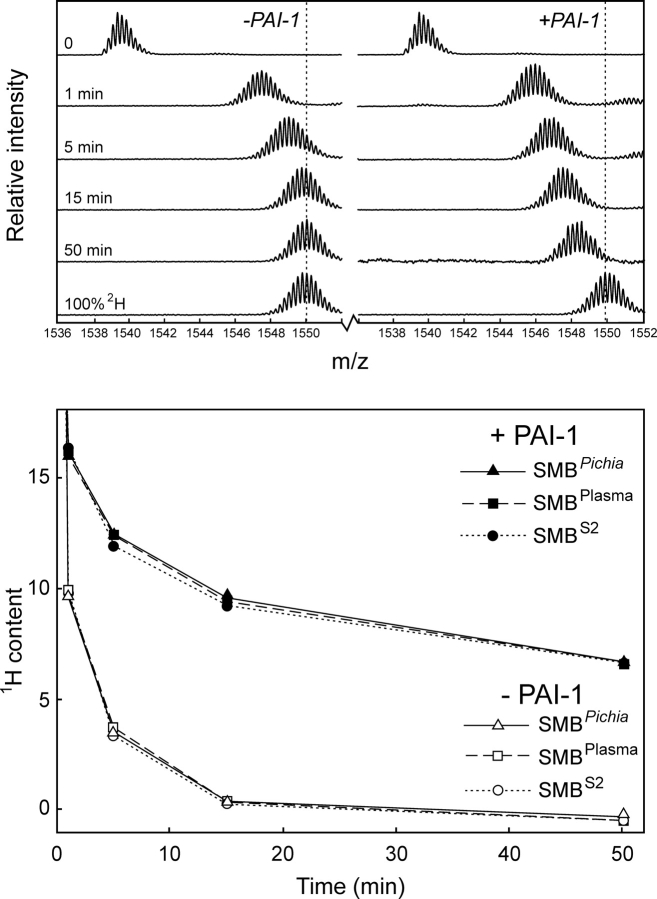
To explore the possible disulfide connectivity of our recombinant SMBPichia, we compared its solvent exposure to those of three synthetic SMB polypeptides with predefined disulfide bonding as in SMB-1, SMB-2, and SMB-3 (▶) by hydrogen (1H/2H) exchange. These SMB preparations were synthesized chemically using native chemical ligation and orthogonal cysteine protection (Li et al. 2007). As shown in ▶ (upper panel), only one of the three synthetic SMB samples tested exhibited a solvent exposure comparable to the one recorded for SMBPichia. This suggests that only the disulfide connectivity found in SMB-2 provides a domain structure and stability similar to that for SMBPichia. Before the first data points were recorded, both synthetic SMB-1 and SMB-3 exchanged completely, as expected for misfolded polypeptides.
Figure 4.
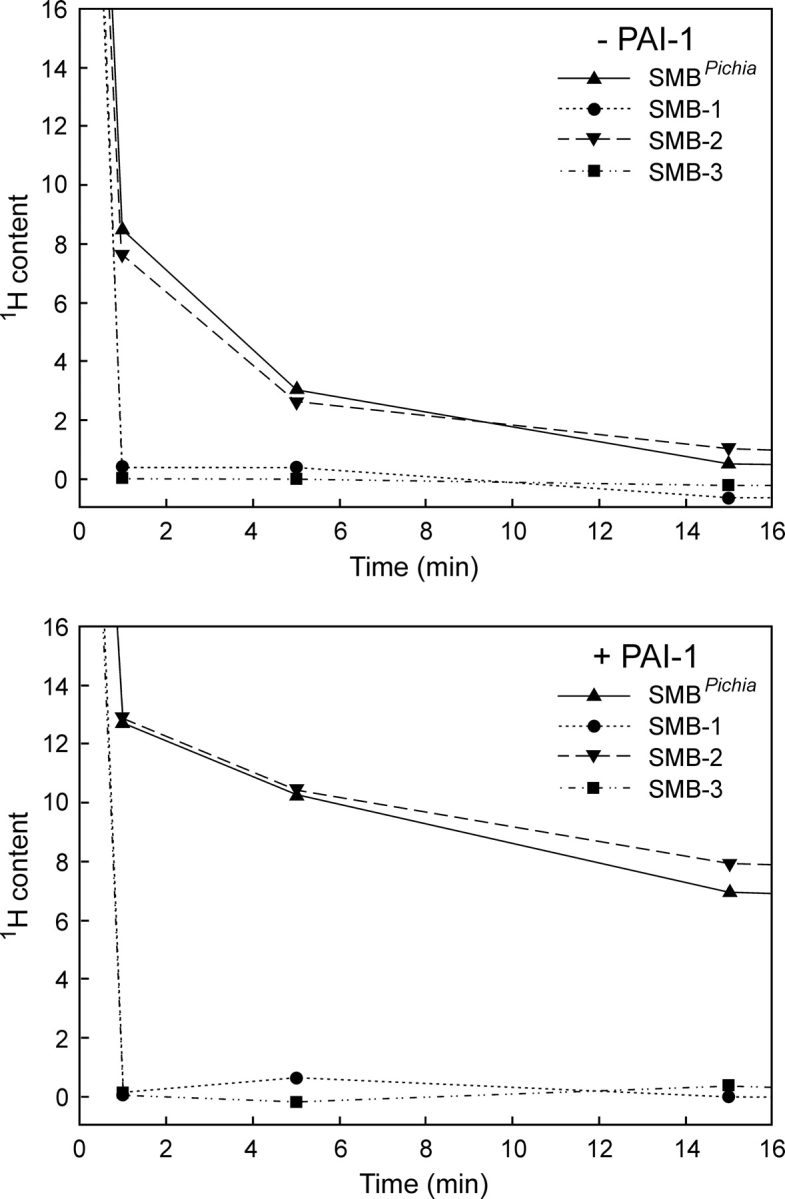
Comparison of synthetic SMB domains with predefined disulfide connectivities and SMBPichia reveals different global hydrogen (1H/2H) exchange kinetics. The solvent exposure of three different synthetic SMB domains with predefined disulfide connectivities (▶) was compared pairwise with that of SMBPichia as outlined in ▶. (Upper panel) The global exchange profiles of the free SMB domains reveal that only SMB-2 resembles SMBPichia in this respect. Both SMB-1 and SMB-3 are fully exchanged before the first data point. (Lower panel) The functional integrity of these SMB domains are probed by PAI-1-induced changes in their global hydrogen exchange profiles. Only SMB-2 and SMBPichia bind PAI-1 as assessed by their reduced solvent exposure.
In separate experiments, where hydrogen (1H/2H) exchange was performed in the presence of a molar excess of PAI-1 (▶, lower panel) only the synthetic product representing SMB-2 obtained a protection comparable to that observed for SMBPichia. Under these conditions, neither SMB-1 nor SMB-3 experienced any protection from PAI-1, excluding the formation of stable SMB·PAI-1 complexes consistent with data recorded by surface plasmon resonance as published by Li et al. (2007).
NMR structure determination
The recombinant SMB produced in P. pastoris is ideally suited for structure determination by NMR, as this expression host offers the possibility for stable isotope labeling, high yields, and a product that retains the overall folding and functional properties of the SMBplasma derived from plasma-purified vitronectin.
An examination of various solution conditions using HSQC spectra of 15N-labeled SMB (120 μM) showed that at pH 7.4, three peaks are missing compared to pH 6.5, presumably due to fast exchange with the solvent. The presence of 50 mM NaCl and 20 mM phosphate buffer did not affect the spectrum, compared to water alone. For the subsequent structure determination, we thus used a 13C,15N-labeled sample at pH 6.5 without buffer or salt.
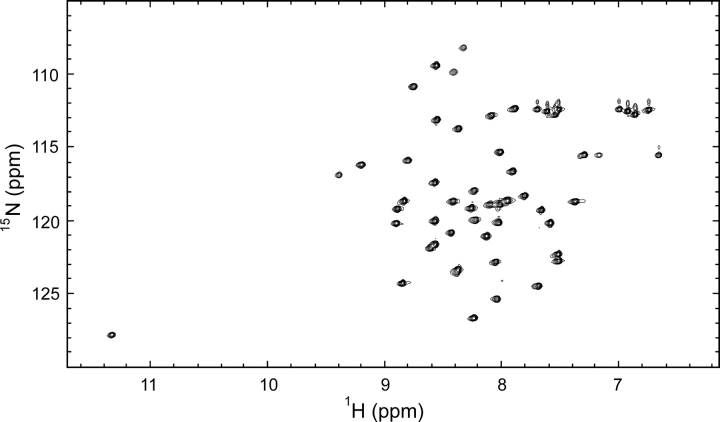
The HSQC spectrum is well dispersed with peaks of comparable line width, which is characteristic for a well-folded monomeric protein (▶). The HSQC showed 46 distinct amide peaks, which presumably originate from peptide bonds, as compared to a theoretical maximum of 51 unique peptide peaks. This is indicative of only one detectable species in the analyzed sample, which is in accordance with data from our previous hydrogen (1H/2H) exchange experiments. Resonance assignments for residues 3–47 were carried out by standard methods using a set of triple resonance multidimensional NMR experiments (Cavanagh et al. 1995). We did not observe any resonances that could be attributed to the two N-terminal residues and the C-terminal His tag. One peak in the HSQC remained unidentified after this assignment. Comparison of HSQC spectra with different sweep widths ruled out the possibility that it could be a folded side-chain peak; thus, presumably it represents one of the unassigned terminal residues. The assignment of the nonlabile side-chain resonances for residues 3–47 is nearly complete as only Hβ of Thr10 and Hζ of Phe13 are lacking. The chemical shift of HN of Cys5 is unusually high (11.3 ppm), as reported also for bacterial and plasma-derived SMB (Kamikubo et al. 2004; Mayasundari et al. 2004).
Figure 5.
The 15PN–1H HSQC spectrum of SMBPichia. This HSQC spectrum was recorded at pH 6.5 in 10% D2O at 25°C and 750 MHz. The peaks are well dispersed with similar intensities as is characteristic for a well-folded protein.
Initially, our structure calculations were carried out without any restraints originating from disulfide bonds. We subsequently introduced constraints corresponding to each of the disulfide configurations shown in ▶. During the initial calculations in CYANA, the SMB-2 disulfide configuration produced fewer violations and better geometry. After water refinement, this preference disappeared, and the statistics for the structure calculations with each of the three disulfide connectivities became acceptable (▶). Since the hydrogen (1H/2H) exchange data unambiguously demonstrates that the solvent exposure of SMBPichia is replicated by only the synthetic domain with the disulfide configuration SMB-2, we proceeded using this disulfide connectivity.
Table 1.
Comparison of structure calculations with different disulfides
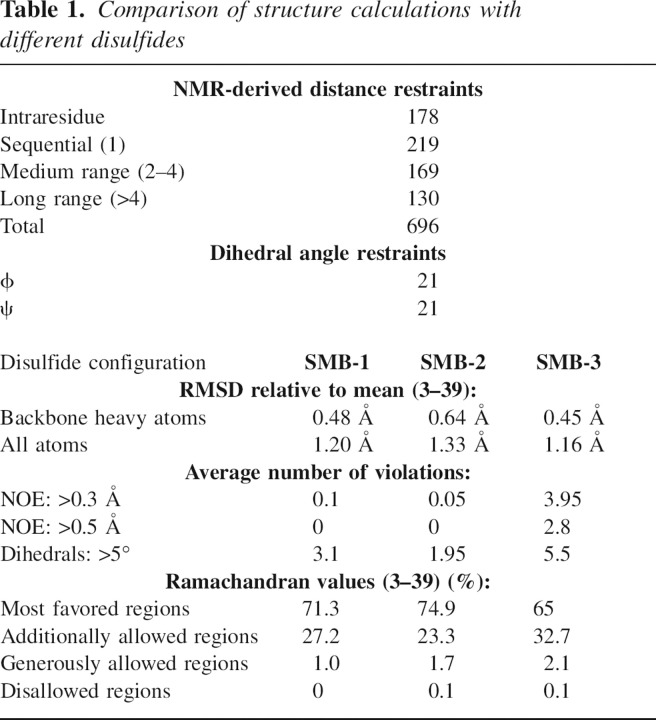
The SMBPichia domain is folded into a roughly globular structure with the disulfide bonds buried in the core (▶). Although this domain primarily consists of loops with no well-defined secondary structure, two very short helical elements are nevertheless present (residues 24–28 and 34–38). The ensemble of NMR structures we obtained for SMBPichia is highly similar to the crystal structure reported for SMB-2 (Zhou et al. 2003) and the NMR structure for SMB-1 (Kamikubo et al. 2004) with backbone heavy atom RMSDs for residues 3–39 of 1.18 Å and 1.71 Å, respectively (▶, left panel). As opposed to this, we were unable to satisfactorily superimpose our structure on that published for SMB-3 (Mayasundari et al. 2004), as shown in ▶ (right panel). The PAI-1-binding residues are found in a contiguous hydrophobic patch on the surface of the domain, as reported previously (Zhou et al. 2003). Interestingly, Gln29 has an unusual geometry, with the backbone occupying a positive ϕ angle. This is confirmed by the presence of a strong NOE between HN and Hα for this residue (Ludvigsen and Poulsen 1992). This geometry allows the backbone to undergo a sharp turn to accommodate the disulfide bridge Cys25–Cys31.
Figure 6.

Schematic representation of the NMR structure of human SMB produced in P. pastoris. (Upper panel) The backbones of the 20 structures with the lowest energy are shown in the cartoon representation for SMB residues 3–47. Side chains of the PAI-1-binding residues (Phe13, Asp22, Leu24, Tyr27, and Tyr28) are shown as sticks and are clustered in a hydrophobic patch on the surface of the SMB domain. Multiple NOEs demonstrate that these side chains interact. Nonetheless, the lack of stereospecific assignments for the aromatic residues suggest that the rings can flip in solution. (Lower panel) The disulfide bonds are found buried in the core of the domain, where they replace a traditional hydrophobic core. The disulfide configuration SMB-2 provides a high degree of cross-linking between regions that are distant in the sequence, thus presumably lowering the entropy of the unfolded state.
Figure 7.
Comparison of the different three-dimensional structures reported for SMB. (Left panel) Overlay of the backbone traces for SMBPichia (residues 3–39) displayed in blue on the structures reported for SMB-1 (red) and SMB-2 (green); (right panel) the overlay on SMB-3 (aquamarine). The structures were fitted in MOLMOL using residues 3–39 for SMB-1 and SMB-2, but only using residues 19–39 for SMB-3. As evident from this comparison, the structure of SMBPichia resembles those reported for SMB-1 and SMB-2, whereas it fails to converge on the structure reported for SMB-3. The PDB coordinates used were: 1SSU for SMB-1 (Kamikubo et al. 2004), 1OC0 for SMB-2 (Zhou et al. 2003), and 1S4G for SMB-3 (Mayasundari et al. 2004).
Discussion
The SMB domain has four disulfide bonds within only 35 residues and is highly resistant to digestion by all the proteases we have tested, that is, trypsin, pepsin, Asn-N, Glu-C, and thermolysin. The proximity of the disulfides combined with the resilience toward proteolytic degradation render the assignment of the disulfide configuration of SMB a methodological challenge. Three different disulfide pairings have been published so far, using SMB from different sources (▶), including SMBplasma generated from vitronectin purified from human blood (Horn et al. 2004). A linear set of disulfide bridges (SMB-1) was reported for a recombinant product produced in E. coli using partial reduction with subsequent alkylation (Kamikubo et al. 2002). Another disulfide connectivity (SMB-2) was derived from SMB produced in E. coli based on electron densities recorded for SMB·PAI-1 complexes by X-ray diffraction (Zhou et al. 2003). A third connectivity (SMB-3) was finally reported for plasma vitronectin-derived SMB, also relying on partial reduction and alkylation (Horn et al. 2004).
Generation of free thiol groups by partial reduction during experimental disulfide assignment calls for great caution in the subsequent analysis, because of the obvious risk of thiol–disulfide interchange. If used, such protocols require corroboration by independent methods. The originally determined connectivity in SMB-1 was therefore sought to be verified by introduction of a CNBr cleavage site by site-directed mutagenesis (Kamikubo et al. 2004). Despite being functionally active, this mutant (Asn14 → Met) did not produce a fragmentation pattern compatible with the original set of disulfide bonds (Kamikubo et al. 2004), leading us to question the validity of the disulfide connectivities in SMB-1. This suspicion was further substantiated by the impaired PAI-1 binding reported for a synthetic SMB with the disulfide bonds of SMB-1 (Li et al. 2007). In accordance, we find that this synthetic SMB-1 behaves as a misfolded domain without detectable PAI-1 binding in hydrogen (1H/2H) exchange experiments. Similar properties were observed for the synthetic SMB-3, demonstrating that neither SMB-1 nor SMB-3 is compatible with proper folding and ligand binding. In contrast, synthetic SMB-2 exhibits an identical hydrogen exchange protection (i.e., structure) as compared to SMBplasma, SMBPichia, and SMBS2. This similarity is also extended to PAI-1 binding. These data are consistent with the observation by Li et al. that either SMB-1 or SMB-3 produces SMB domains with severely compromised PAI-1 binding, whereas SMB-2 produces an SMB domain with an affinity comparable to that of SMBplasma (Li et al. 2007). The lack of amide protection we observe in (1H/2H) exchange experiments provides the structural basis for this observation, as SMB-1 and SMB-3 fail to yield a properly folded domain, thus disproving the proposition that different sets of disulfide connectivities can stabilize the same SMB fold (Kamikubo et al. 2004). Together, these data therefore conclusively demonstrate that only the disulfide connectivity of SMB-2 is compatible with a folded and biologically active SMB domain. As PAI-1 binding was verified for all SMB preparations irrespective of origin, that is, from E. coli (Kamikubo et al. 2002; Zhou et al. 2003) and from plasma-derived vitronectin (▶), they must contain the disulfide pattern represented by SMB-2. The deviating connectivities represented by SMB-1 and SMB-3 most likely stem from thiol–disulfide interchange during partial reduction (Li et al. 2007).
The three-dimensional structure of the SMB domain is also the subject of some controversy, which partly arises because of the abovementioned ambiguity on the disulfide arrangements. The first structure was solved by X-ray crystallography for E. coli produced SMB in complex with PAI-1 (Zhou et al. 2003) defining the SMB-2 disulfide connectivity. Subsequently, the NMR structure of free SMB produced by E. coli was solved using the SMB-1 connectivity (Kamikubo et al. 2004). These two structures present a comparable backbone fold of SMB despite the fact that they encompass different sets of disulfide bonds. Finally, the NMR structure of plasma-derived SMB was reported using the SMB-3 connectivity (Mayasundari et al. 2004). This structure is more loosely defined and heterogeneous compared to the previous structures and mainly agrees on the location of the short α-helical stretch (residues 24–28).
In the present study, we have expressed and characterized recombinant SMB produced in two eukaryotic hosts. Prior to structure determination of the recombinant SMBPichia domain, we verified that its overall structure and dynamics are equal to those of SMBplasma by comparison of their hydrogen exchange kinetics. Our data clearly show that the three forms are indistinguishable in their global exchange kinetics (▶, lower right panel). Furthermore, an identical change in the exchange kinetics is introduced in all SMB preparations upon the high-affinity binding to PAI-1 (▶, lower panel). On average, nine backbone amides are shifted toward slow exchange in the presence of PAI-1, which demonstrates that the binding of the SMB·PAI-1 complex is sufficiently tight to exclude water from the interface. Apart from demonstrating the structural similarity between the SMB preparations, this analysis also establishes the biological activity of both the recombinant and the plasma-derived SMB. This experiment emphasizes that the SMBplasma is, indeed, biologically active, eliminating this property as an explanation for the difference in the solved structures, as proposed previously (Kamikubo et al. 2006). Since the isotope exchange experiment is sensitive to conformational heterogeneity in the SMB preparation, subpopulations would produce bimodal exchange patterns in the isotope envelopes. Importantly, the mass distribution of the recombinant SMBPichia is shifted as a single isotope envelope both in the presence and absence of PAI-1. This demonstrates that the preparation is homogeneous and that the entire population is binding active. As alluded to above, the hydrogen (1H/2H) exchange kinetics recorded for SMBPichia alone or in complex with PAI-1 are compatible only with the SMB-2 disulfide connectivity.
Having verified that the SMB expressed in Pichia is biologically active and in a conformation identical to the SMBplasma, we have set the stage for the structure determination of SMBPichia by NMR spectroscopy. The models calculated from the NMR data reveal a well-folded structure of the core region of the SMB domain that is cross-linked by disulfide bonds. A classic hydrophobic core is absent in the SMB domain, as it contains very few hydrophobic residues. Multiple crossed disulfide bonds may, nevertheless, provide such small domains with sufficient structural stability, alleviating the necessity of a hydrophobic core. The majority of the hydrophobic residues in the free SMB domain are found in a surface-exposed patch (▶) that is involved in ligand binding, as demonstrated for PAI-1 binding by X-ray crystallography (Zhou et al. 2003) and for uPAR by alanine-scanning mutagenesis. We detected multiple NOEs between Phe13, Val15, Leu24, Tyr27, and Tyr28, suggesting that these surface-exposed hydrophobic residues interact to reduce their contact with water.
The present NMR structure for free SMBPichia is very similar to the structures described previously for E. coli produced SMB by X-ray crystallography (Zhou et al. 2003) or NMR (Kamikubo et al. 2004), despite the fact that the latter assumed a disulfide connectivity now known to produce a nonfunctional, misfolded domain. Notably, the structure published for SMBplasma differs markedly from the present structure of SMBPichia. This discrepancy is unexpected since our hydrogen (1H/2H) exchange experiments cannot distinguish between the solvent exposures of the two SMB preparations. It should, however, be emphasized that the structure solved for SMBplasma relied on data recorded on a very dilute (0.09 mM), unlabeled protein sample, owing to the inherent difficulty in preparing large amounts of SMB from vitronectin purified from plasma. This compromise resulted in rather few long-range NOE effects, which constitute the pivotal data for subsequent structure calculation (Mayasundari et al. 2004). In the absence of sufficient numbers of such long-range NOEs, assumed disulfide restraints are expected to have a disproportionately large and undesirable impact on structure calculations.
To provide an explanation for the apparent ambiguity between structure and disulfide connectivity for E. coli produced SMB, it was previously proposed that a functional and folded SMB domain could be achieved with different sets of disulfide bridges (Kamikubo et al. 2004). A similar proposition was advanced in a recent bioinformatics study on disulfide-rich protein domains based solely on the different structures and disulfide connectivities reported for SMB (Cheek et al. 2006). This proposal was apparently corroborated by molecular dynamics simulations revealing that the SMB structure solved by NMR has comparable conformational energies whether using the disulfide connectivities of SMB-1 or SMB-2 (Kamikubo et al. 2004). These speculations were even further advanced by the proposition that this alleged ability of SMB to accommodate more than one set of disulfide-bond connectivities could be functionally important for the regulation of the multiple functions of vitronectin (Chen and Hogg 2006). Our structure calculations produce a similar backbone fold irrespective of the disulfide connectivity used, without significant violations of the NMR data and the geometric properties imposed by the force field. This indicates that the fold of the SMB domain is theoretically capable of accommodating all three published disulfide connectivities without steric clashes. Nevertheless, the hydrogen (1H/2H) exchange experiments unambiguously demonstrate that SMB domains with disulfide connectivities corresponding to SMB-1 and SMB-3 are misfolded and inactive. Intriguingly, this highlights that only one of the three connectivities is capable of stabilizing the structure in its active conformation despite the fact that all three connectivities can be accommodated in the SMB fold. It is noteworthy that the preferred disulfide connectivity (SMB-2) provides the highest degree of cross-linking between the termini of the SMB domain, thus limiting the conformational freedom of these regions. This is in accordance with the proposition that multiple disulfide bonds may substitute the traditional hydrophobic core to introduce the required stability of small domains, presumably by lowering the entropy of the unfolded state (Thornton 1981).
In conclusion, we have solved the structure of the SMB domain of vitronectin produced in P. pastoris. This recombinant domain is ideally suited for further interaction studies as revealed for PAI-1 binding by hydrogen (1H/2H) exchange (present study) and for uPAR binding by surface plasmon resonance studies (Gårdsvoll and Ploug 2007).
Materials and Methods
Materials and protein preparations
Deuterium oxide (99.9 atom % D) was obtained from Cambridge Isotope Laboratories. Ultra-high quality water was obtained from a PURELAB Ultra unit (ELGA Labwater). Deuterated PBS buffer (pH 7.7, uncorrected value) was prepared from PBS buffer by two cycles of lyophilization and redissolution in D2O.
Purified SMB (1–51), derived from plasma vitronectin by CNBr cleavage, was generated as described (Mayasundari et al. 2004) and denoted SMBplasma. Purified SMB preparations with three different, predefined disulfide connectivities (SMB-1, SMB-2, and SMB-3) (▶), controlled by Acm-protection of selected cysteines during peptide synthesis, were kind gifts from Dr. W. Lu (Li et al. 2007). Recombinant enterokinase (EC 3.4.21.9) was from Stratagene. Recombinant active PAI-1 was produced in E. coli and was a kind gift from Dr. M.D. Andersen (Novo Nordisk, Bagsværd, Denmark). The penta anti-His monoclonal antibody was purchased from Invitrogen.
Expression of SMB in Drosophila S2 cells
Recombinant SMB constructs covering residues 1–44 (SMB1–44, SMB-D22R1–44) and 1–47 (SMB1–47) of human vitronectin were expressed and purified as fusion proteins with the domain III of uPAR as C-terminal purification tag in Drosophila S2 cells using the Drosophila expression system (Invitrogen) as described in detail elsewhere (Gårdsvoll et al. 2007).
Expression and isotope labeling in P. pastoris
To enable subsequent biosynthetic 15N and/or 13C labeling, recombinant SMB1–47 with a C-terminal 6-histidine tag (SMBPichia) was produced using a P. pastoris expression system (Invitrogen). The plasmid HSP-pUC18 with cDNA encoding human vitronectin was obtained from American Type Culture Collection, no. 65450. Two synthetic oligonucleotide primers were used for PCR amplification of the DNA fragment encoding residues 1–47 from vitronectin, together with a C-terminal His-tag, a stop codon, and appropriate restriction sites in bold: 5′-CGCCTCGAGAAAAGAGACCAAGAGTCATGCAAGGGCCGCTGCAC-3′ and 5′-TAGTCTAGATTAATGATGATGATGATGATGGTCCCCGCGAGTCACTTGGGGCTTGC-ACTC-3′. Subsequently, the amplified DNA was cloned into the vector pPICZα A. The resulting construct pPICZα A SMB1–47-His thus encodes the secretion signal of the α-mating factor from Saccharomyces cerevisiae fused to His6-tagged SMB1–47, under the control of the alcohol oxidase 1 promoter. A single Kex2 signal cleavage site is contained within the C-terminal end of the α-factor sequence, which ensures a recombinant product with a free native N terminus of vitronectin.
Electro-competent cells of P. pastoris strain X-33 were transfected with 10 μg of PmeI-linearized vector by electroporation, plated on YPDS agar containing Zeocin (100 μg/mL), and incubated for 5 d at 30°C. Sixty Zeocin-resistant colonies were then selected and screened for expression in BMMY using daily additions of 0.5% (v/v) methanol. Expression levels were monitored by SDS-PAGE followed by silver staining or immunoblotting using an anti-His monoclonal antibody. The most strongly expressing clone was selected for large-scale production in BMMY. Owing to an extraordinary protease resistance of the recombinant SMB, we used an unusually long induction period of up to 10 d for production in Pichia.
For 15N and 13C labeling, the abovementioned Pichia clone was expanded by growth in 2% (w/v) 13C glucose and 1.2% (w/v) (15NH4)2SO4. The yeast was pelleted by centrifugation (5000g), resuspended in a medium containing 1.2% (w/v) (15NH4)2SO4 and supplemented daily with 1% (v/v) (13C) methanol for 5 d while grown at 30°C during orbital shaking at 300 rpm.
SMBPichia was purified by Ni2+ NTA affinity chromatography followed by reversed-phase chromatography on a VYDAC C4 column using a 50-min linear gradient from 0% to 70% (v/v) acetonitrile in 0.1% (v/v) TFA. This protocol led to a purified SMBPichia with an average isotope incorporation close to 100% for the doubly labeled product as judged by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry, MALDI-TOF MS (average mass 6481.3 Da). The yield of purified SMB was 3–4 mg/L when Pichia was cultured on minimal medium.
Global amide hydrogen (1H/2H) exchange analysis of SMBS2, SMBPichia, and SMBplasma monitored by mass spectrometry
Prior to the isotopic exchange experiment, a solution containing SMBS2, SMBPichia, and SMBplasma was prepared, in which their concentrations were adjusted to yield similar relative ion abundances when analyzed concomitantly by electrospray ionization mass spectrometry. The concentrations were 20 μM SMBS2, 30 μM SMBPichia, and 60 μM SMBplasma. This difference reflects the distinct ionization efficiencies of the three SMB variants. Isotope exchange was initiated by a 50-fold dilution of the SMB mixtures into deuterated PBS at 25°C or 4°C. For time-course experiments, 75-μL aliquots were withdrawn after 1 min, 5 min, 15 min, and 50 min of deuterium exchange, and further isotope exchange was quenched by addition of 3 μL of 2.5% (v/v) TFA followed by snap freezing in liquid nitrogen. Fully deuterated SMB (100% 2H control) was prepared by 60 min of incubation in deuterated solvents at 25°C and 15 min at 55°C. Complex formation with PAI-1 was achieved by adding a ninefold molar excess of PAI-1 to one aliquot of the SMB mixture and incubating for 1 h at 25°C prior to isotope exchange. Protein concentrations were determined by amino acid analysis (Biochrom 30 amino acid analyzer). Pairwise comparison of recombinant SMBPichia and the three synthetic SMB products with predefined disulfide connectivities were performed in a similar manner.
Rapid desalting of SMB was performed with the liquid-chromatography system described previously (Jørgensen et al. 2004). Solvents were delivered by two HPLC syringe pumps, one for desalting and one for elution directly into the electrospray ion source of the mass spectrometer. Samples were manually thawed and injected with an ice-cold glass syringe into a stainless steel loop mounted on an injection valve. By switching the position of this valve, the sample was flushed from the loop to another valve equipped with a reversed-phase C18 microcolumn (Rist et al. 2005) and was desalted at a 400 μL/min flow of 0.05% (v/v) TFA for 1 min. A step gradient was used to separate the SMB variants from PAI-1, as the mass spectrometric peaks from PAI-1 interfered with those of the SMB variants. Elution of the SMB variants was achieved with 21% (v/v) acetonitrile (30 μL/min), 0.05% (v/v) TFA, and PAI-1 with 56% (v/v) acetonitrile. The total time from injection to mass spectrometric detection was 2 min for SMB and 6 min for PAI-1. The solvents, valves, and C18 microcolumn were maintained at 0°C in an ice-water bath. The liquid-chromatography system was coupled to an electrospray ionization quadrupole time-of-flight mass spectrometer (QToF Ultima, Micromass). The capillary voltage was set to 3.5 kV, the cone voltage to 55 V, the RF lens 1 voltage to 70 V, and the ion source block temperature to 80°C with a desolvation gas flow of 400 L/h at 200°C and a nebulizing gas flow of 20 L/h at room temperature. The deuterium content was determined by the average mass difference between the nondeuterated protein and the partly deuterated protein. The residual proton content after isotope exchange was attained using the average mass difference between the partly deuterated protein and the 100% 2H control. Average masses were calculated by two independent methods. In the first, average mass shifts were determined from deconvoluted mass spectra generated by the maximum entropy algorithm (MaxEnt 1) provided with the Masslynx software. In the second, the deuterium contents were determined using the HXexpress software (Weis et al. 2006). Nearly identical results were achieved by either method, and the data shown were obtained with HXexpress.
NMR spectroscopy
The protein concentration of the doubly labeled preparation was measured by absorbance at 280 nm prior to lyophilization using an extinction coefficient of 4470 M−1 cm−1. The NMR sample contained ∼0.4 mM 13C,15N-labeled SMB adjusted to pH 6.5 in 10% (v/v) D2O and 3 mM NaN3. All spectra were recorded at 298°K on a Varian Unity Inova 750 MHz NMR spectrometer equipped with a cold probe and Z-field gradient. The spectra were processed in nmrDraw (Delaglio et al. 1995) and analyzed in Pronto3D (Kjær et al. 1994) or Sparky (Goddard and Kneller). The backbone assignments were obtained for residues 3–47 by a standard procedure (Cavanagh et al. 1995) using HSQC, HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra (all pulse sequences were from the Varian Protein Pack version 1.6c). Side-chain assignments were obtained using N-edited TOCSY-HSQC and HCCH-TOCSY spectra. The chemical shifts have been deposited in BMRB (acc. code 15271). NOE restraints were generated from 2D NOESY, N-edited NOESY-HSQC, and C-edited NOESY-HSQC with mixing times of 150 ms. NOESY peaks of at least twice the noise were picked by visual inspection and compiled into NOE peak lists for subsequent structure calculation. Twenty-one pairs of ϕ and φ angle intervals were generated using TALOS (Cornilescu et al. 1999) from the list of chemical shifts and added as restraints in the structure calculations. Automated NOE assignment and initial structure calculations were carried out using the noeassign module of CYANA 2.21 (Güntert et al. 1997; Herrmann et al. 2002) with a maximal upper NOE distance set at 6.5 Å and calibrated using a median NOE length of 4.5 Å. In each of the seven rounds of iterative NOE assignment and structure calculation, 100 structures were calculated during 10,000 steps of torsion angle dynamics. Prior to the NOE assignments, the methyl and aromatic regions were interactively assigned, and these were kept fixed during the automatic assignments. No restraints representing the disulfide bonds were introduced, to avoid bias toward a specific disulfide pairing. In the last four calculations, the initial distance assignments were allowed an elasticity of 50% within the fixed upper limit. The C-terminal His6-tag was excluded from the structure calculations, since no resonances were attributed to this part of the molecule. Several undefined resonances were introduced into the automatic NOE assignments as free-floating proxy atoms (Ab et al. 2006). This led to assignment of the ɛ-proton of Arg8, the δ-protons of Asn14, and the γ-proton of Gln20 by inspection of the NOEs of the proxy atoms, and these assignments were used in subsequent calculations. The CYANA calculations resulted in a set of structures with RMSD <3 Å in the first round, and the overall structure did not change in subsequent rounds in compliance with the quality criteria recommended (Herrmann et al. 2002). Subsequently, restraints representing the three different disulfide configurations published previously were introduced in three different steps of calculations, and 100 structures were calculated during 5000 steps of torsion angle dynamics in CYANA. The 20 best structures in terms of number of violations were transferred to CNS (Brunger et al. 1998) using FormatConverter (Vranken et al. 2005). In CNS, a brief restrained molecular dynamics run was performed in a thin layer of explicit solvent in order to improve the geometry of the models as described (Linge et al. 2003). The structures were analyzed using Procheck-NMR (Laskowski et al. 1996). The statistics for the water-refined structures can be seen in ▶. The 20 structures with the lowest energy and the Cys5–Cys21, Cys9–Cys39, Cys19–Cys32, Cys25–Cys31 disulfide configuration (SMB-2 in ▶) were selected and submitted to the PDB (acc. code: 2jq8).
Acknowledgments
We thank Dr. W. Lu (Institute of Human Virology, University of Maryland Biotechnology Institute) for the kind gift of synthetic SMB domains with predefined disulfide connectivities. We thank Gitte Juhl Funch, John Post, and Yvonne DeLotto for excellent technical assistance and Dr. Birthe B. Kragelund and Benedikte Jacobsen for helpful comments. We thank “The Biocampus Program of The University of Copenhagen” for a scholarship to M.K. and The John and Birthe Meyer Foundation, The Lundbeck Foundation, and The Danish Cancer Society for financial support.
Footnotes
Reprint requests to: Michael Ploug, Finsen Laboratory, Rigshospitalet section 3735, Copenhagen Biocenter Room 3.3.31, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark; e-mail: m-ploug@finsenlab.dk; fax: 45-35453797.
Abbreviations: HSQC, heteronuclear single quantum coherence; MALDI-TOF MS, matrix assisted laser desorption ionization time-of-flight mass spectrometry; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; PAI-1, plasminogen activator inhibitor-1; RMSD, root mean square deviation; SMB, somatomedin B; uPA, urokinase-type plasminogen activator; uPAR, urokinase-type plasminogen activator receptor.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072949607.
References
- Ab E., Pugh, D.J.R., Kaptein, R., Boelens, R., and Bonvin, A.M. 2006. Direct use of unassigned resonances in NMR structure calculations with proxy residues. J. Am. Chem. Soc. 128 7566–7571. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1995. Protein NMR spectroscopy: Principles and practice. Academic Press, New York.
- Cheek S., Krishna, S.S., and Grishin, N.V. 2006. Structural classification of small, disulfide-rich protein domains. J. Mol. Biol. 359 215–237. [DOI] [PubMed] [Google Scholar]
- Chen V.M. and Hogg, P.J. 2006. Allosteric disulfide bonds in thrombosis and thrombolysis. J. Thromb. Haemost. 4 2533–2541. [DOI] [PubMed] [Google Scholar]
- Cheresh D.A., Smith, J.W., Cooper, H.M., and Quaranta, V. 1989. A novel vitronectin receptor integrin (αvβx) is responsible for distinct adhesive properties of carcinoma cells. Cell 57 59–69. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Deng G., Curriden, S.A., Wang, S., Rosenberg, S., and Loskutoff, D.J. 1996a. Is plasminogen activator inhibitor-1 the molecular switch that governs urokinase receptor-mediated cell adhesion and release? J. Cell Biol. 134 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G., Royle, G., Wang, S., Crain, K., and Loskutoff, D.J. 1996b. Structural and functional analysis of the plasminogen activator inhibitor-1 binding motif in the somatomedin B domain of vitronectin. J. Biol. Chem. 271 12716–12723. [DOI] [PubMed] [Google Scholar]
- Ekmekci H., Sonmez, H., Ekmekci, O.B., Ozturk, Z., Domanic, N., and Kokoglu, E. 2002. Plasma vitronectin levels in patients with coronary atherosclerosis are increased and correlate with extent of disease. J. Thromb. Thrombolysis 14 221–225. [DOI] [PubMed] [Google Scholar]
- Gårdsvoll H. and Ploug, M. 2007. Mapping of the vitronectin binding site on the urokinase receptor. Involvement of a coherent receptor interface comprising residues from both domain I and the flanking interdomain region. J. Biol. Chem. 282 13561–13572. [DOI] [PubMed] [Google Scholar]
- Gårdsvoll H., Werner, F., Søndergaard, L., Danø, K., and Ploug, M. 2004. Characterization of low-glycosylated forms of soluble human urokinase receptor expressed in Drosophila Schneider 2 cells after deletion of glycosylation-sites. Protein Expr. Purif. 34 284–295. [DOI] [PubMed] [Google Scholar]
- Gårdsvoll H., Gilquin, B., Le Du, M.H., Menéz, A., Jørgensen, T.J., and Ploug, M. 2006. Characterization of the functional epitope on the urokinase receptor. Complete alanine scanning mutagenesis supplemented by chemical cross-linking. J. Biol. Chem. 281 19260–19272. [DOI] [PubMed] [Google Scholar]
- Gårdsvoll H., Hansen, L.V., Jørgensen, T.J., and Ploug, M. 2007. A new tagging system for production of recombinant proteins in Drosophila S2 cells using the third domain of the urokinase receptor. Protein Expr. Purif. 52 384–394. [DOI] [PubMed] [Google Scholar]
- Goddard T.D. and Kneller, D.G. SPARKY 3. University of California, San Francisco.
- Güntert P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert, P., and Wüthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319 209–227. [DOI] [PubMed] [Google Scholar]
- Horn N.A., Hurst, G.B., Mayasundari, A., Whittemore, N.A., Serpersu, E.H., and Peterson, C.B. 2004. Assignment of the four disulfides in the N-terminal somatomedin B domain of native vitronectin isolated from human plasma. J. Biol. Chem. 279 35867–35878. [DOI] [PubMed] [Google Scholar]
- Jørgensen T.J.D., Gårdsvoll, H., Danø, K., Roepstorff, P., and Ploug, M. 2004. Dynamics of urokinase receptor interaction with peptide antagonists studied by amide hydrogen exchange and mass spectrometry. Biochemistry 43 15044–15057. [DOI] [PubMed] [Google Scholar]
- Kamikubo Y., Okumura, Y., and Loskutoff, D.J. 2002. Identification of the disulfide bonds in the recombinant somatomedin B domain of human vitronectin. J. Biol. Chem. 277 27109–27119. [DOI] [PubMed] [Google Scholar]
- Kamikubo Y., DeGuzman, R., Kroon, G., Curriden, S., Neels, J.G., Churchill, M.J., Dawson, P., Oldziej, S., Jagielska, A., Scheraga, H.A., et al. 2004. Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin. Biochemistry 43 6519–6534. [DOI] [PubMed] [Google Scholar]
- Kamikubo Y., Kroon, G., Curriden, S.A., Dyson, H.J., and Loskutoff, D.J. 2006. The reduced, denatured somatomedin B domain of vitronectin refolds into a stable, biologically active molecule. Biochemistry 45 3297–3306. [DOI] [PubMed] [Google Scholar]
- Kjær M., Andersen, K.V., and Poulsen, F.M. 1994. Automated and semiautomated analysis of homo- and heteronuclear multidimensional nuclear magnetic resonance spectra of proteins: The program Pronto. Methods Enzymol. 239 288–307. [DOI] [PubMed] [Google Scholar]
- Kjøller L. and Hall, A. 2001. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J. Cell Biol. 152 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche Y., Storme, V., De Meutter, J., Messens, J., and Lauwereys, M. 1994. High-level secretion and very efficient isotopic labeling of tick anticoagulant peptide (TAP) expressed in the methylotrophic yeast, Pichia pastoris . Biotechnology (N. Y.) 12 1119–1124. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Li X., Zou, G., Yuan, W., and Lu, W. 2007. Defining the native disulfide topology in the somatomedin B domain of human vitronectin. J. Biol. Chem. 282 5318–5326. [DOI] [PubMed] [Google Scholar]
- Linge J.P., Williams, M.A., Spronk, C.A.E.M., Bonvin, A.M.J.J., and Nilges, M. 2003. Refinement of protein structures in explicit solvent. Protein Struct. Funct. Genet. 50 496–506. [DOI] [PubMed] [Google Scholar]
- Ludvigsen S. and Poulsen, F.M. 1992. Positive theta-angles in proteins by nuclear magnetic resonance spectroscopy. J. Biomol. NMR 2 227–233. [DOI] [PubMed] [Google Scholar]
- Lynn G.W., Heller, W.T., Mayasundari, A., Minor, K.H., and Peterson, C.B. 2005. A model for the three-dimensional structure of human plasma vitronectin from small-angle scattering measurements. Biochemistry 44 565–574. [DOI] [PubMed] [Google Scholar]
- Mayasundari A., Whittemore, N.A., Serpersu, E.H., and Peterson, C.B. 2004. The solution structure of the N-terminal domain of human vitronectin: Proxymal sites that regulate fibrinolysis and cell migration. J. Biol. Chem. 279 29359–29366. [DOI] [PubMed] [Google Scholar]
- Molinari M. 2007. N-Glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3 313–320. [DOI] [PubMed] [Google Scholar]
- Preissner K.T. and Seiffert, D. 1998. Role of vitronectin and its receptors in haemostasis and vascular remodeling. Thromb. Res. 89 1–21. [DOI] [PubMed] [Google Scholar]
- Reheman A., Gross, P., Yang, H., Chen, P., Allen, D., Leytin, V., Freedman, J., and Ni, H. 2005. Vitronectin stabilizes thrombi and vessel occlusion but plays a dual role in platelet aggregation. J. Thromb. Haemost. 3 875–883. [DOI] [PubMed] [Google Scholar]
- Rist W., Mayer, M.P., Andersen, J.S., Roepstorff, P., and Jørgensen, T.J.D. 2005. Rapid desalting of protein samples for online microflow electrospray ionization mass spectrometry. Anal. Biochem. 342 160–162. [DOI] [PubMed] [Google Scholar]
- Seiffert D. and Loskutoff, D.J. 1991. Evidence that type 1 plasminogen activator inhibitor binds to the somatomedin B domain of vitronectin. J. Biol. Chem. 266 2824–2830. [PubMed] [Google Scholar]
- Seiffert D. and Smith, J.W. 1997. The cell adhesion domain in plasma vitronectin is cryptic. J. Biol. Chem. 272 13705–13710. [DOI] [PubMed] [Google Scholar]
- Seiffert D., Ciambrone, G., Wagner, N.V., Binder, B.R., and Loskutoff, D.J. 1994. The somatomedin B domain of vitronectin. Structural requirements for the binding and stabilization of active type 1 plasminogen activator inhibitor. J. Biol. Chem. 269 2659–2666. [PubMed] [Google Scholar]
- Stefanson S. and Lawrence, D.A. 1996. The serpin PAI-1 inhibits cell migration by blocking integrin (αvβ3) binding to vitronectin. Nature 383 441–443. [DOI] [PubMed] [Google Scholar]
- Thornton J.M. 1981. Disulphide bridges in globular proteins. J. Mol. Biol. 151 261–287. [DOI] [PubMed] [Google Scholar]
- Vranken W.F., Boucher, W., Stevens, T.J., Fogh, R.H., Pajon, A., Llinas, M., Ulrich, E.L., Markley, J.L., Ionides, J., and Laue, E.D. 2005. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59 687–696. [DOI] [PubMed] [Google Scholar]
- Wayner E.A., Orlando, R.A., and Cheresh, D.A. 1991. Integrins αvβ3 and αvβ5 contribute to cell attachment to vitronectin but differentially distribute on the cell surface. J. Cell Biol. 113 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Waltz, D.A., Rao, N., Drummond, R.J., Rosenberg, S., and Chapman, H.A. 1994. Identification of the urokinase receptor as an adhesion receptor for vitronectin. J. Biol. Chem. 269 32380–32388. [PubMed] [Google Scholar]
- Weis D.D., Engen, J.R., and Kass, I.J. 2006. Semi-automated data processing of hydrogen exchange mass spectra using HX-express. J. Am. Soc. Mass Spectrom. 17 1700–1703. [DOI] [PubMed] [Google Scholar]
- White C.E., Kempi, N.M., and Komives, E.A. 1994. Expression of highly disulfide-bonded proteins in Pichia-pastoris . Structure 2 1003–1005. [DOI] [PubMed] [Google Scholar]
- Zhou A., Huntington, J.A., Pannu, N.S., Carrell, R.W., and Read, R.J. 2003. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 10 541–544. [DOI] [PubMed] [Google Scholar]