Abstract
Quantum mechanical optimizations of theoretical enzymes (theozymes), which are predicted catalytic arrays of biological functionalities stabilizing a transition state, have been carried out for a set of nine diverse enzyme active sites. For each enzyme, the theozyme for the rate-determining transition state plus the catalytic groups modeled by side-chain mimics was optimized using B3LYP/6–31G(d) or, in one case, HF/3–21G(d) quantum mechanical calculations. To determine if the theozyme can reproduce the natural evolutionary catalytic geometry, the positions of optimized catalytic atoms, i.e., covalent, partial covalent, or stabilizing interactions with transition state atoms, are compared to the positions of the atoms in the X-ray crystal structure with a bound inhibitor. These structure comparisons are contrasted to computed substrate–active site structures surrounded by the same theozyme residues. The theozyme/transition structure is shown to predict geometries of active sites with an average RMSD of 0.64 Å from the crystal structure, while the RMSD for the bound intermediate complexes are significantly higher at 1.42 Å. The implications for computational enzyme design are discussed.
Keywords: theozyme, active site structure, density functional theory, enzyme, biological catalysis
The computational investigation of mechanism and catalysis delivered by biomolecules is a rich and rapidly growing field of computational chemistry (Warshel and Levitt 1976; Sherwood 2000; Lin and Truhlar 2006; Senn and Thiel 2007). Progress in this area has been facilitated by the application of theoretical models representing a truncated active site. Such a model has been called a theozyme, short for theoretical enzyme, which embodies the predicted catalytic array of biological functionalities stabilizing a particular transition state (TS) (Na and Houk 1996; Tantillo et al. 1998). The active site is limited to the necessary catalytic residues, which are then further reduced to include hydrogen-capped mimics for the side-chain or peptide backbone functionalities.
Our group has utilized the theozyme model to gain valuable insights into the origin of catalysis and stereoselectivity delivered by catalytic antibodies (Na and Houk 1996; Na et al. 1996; Tantillo et al. 1998; Ujaque et al. 2002; Zhang et al. 2002; Cannizzaro et al. 2003; Leach et al. 2004). Domingo and coworkers applied similar models to design inhibitors for an aldol reaction (Arnó and Domingo 2001, 2003) and Landry and coworkers used this model to elucidate the mechanism of cocaine hydrolysis in human butyrylcholinesterase (Zhan et al. 2003). Venturini et al. applied theozymes to study the catalytic mechanism of HIV-1 protease (Venturini et al. 1998), which sparked experimental studies to develop a transition state analog (TSA) inhibitor (Hyland et al. 1991). The theozyme concept has been applied to study various metalloenzymes, such as cytochrome P450 (Harris et al. 2001), B12 cofactors (Dölker et al. 2003), iron hydrogenases (Cao and Hall 2001), and the selenoprotein glutathione peroxidase (Prabhakar et al. 2005). Siegbahn and coworkers have carried out the most extensive and effective use of the theozyme model in the study of mechanism and catalysis of several metal-dependent enzymatic processes such as nitrogenases (Siegbahn et al. 1998), heme peroxidases (Wirstam et al. 1999), methane monooxygenase (Siegbahn and Borowski 2006), ribonucleotide reductase (Cho et al. 2004), galactose oxidase (Himo et al. 2000), hydrogenases (Siegbahn et al. 2001), cytochrome c oxidase (Blomberg and Siegbahn 2003; Margareta and Siegbahn 2006), copper amine oxidases (Prabhakar and Siegbahn 2003), tyrosinase (Siegbahn and Wirstam 2001), and the manganese-based water-splitting complex in photosystem II (Lundberg et al. 2003).
Another application of the theozyme model is the prediction and design of enzymatic active sites that can deliver dramatic rate enhancements and stereoselectivity to nonnatural reactions. The design of active sites utilizing a transition state model has been pioneered for computational enzyme design methods by Baker (Zanghellini et al. 2006), Mayo (Lassila et al. 2006), and Hellinga (Dwyer et al. 2004). Although each design method is unique in detail, all rely on a model for the three-dimensional active site structure. The theozyme model provides a theoretical method to predict the geometry of the active site, and the merger with protein design methodology holds great potential for a general means to design enzymes to accelerate any target reaction (Zanghellini et al. 2006).
The connections between theoretical treatments of enzyme catalysts and experiment generally involve energetics and structural comparisons. The activation barriers of catalyzed reactions can be computed and measured. Such information is very course grained, as a multitude of mechanistic steps is manifested in only one (ΔG ‡ corresponding to k cat/Km) or two (ΔG ‡ and ΔG 0 corresponding to k cat and Km, respectively) energetic comparisons.
Structural data are much more detailed, since the position of all the atoms predicted by theory and measured experimentally can be compared. However, whereas theoretical methods such as QM/MM or even QM are able to locate transition states of enzyme-catalyzed reactions (Gao and Thompson 1998), no transition structures are available experimentally, since a transition state has a lifetime of ∼10−13 s. Instead, enzymes liganded with transition state analogs, stable structures thought to be similar to transition states, are available by X-ray crystallography. It is assumed, but has not been verified for many cases, that the arrangement of side-chain atoms in the active site with an inhibitor/TSA bound is related to the arrangement of the groups in the TS of catalyzed reactions (Siegbahn and Borowski 2006). Here we test whether a QM theozyme actually predicts structures related to these functional protein crystal structures.
Many factors can cause a theozyme to differ from that of the crystal structure: (1) A theozyme omits the protein backbone, necessarily eliminating the influence of the fold of the protein scaffold to hold, fix, or orient the catalytic residues in the active site. In addition, catalytic residues generally make hydrogen-bonding interactions with second-shell protein residues that are usually excluded in the theozyme model. Thornton et al. noted that catalytic residues have limited conformational freedom due to the extensive hydrogen bonding network involved with both the main chain and side chain of these residues (Bartlett et al. 2002). In this work, only the aspartic acid proteinase theoyzme includes the secondary residues that are hypothesized to aid in orientation of the catalytic residues (Gutteridge and Thornton 2005). Furthermore, a theozyme involves truncation of residues, and where this can be done without altering the active geometries is not known in advance. (2) Theozyme optimizations are carried out in the gas phase in the absence of any constraints. Although enzymatic active sites are often believed to have relatively low dielectric constants (ɛ ∼ 4) compared to that of the surrounding bulk water (ɛ ∼ 80) (Simonson and Brooks III 1996), the active site might be better mimicked by a dielectric closer to water rather than to a vacuum. (3) The theozyme involves a transition state, often involving partial covalent bonds to the enzyme; the crystal structure contains a stable intermediate that is, at best, an analog of the transition state and is usually bound noncovalently.
To assess the predictive accuracy of theozymes, a diverse set of enzymes with differing mechanisms of catalysis was assembled. The active site structure predicted from the QM model was compared to the corresponding crystal structure. To better interpret the magnitude of the theozyme structural comparison results, the bound substrates were optimized with the catalytic residues and similarly compared to crystal structures. The goal is to assess quantitatively how well the predicted geometries of a theozyme and of a bound substrate correlate to the crystal structure including a bound stable inhibitor that only roughly mimics the transition state.
Results
Theozymes/transition states and substrate–active site complexes have been located for nine enzymes consisting of lyases, hydrolyases, and mutases summarized in ▶. ▶ gives the optimized theozyme structures, overlays with the corresponding X-ray structures, and calculated values of root-mean-square deviations for catalytic atoms, RMSDTS. ▶ gives the optimized substrate–active site complexes, overlays with the corresponding X-ray structures, and calculated values of root-mean-square deviation of the catalytic atoms, RMSDSUB. The catalytic atoms are depicted in the sphere representation in ▶ and ▶, column 3.
Table 1.
Enzymes used for benchmarks, PDB designations, and functions
Table 2.
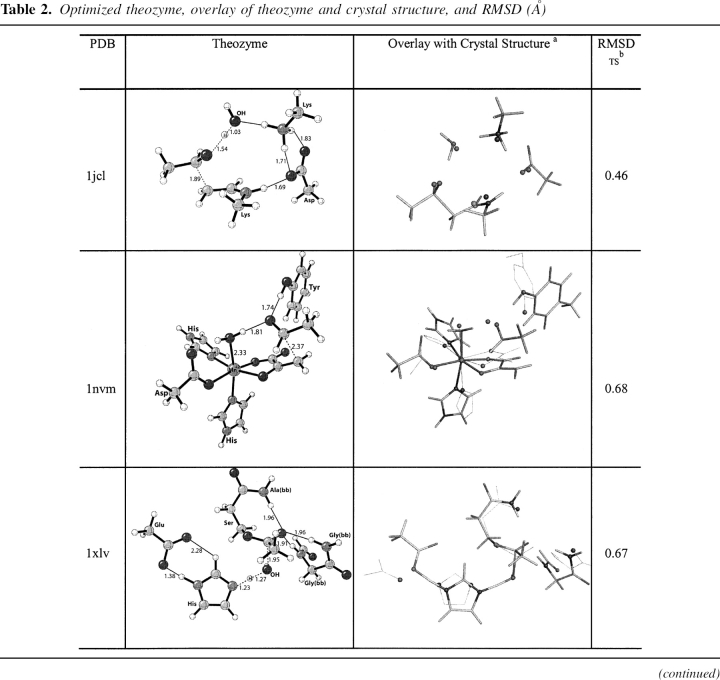
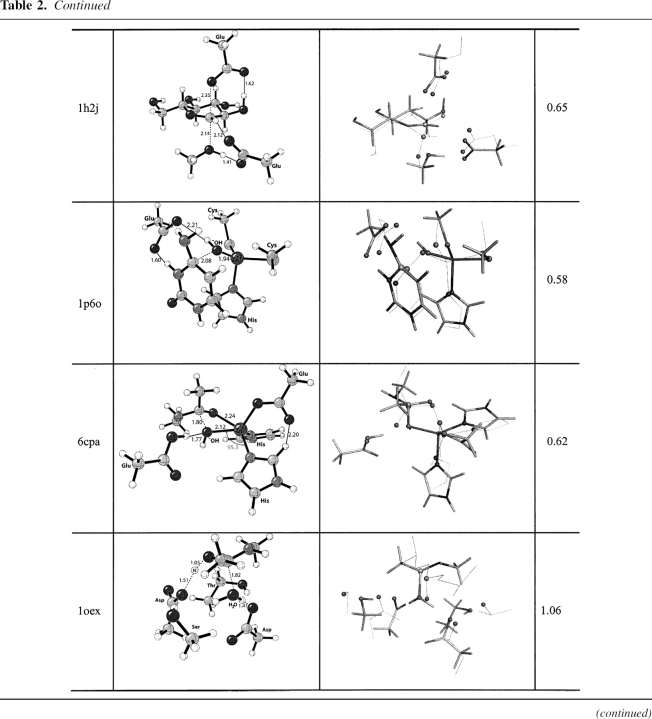
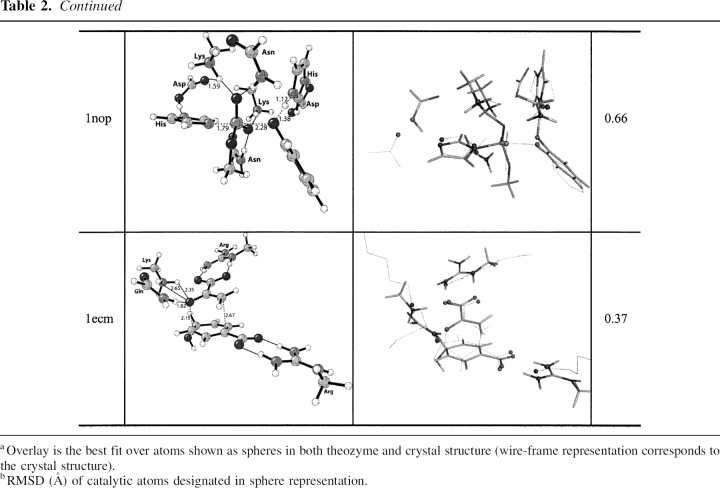
Optimized theozyme, overlay of theozyme and crystal structure, and RMSD (Å)
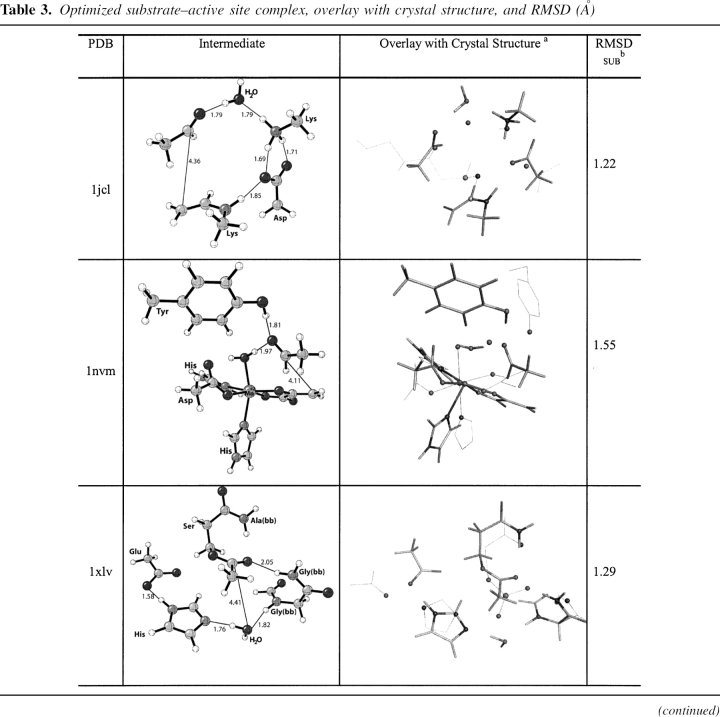
Table 3.
Optimized substrate–active site complex, overlay with crystal structure, and RMSD (Å)
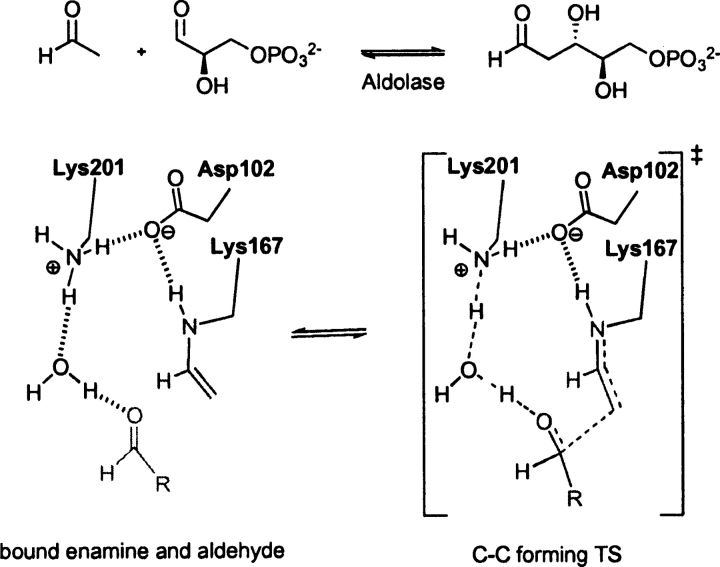
Escherichia coli D-2-deoxyribose-5-phosphate aldolase (PDB code 1jcl)
DERA, EC 4.1.2.4, a type I aldolase, catalyzes the aldol reaction of acetaldehyde and D-glyceraldehyde-3-phosphate (Heine et al. 2001). The enzymatic reaction mechanism involves three catalytic residues and a conserved active site water. The NH of Lys 167 functions as a nucleophile and attacks the substrate carbonyl carbon to form the enamine intermediate. Asp 102 and Lys 201 and a water molecule catalyze the reaction as general acid/general base. In the unbound state of the enzyme, Lys 167 exists in the neutral form. It is believed the close proximity (ɛ-N atom distance) to Lys 201 at 3.4 Å controls the pKa of this residue, rendering the side chain nucleophilic (Heine et al. 2001). The optimized theozyme corresponding to the C–C bond formation step (▶) is shown in ▶, PDB 1jcl.
Scheme 1.
Bound substrates (substrate–active site complex) and transition state (theozyme) for the DERA-catalyzed aldol reaction.
The theozyme structure geometry corresponds clearly to the crystal structure resulting in an RMSDTS of 0.46 Å, ▶, PDB 1jcl. The only structural difference between the theozyme and crystal structure is the two hydrogen-bonding interactions between Asp 102 and Lys 201 in the theozyme, whereas only one is observed in the crystal. This minor difference is likely due to the lack in the theozyme of the surrounding secondary hydrogen bonding interactions to lock the second carboxylate oxygen of Asp 102 away from the favorable second interaction with the positively charged lysine side chain. In the substrate–active site complex, however, the distant association of the carbon atoms of the aldehyde and enamine products at 4.36 Å creates a drastic shift in the positions of the side-chain mimics and the catalytic water, resulting in the relatively larger RMSDSUB of 1.22 Å (▶, PDB 1jcl).
Pseudomonas sp. 4-hydroxy-2-oxovalerate aldolase (PDB code 1nvm)
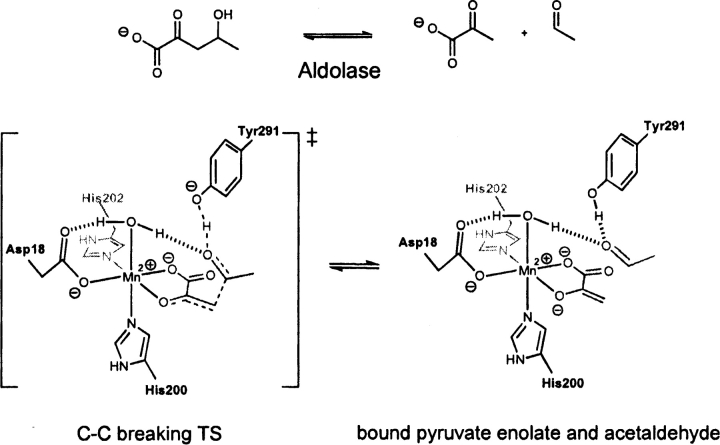
DmpG, EC 4.1.3.-, a type II aldolase, catalyzes the retro-aldol reaction of 4-hydroxy-2-ketovalerate (Manjasetty et al. 2003). The active site of DmpG binds Mn+2 in an octahedral fashion, utilizing the side chains of Asp 18, His 200, His 202, and three water molecules. The enolate substrate binds to the metal ion by displacing two waters occupying the in-plane positions of the octahedral, leaving the remaining water coordinated in an axial position. The metal ion then accelerates the subsequent retro-aldol reaction by Lewis-acid catalysis (▶). The other major component of catalysis is the side chain of Tyr 291, which assists as a general acid/base (Manjasetty et al. 2003). The optimized theozyme corresponding to the retro-aldol step is shown in ▶, PDB 1nvm.
Scheme 2.
Bound substrates (substrate–active site complex) and transition state (theozyme) for the DmpG-catalyzed retro-aldol reaction.
The crystal structure does not contain an inhibitor molecular structure to mimic the aldehyde functionality resulting from the retro-aldol reaction. However, a water molecule is observed in the crystal structure in a comparable position to the oxygen of this moiety in the theozyme. Therefore, this crystallographic water was used in the calculation of the RMSDTS of 0.68 Å reported in ▶, PDB 1nvm. Overall, the theozyme structure displays excellent correlation around the Mn+2 coordination geometry. The only notable geometric difference between the theozyme and the crystal structure is a small deviation in the position of the tyrosine (oxygen) side-chain catalytic atom.
As in the case in DERA, the substrate–active site complex of DmpG displays a relatively long distance between the retro-aldol products at 4.11 Å (▶, PDB 1nvm). Additionally, the aldehyde is positioned perpendicular to the Mn+2-bound enolate, causing the catalytic tyrosine mimic to invert its position relative to the crystal structure. These geometric divergences result in the larger RMSDSUB of 1.55 Å compared to the theozyme.
Human butyrylcholinesterase (PDB code 1xlv)
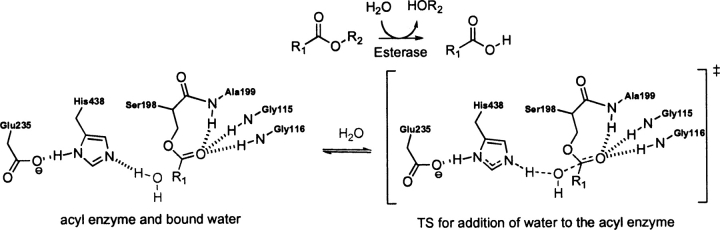
BChE, EC 3.1.1.8, catalyzes the hydrolysis of butyrylcholine to choline and butyric acid. The catalytic mechanism of butyrylcholinesterase involves a Ser–His–Glu catalytic triad acting as a nucleophile, attacking the carbonyl carbon of the substrate ester and generating an oxyanionic tetrahedral intermediate, which is stabilized by three backbone NH groups forming the oxyanion hole (Zhan et al. 2003).
The rate-determining step (RDS) for BChE-catalyzed hydrolysis of cocaine is the addition of water to the acyl enzyme (▶; Zhan et al. 2003). Presumably this is the case for any ester substrate. Therefore, the optimized theozyme corresponding to this TS was optimized (▶, PDB 1xlv) and compared to the crystal structure (Nachon et al. 2005). Ser 198 is modeled by N-methyl-3-propanolamide to best represent the dual role of this residue as both nucleophile and stabilizing group member of the oxyanion hole.
Scheme 3.
Bound acyl-enzyme intermediate and substrate (water) (substrate–active site complex) and transition state (theozyme) for the BChE-catalyzed hydrolysis of an ester.
The theozyme and crystal structure are similar, with an RMSDTS of 0.67 Å. The aspartate, however, is positioned very differently in the two structures: The theozyme predicts a syn Asp–His hydrogen bond as opposed to the anti-orientation observed in the crystal structure. There are smaller differences in the oxyanion hole atoms in the two structures.
The optimized substrate (water)-active site complex (▶, PDB 1xlv) is highly divergent from the crystal structure geometry, giving an RMSDSUB of 1.29 Å. In the bound-substrate structure, the water oxygen is quite far from the electrophilic carbonyl carbon of 4.41 Å, 2.46 Å longer than the distance in the transition structure. The glycine of the oxyanion hole is predicted to hydrogen bond to the water oxygen at 1.82 Å instead of the carbonyl oxygen in the substrate (▶, PDB 1xlv). The absence of this interaction in the ground state suggests an inherit instability of three hydrogen-bonding interactions to an unpolarized carbonyl oxygen.
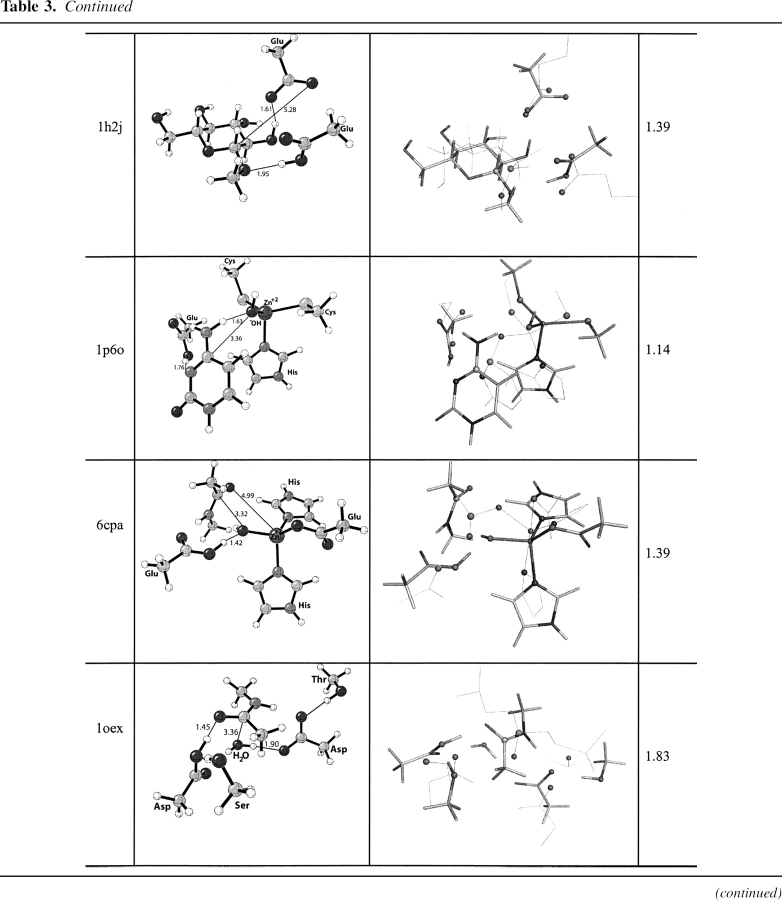
Bacillus agaradhaerens endoglucanase Cel5 A (PDB code 1h2j)
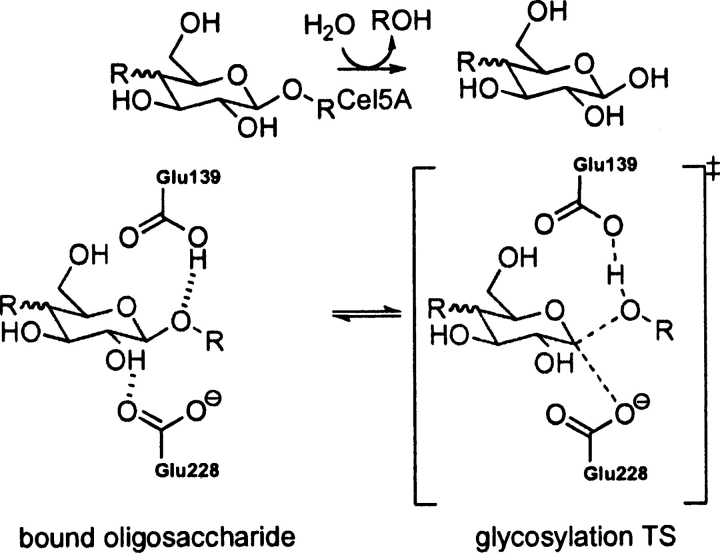
EC 3.2.1.4, catalyzes the cleavage of the β-1–4 glycosidic bond in cello-oligosaccharides with retention of stereochemistry (Davies et al. 1998; Blanchard and Withers 2001; Skopec et al. 2003; Varrot and Davies 2003). The catalytic residues are Glu 228 (nucleophile) and Glu 139 (general acid/base) (Davies et al. 1998; Skopec et al. 2003; Varrot and Davies 2003). The theozyme was optimized for the TS corresponding to glycosylation (▶) and compared to the crystal structure (Varrot and Davies 2003).
Scheme 4.
Bound substrate (substrate–active site complex) and transition state (theozyme) for the Cel5 A-catalyzed hydrolysis of oligosaccaharides.
Overlay of this theozyme with the crystal structure results in an RMSDTS of 0.65 Å (▶, PDB 1h2j). As displayed in the X ray, the theozyme predicts a hydrogen bond between Glu 228 and the appended alcohol functionality of the substrate C2 atom at 1.62 Å (▶, PDB 1h2j). This interaction is known to be critical for proficient catalysis (Notenboom et al. 1998). Additional electrostatic stabilization is observed in the theozyme structure involving Glu 139 and the anomeric carbon of the substrate. Rotation about the O—HO–Glu 228 partial bond in the transition structure predicts the energetic stabilization gained by this interaction to be ∼3.0 kcal/mol. The likely catalytic benefit is the relatively strong C+H—O interaction in the TS compared to the energetically benign CH—O interaction in the ground-state complex. Interestingly, the Glu 139 in the crystal structure is identically poised to make such an interaction. Similar stabilizing CH—O interactions on the order of a strong hydrogen bond have been reported by Corey and coworkers in complexes involving oxazaborolidinium salts and α,β-enals and α,β-unsaturated carbonyl compounds (Ryu et al. 2005).
The optimized substrate–active site complex (▶, PDB 1h2j) gives an RMSDSUB of 1.39 Å. The main difference between this structure and the crystal is the outward rotation of the nucleophilic carboxylate oxygen of Glu 228 from the electrophilic anomeric carbon to a distance of 5.38 Å, 2.93 Å longer than the distance in the transition structure (▶, PDB 1h2j).
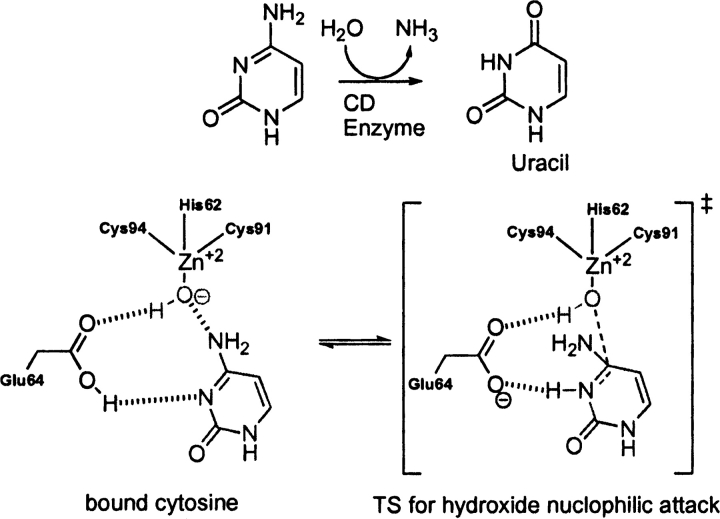
Yeast cytosine deaminase (PDB code 1p6o)
EC 3.5.4.1 catalyzes the deamination of cytosine to uracil and of 5-methylcytosine to thymine (Ko et al. 2003). The deamination of cytosine is initiated by nucleophilic attack of an activated water molecule bound to zinc, where His 62, Cys 91, and Cys 94 ligate the Zn+2 ion in a tetrahedral fashion. An adjacent Glu 64 participates in the reaction mechanism as a general acid/base catalyst. The likely RDS corresponds to nucleophilic attack of the zinc bound hydroxide to cytosine (▶). The overlay of the theozyme corresponding to this step and the crystal structure (Ireton et al. 2003) gives an RMSDTS of 0.58 Å, shown in ▶, PDB 1p6o.
Scheme 5.
Bound substrate (substrate–active site complex) and transition state (theozyme) in the cytosine deaminase-catalyzed hydrolysis of cytosine to uracil.
The theozyme catalytic atoms are in excellent geometrical agreement with the crystal structure. The bound cytosine-active site complex prior to nucleophic attack of the zinc-bound hydroxide (▶, PDB 1p6o), however, results in an RMSDSUB of 1.14 Å. The appreciable difference between RMSDTS and RMSDSUB is largely due to the distance of the hydroxide oxygen and electrophilic carbon of the substrate at 3.36 Å and the consequential geometrical adjustment of Glu 64.
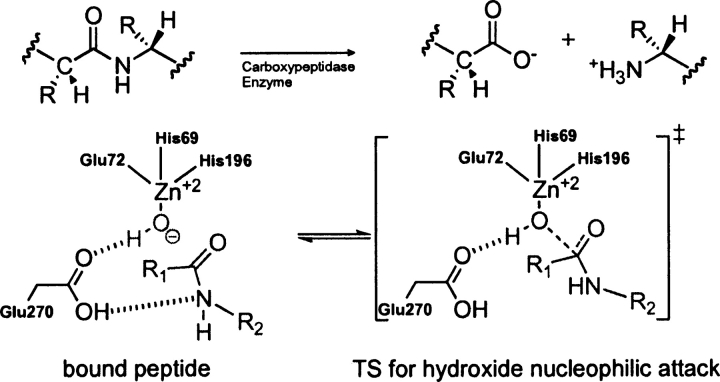
Bos Taurus carboxypeptidase (PDB code 6cpa)
EC 3.4.17.1, a zinc proteinase enzyme, cleaves amino acids successively from the C terminus of polypeptides (Mock 1997). The zinc-binding site is composed of two histidine residues and one glutamic acid that coordinates the metal ion in a tetrahedral geometry. A water molecule occupies the fourth tetrahedral position and acts as a nucleophile in the catalytic mechanism (▶). Glu 270 participates in the reaction as a general acid/base (Breslow et al. 1983; Christianson et al. 1987).
Scheme 6.
Bound substrate (substrate–active site complex) and transition state (theozyme) in the carboxypeptidase-catalyzed hydrolysis of a peptide.
A theozyme was optimized for the TS corresponding to nucleophilic attack of the zinc-bound hydroxide to the carbonyl carbon of the substrate, resulting in a RMSDTS of 0.62 Å (▶, PDB 6cpa) to the crystal structure (Kim and Lipscomb 1990). The most outstanding feature of the overlaid structures is the pronounced trigonal pyramidal geometry about the zinc center in the theozyme compared to the more tetrahedral-like zinc geometry in the crystal structure. This geometry is established in the theozyme by the angle of the attacking hydroxide–Zn–His(nitrogen) at 95.3°. The trigonal pyramidal geometry of zinc in the theozyme is due to strong interactions with five atoms in the transition structure, two nitrogens from His 69/196, oxygen from Glu 72, the amide carbonyl oxygen of the substrate that is developing a negative charge in the TS, and the oxygen of the hydroxide nucleophile.
Unlike the theozyme structure, the optimized substrate–active site complex (▶, PDB 6cpa) clearly predicts a tetrahedral geometry about the zinc ion, in better agreement with the crystal structure. However, the calculated RMSDSUB of 1.39 Å is significantly higher than that for the theozyme because the interaction between the zinc ion and the oxygen of the substrate amide is nonexistent at the distance of 4.99 Å. Therefore, the distance between the zinc-bound hydroxide and the electrophilic carbon of the substrate is subsequently increased by 1.52 Å more than that in the transition structure (▶, PDB 6cpa).
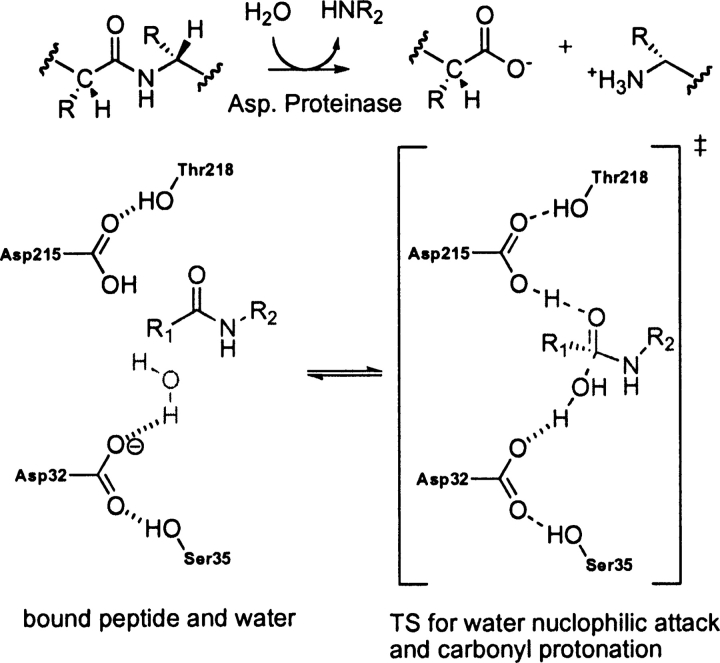
Cryphonectria parasitica aspartic proteinase Endothiapepsin (PDB code 1oex)
EC 3.4.23.- catalyzes the hydrolysis of a peptide bond utilizing an aspartate catalytic dyad (Tang and Wong 1987; Cascella et al. 2005). The two catalytic residues (Asp 32 and Asp 215) alternate between catalytic acid and base (Hofmann et al. 1984; James and Sielecki 1986; Brik and Wong 2003). A water molecule activated by the base asparate (Asp 32) attacks the carbonyl oxygen of substrate that is protonated by the acid aspartate (Asp 215) as shown in ▶ (Beveridge 1998; Frazao et al. 1999). The crystal structure shows that the catalytic residues also interact with side chains of Ser 35 and Thr 218 (Frazao et al. 1999). These secondary residues are thought to orient of the aspartate residues in the active site and to control the local pKa (Gutteridge and Thornton 2005). The theozyme compared to the crystal structure gives an RMSDTS of 1.06 Å (▶, PDB 1oex).
Scheme 7.
Bound substrate (substrate–active site complex) and transition state (theozyme) in the aspartic proteinase-catalyzed hydrolysis of a peptide.
The geometry of the aspartate residues, which are involved in partial covalent interactions in the transition structure, display good agreement with the crystal structure, particularly the one responsible for activating the water molecule for nucleophile attack (Asp 32), as shown in ▶, PDB 1oex. An RMSDTS improves to 0.74 Å when excluding the secondary residues.
The optimized substrate–active site complex (▶, PDB 1oex) predicts the nucleophilic water to be quite far from the substrate electrophilic amide carbon at 3.36 Å, 1.54 Å longer than the distance in the transition structure. As expected, the RMSDSUB is significantly higher at 1.83 Å, due to the reconfiguration of the catalytic aspartic acid models around this water molecule.
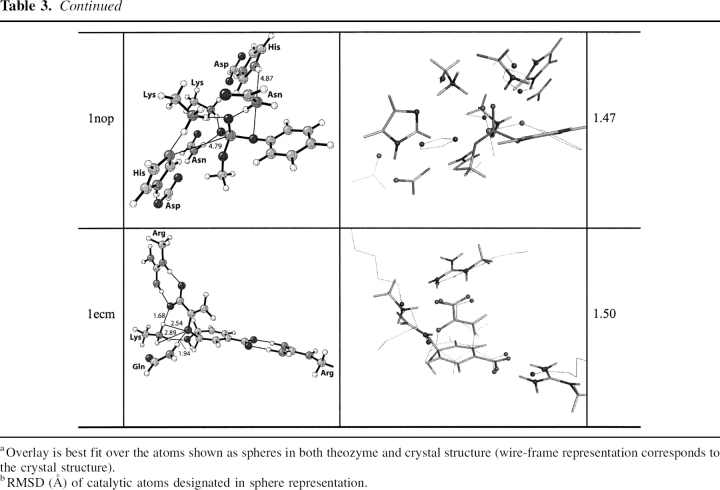
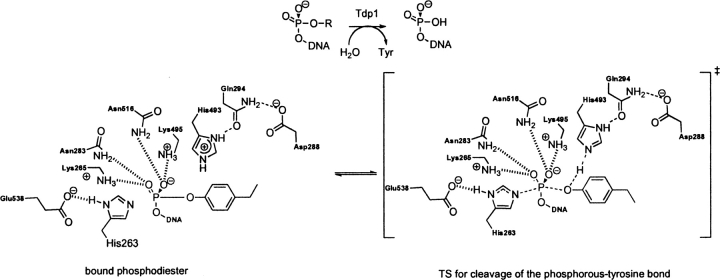
Human tyrosyl-DNA-phosphodiesterase (PDB code 1nop)
Tdp1, EC 4.6.1.13, a DNA repair enzyme, catalyzes the hydrolysis of a phosphodiester bond between a tyrosine residue and a DNA 3′ or 5′ phosphate (Yang et al. 1996; Nitiss et al. 2006). The catalytic mechanism involves first, a nucleophilic attack of His 263 on the phosphate, displacing the tyrosine in an SN2 fashion. The tyrosine is then protonated by the nearby general acid/base His 493. The nucleophilic His 263 is stabilized by Glu 538, and the tautomeric state of the general acid/base His 493 is controlled via a hydrogen bond to the carbonyl oxygen of the nearby Gln 294 residue. Lys 265/495 and Asn 283/516 residues stabilize a negative charge on the oxygen–phosphorous bonds in the substrate, facilitating cleavage of the phosphorous–tyrosine bond. The second step of the mechanism involves nucleophile attack of an activated water molecule to regenerate the catalyst (Davies et al. 2002b). It remains unclear which nucleophilic step corresponds to the RDS. In the crystal structure the TSA–protein complex is designed to mimic the TS for the nucleophilic attack of His 263 (▶; Davies et al. 2002a). The optimized theozyme corresponding to this step is shown in ▶, PDB 1nop. A modification was applied to the theozyme model to approximate the Gln294–Asp288–His493 hydrogen-bond network by eliminating the mediating Gln 294 residue. Therefore, RMSD calculations exclude residues Gln 294 and Asp 288. Overlay of the theozyme with the crystal structure results in an RMSDTS of 0.66 Å (▶, PDB 1nop).
Scheme 8.
Bound substrate (substrate–active site complex) and transition state (theozyme) in the Tdp1-catalyzed hydrolysis of a phosphodiester.
The largest geometrical deviation in the theozyme compared to the crystal occurs with the Glu 538–Lys 265 hydrogen-bonding interaction at 1.59 Å. Nevertheless, the geometry of the residues involved in the partial covalent interactions in the transition structure, namely His 216 which is forming a covalent bond with the substrate and His 493 which is protonating the leaving group, exhibit excellent agreement with the crystal structure, as shown in ▶, PDB 1nop.
The optimized substrate–active site complex is shown in ▶, PDB 1nop. Similar to the previous benchmark enzymes, the RMSDSUB of 1.47 Å is higher than the RMSDTS. The large deviation here is due to the distance between His 216 (nitrogen)–phosphorous (substrate) at 4.79 Å and the resultant amino acid geometrical perturbations.
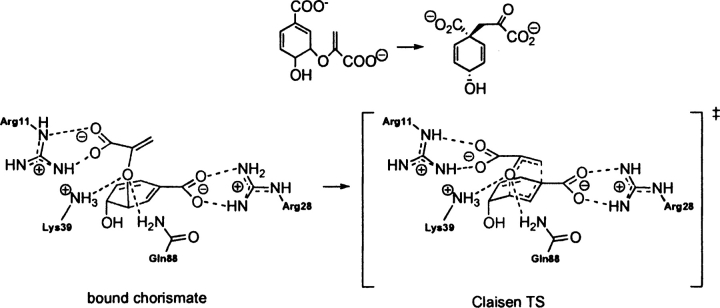
E. coli chorismate mutase (PDB 1ecm)
EC 5.4.99.5 catalyzes the Claisen rearrangement of chorismate to prephenate (▶). Several catalytic antibodies have been raised against Bartlett's stable TSA. This system has been the subject of numerous experimental (Kast and Hilvert 1996; Gamper et al. 2000) and theoretical investigations (Wiest and Houk 1994; Khanjin et al. 1999; Guo et al. 2001; Lee et al. 2002; Woodcock et al. 2003; Ranaghan et al. 2004; Guimarães et al. 2005). In recent years the origin of enzymatic rate enhancement has been the subject of heated debate (Guimarães et al. 2003; Martí et al. 2003; Štrajbl et al. 2003) sparked by the near attack conformer (NAC) proposal advocated by Hur and Bruice (2003a,b,c).
Scheme 9.
Bound reactant (substrate–active site complex) and transition state (theozyme) in the conversion of chorismate to prephenate in chorismate mutase.
The theozyme structure corresponding to the transition state for chorismate mutase, a [3,3]-sigmatropic shift is shown in ▶, PDB 1ecm. The transition state is stabilized by an array of noncovalent interactions, which include arginine residues coordinated to each carboxylate group and lysine and glutamine residues stabilizing the ether oxygen.
The RMSDTS for the theozyme structure compared to the X-ray structure (Lee et al. 1995) is 0.37 Å (▶, PDB 1ecm). The theozyme accurately reproduces the catalytically critical Lys 39 and Gln 88 hydrogen bonds to the developing negative charge to the ether oxygen in the transition structure. In the chorismate-bound geometry (▶, PDB 1ecm), however, the negative charge of this oxygen is diminished, rendering hydrogen-bonding interactions weaker, as supported by the increased hydrogen-bonding distances of 0.19 Å and 0.24 Å to Lys 39 and 0.12 Å to Gln 88. The geometrical positions of these hydrogen bonds are guided by the lone pair projections on the ether oxygen in the reactant, which differ from that in the transition structure. Thus, the geometry of the reactant complex leads to a higher RMSDSUB of 1.50 Å compared to the theozyme.
Discussion
The theozyme model optimizes positions of the protein side chains that stabilize a particular transition state. In contrast, the crystal structure is a stable ground state structure involving a stable transition state analog. At the onset of this study it was not known how close the geometry of the catalytic residues in the transition state would be to those binding an inhibitor/TSA, considering the inherent differences in the complexes, especially, the atoms that compose the inhibitor or TSA molecule do not exactly match the geometry of the transition structure for the actual substrate. However, all benchmark theozymes achieved an RMSDTS to the crystal structure of ≤1.06 Å with an average of 0.64 Å. The critical catalytic atom locations of the residues involved in each theozyme remain remarkably consistent with the corresponding crystal structure geometry, despite the many approximations used in the truncated active site/theozyme model. While the crystal structure determination involves errors, as does the quantum mechanical method used for the theozyme optimizations, the correspondence between these two structures does not imply coincidence. Consequently, the exclusion of the active site cavity backbone and secondary hydrogen-bonding interactions in all of the benchmark enzymes does not impede the ability of the theozyme model to recapitulate naturalistic active site structure. The results support the notion that the enzyme has evolved to optimize the positions of catalytic groups for the act of catalysis.
In contrast to the transition state/theozyme model, the optimized geometries of the active site and a bound substrate often deviate from the geometries of the active site structure observed in the X ray. The ground state structures exhibit weaker associations of the atoms involved in the bond-making/-breaking interactions in the TS. Indeed, the average enzyme substrate association is 106 M−1, while that of the enzyme and transition state is 1017 M−1 (Houk et al. 2003). The adjustment of the amino acid side chains to the substrate-binding geometry results in relatively larger RMSDSUB values to an average of 1.43 Å. The values for the RMSDTS and RMSDSUB recalculated excluding the specific catalytic atoms from the transition state and substrate yield similar results (see Supplemental material).
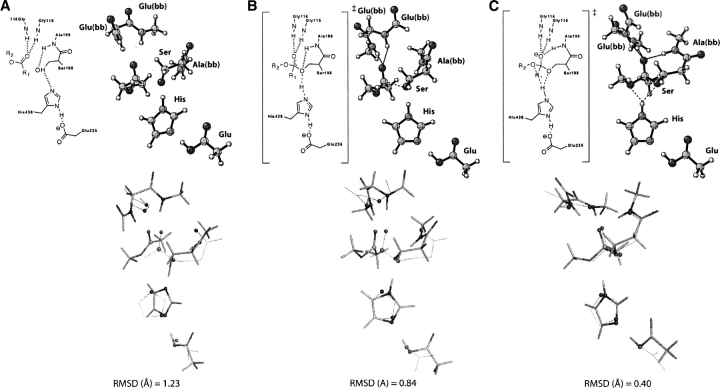
The enzyme must stabilize all of the transition structures involved in the mechanism. ▶ shows theozymes for two transition states and the bound ester substrate involved in the hydrolysis of an ester by the serine protease, butyrylcholinesterase. These are in addition to that shown earlier in ▶. As before, each structure is optimized in the presence of the catalytic functional groups, and RMSD values are determined with respect to the crystal structure. The general trend that the transition states give low RMSDs while the substrate–active site structures yield relatively higher RMSDs is reproduced. Notably, the theozyme corresponding to the nucleophilic attack of the serine side chain gives a larger RMSD (0.84 Å); (▶) compared to the other calculated theozymes for histidine hydrogen bond migration (0.40 Å); (▶) and attack of water to the acyl enzyme (0.67 Å); (▶, PDB 1xlv). The outstanding structural characteristic of the lower RMSD theozymes is the recapitulation of the three-residue oxyanion hole observed in the crystal structure.
Figure 1.
Optimized structures for butyrylcholinesterase and catalytic atom (sphere representation) overlays with the crystal structure (wire-frame representation) for (A) ester substrate bound, (B) TS for nucleophilic attack of the serine side chain to carbonyl carbon of ester, and (C) TS for histidine hydrogen-bond migration from the serine side-chain oxygen to the ester oxygen of the substrate.
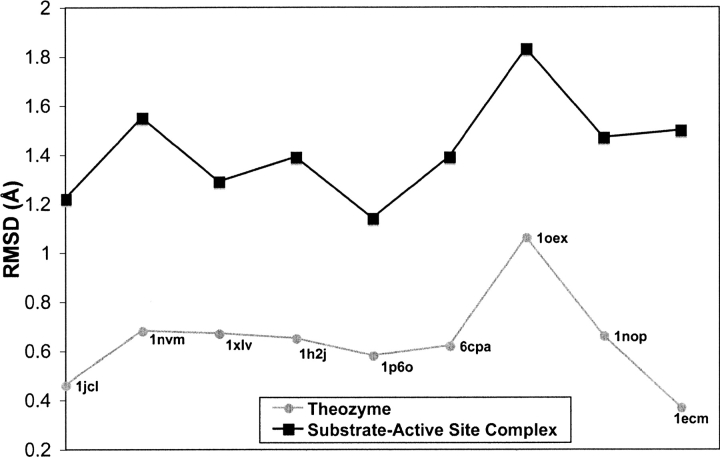
The close structural union of the crystal structure with the transition state/theozyme rather than the bound substrate complexes is clearly established in ▶, which summarizes the results from RMSDTS and RMSDSUB calculations (▶ and ▶). This plot implies that the optimization of the transition state in the presence of the catalytic biological functionality best mimics the geometrical design observed in nature, since optimal stabilization of the transition state(s) determines the geometry of the active site.
Figure 2.
Summary of RMSD (Å) for optimized theozymes and active site–substrate complexes compared to the corresponding crystal structure.
In summary, theozymes (transition structures optimized in the presence of enzyme catalytic functionality) obtained with B3LYP density functional in the gas phase can predict active site geometries involving a variety of common enzymatic functionalities evolved to efficiently stabilize transition states. This close structural agreement between the theozyme and enzyme structure is in accord with the paradigm wherein the enzyme structure assumes maximum free energy lowering of the transition state through covalent (Houk et al. 2003; Zhang and Houk 2005) and noncovalent (Pauling 1946, 1948; Houk et al. 2003; Zhang and Houk 2005) stabilization. The results of this benchmark support the quality of the theozyme model for active site design.
Materials and Methods
All optimizations were carried using the (U)B3LYP/6–31G(d) level of theory except for Human tyrosyl-DNA-phosphodiesterase, which was optimized at the HF/3–21G(d) level. The atoms of the protein that are retained in the theozyme and substrate–active site complexes are determined from the specific residues involved in the known catalytic mechanism. Theozyme calculations employ hydrogen-capped functionality mimics, such as phenol or methanol for a tyrosine and serine side chain, respectively.
The RDS for each benchmark enzymatic mechanism was subject to a QM transition state optimization. The RDS in the reaction mechanism has been identified, where possible, by previous experimental and theoretical work. The knowledge of the RDS and participating catalytic residues determines which residues are kept or removed from the corresponding crystal structure. For example, the catalytic diad functionality of histidine backed by asparate was mimicked in the theozyme by imidazole and acetate, respectively. The TSA or inhibitor was replaced by the actual substrate, and hydrogens were manually added, followed by a TS optimization. A ground state substrate–active site complex adjacent to the TS on the potential energy surface was also optimized using identical catalytic side-chain mimics. All calculations were carried out with Gaussian 03 (Frisch et al. 2004) starting from PDB coordinates of the enzyme.
The RMS deviations (Å) of each theozyme or the associated substrate–active site complex with respect to the X-ray structure were calculated with particular attention on the positions of the catalytic atoms. The root-mean-square deviation between the crystal structure and theozyme catalytic atoms is defined as RMSDTS, to signify comparison to a transition structure plus the catalytic residues. The root-mean-square deviation between the crystal structure and the substrate–active site complex is defined as RMSDSUB, to denote comparison to a substrate plus the catalytic residues. For RMSDTS, catalytic atoms are non-hydrogen atoms that interact with the transition structure atoms through either covalent, partial covalent, or noncovalent fashion. Equivalently, the nonprotein substrate atoms interacting with enzyme catalytic atoms are included in the RMSD calculations. For RMSDSUB, the atoms used in the calculation are identical to those used in RMSDTS. A theozyme is defined as a transition structure optimized in the presence of catalytic functional groups, while the substrate–active site complex represents just a ground state optimized along with the catalytic functional groups.
Electronic supplementary material
All PDB coordinates for reported theozymes, RMSDTS, and RMSDSUB excluding transition state and substrate catalytic atoms, and the complete reference for Gaussian 03 are provided.
Acknowledgments
We are grateful to the National Science Foundation (NSF) and Defense Advanced Research Projects Agency (DARPA) for financial support of this research. This research was facilitated through the Partnerships for Advanced Computational Infrastructure (PACI) through the support of the NSF. The computations were performed on the NSF Terascale Computing System at the Pittsburgh Supercomputing Center (PSC) and on the UCLA Academic Technology Services (ATS) Hoffman Beowulf cluster. J.D. and A.S. acknowledge the National Institutes of Health, UCLA Chemistry-Biology Interface training program for financial support.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: K.N. Houk, Department of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095-1569, USA; e-mail: houk@chem.ucla.edu; fax: (310) 206-1843.
Abbreviations: TS, transition structure or transition state; RMSD, root-mean-square deviation; QM, quantum mechanics; QM/MM, combined quantum mechanics and molecular mechanics; TSA, transition state analog.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072963707.
References
- Arnó M. and Domingo, L.R. 2001. Using theozymes for designing transition-state analogs for the intramolecular aldol reaction of δ-diketones. Int. J. Quantum Chem. 83 338–347. [Google Scholar]
- Arnó M. and Domingo, L.R. 2003. Theozyme for antibody aldolases. Characterization of the transition-state analogue. Org. Biomol. Chem. 1 637–643. [DOI] [PubMed] [Google Scholar]
- Bartlett G.J., Porter, G.T., Borkakoti, N., and Thornton, J.M. 2002. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 324 105–121. [DOI] [PubMed] [Google Scholar]
- Beveridge A.J. 1998. A theoretical study of the initial stages of catalysis in the aspartic proteinases. J. Mol. Struct. 453 275–291. [Google Scholar]
- Blanchard J.E. and Withers, S.G. 2001. Rapid screening of the aglycone specificity of glycosidases: Applications to enzymatic synthesis of oligosaccaharides. Chem. Biol. 8 627–633. [DOI] [PubMed] [Google Scholar]
- Blomberg M.R.A. and Siegbahn, P.E.M. 2003. Metal-bridging mechanism for O–O bond cleavage in cytochrome c oxidase. Inorg. Chem. 42 5231–5243. [DOI] [PubMed] [Google Scholar]
- Breslow R., Chin, J., Hilvert, D., and Trainor, G. 1983. Evidence for the general base mechanism in carboxypeptidase A-catalyzed reactions: Partitioning studies on nucleophiles and H2 18O kinetic istope effects. Proc. Natl. Acad. Sci. 80 4585–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brik A. and Wong, C.-H. 2003. HIV-1 protease: Mechanism and drug discovery. Org. Biomol. Chem. 1 5–14. [DOI] [PubMed] [Google Scholar]
- Cannizzaro C.E., Ashley, J.A., Janda, K.D., and Houk, K.N. 2003. Experimental determination of the absolute enantioselectivity of an antibody-catalyzed Diels–Alder reaction and theoretical explorations of the origins of stereoselectivity. J. Am. Chem. Soc. 125 2489–2506. [DOI] [PubMed] [Google Scholar]
- Cao Z. and Hall, M.B. 2001. Modeling the active sites in metalloenzymes. 3. Density functional calculations on models for [Fe]-hydrogenase: Structures and vibrational frequencies of the observed redox forms and the reaction mechanism at the Diiron Active Center. J. Am. Chem. Soc. 123 3734–3742. [DOI] [PubMed] [Google Scholar]
- Cascella M., Micheletti, C., Rothlisberger, U., and Carloni, P. 2005. Evolutionarily conserved functional mechanics across pepsin-like and retroviral aspartic proteases. J. Am. Chem. Soc. 127 3734–3742. [DOI] [PubMed] [Google Scholar]
- Cho K.-B., Pelmenschikov, V., Gräslund, A., and Siegbahn, P.E.M. 2004. Density functional calculations on class III ribonucleotide reductase: Substrate reaction mechansim with two formates. J. Phys. Chem. B 108 2056–2065. [Google Scholar]
- Christianson D.W., David, P.R., and Lipscomb, W.N. 1987. Mechanism of carboxypeptidase A: Hydration of a ketonic substrate analogue. Proc. Natl. Acad. Sci. 84 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G.J., Mackenzie, L., Varrot, A., Dauter, M., Brzozwski, M., Schulein, M., and Withersw, S.G. 1998. Snapshots along an enzymatic reaction coordinate: Analysis of a retaining β-glycoside. Biochemistry 37 11707–11713. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Interthal, H., Champoux, J.J., and Hol, W.G.J. 2002a. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure 10 237–248. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Interthal, H., Champoux, J.J., and Hol, W.G.J. 2002b. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J. Mol. Biol. 324 917–932. [DOI] [PubMed] [Google Scholar]
- Dölker N., Maseras, F., and Lledó, A. 2003. Density functional study on the effect of the trans axial ligand of B12 cofactors on the heterolytic cleavage of the Co–C bond. J. Phys. Chem. B 107 306–315. [Google Scholar]
- Dwyer M.A., Looger, L.L., and Hellinga, H.W. 2004. Computational design of a biologically active enzyme. Science 304 1967–1971. [DOI] [PubMed] [Google Scholar]
- Frazao C., Bento, I., Costa, J., Soares, C.M., Verissimo, P., Faro, C., Pires, E., Cooper, J., and Carrondo, M.A. 1999. Crystal structure of cardosin A, a glycosylated and arg-gly-asp-containing aspartic proteinase from the flowers of Cynara cardunculus L. J. Biol. Chem. 274 27694–27701. [DOI] [PubMed] [Google Scholar]
- Frisch M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Montgomery Jr, J.A., Vreven, T., Kudin, K.N., Burant, J.C., et al. 2004. Gaussian 03 revision C.02. Gaussian, Inc, Wallingford, CT.
- Gamper M., Hilvert, D., and Kast, P. 2000. Probing the role of the C-terminus of Bacillus subtilis chorismate mutase by a novel random protein-termination strategy. Biochemistry 39 14087–14094. [DOI] [PubMed] [Google Scholar]
- Gao J. and Thompson, M.A. 1998. Methods and applications of hybrid quantum mechanical and molecular mechanical methods. ACS Symposium Series, Vol. 712. American Chemical Society, Washington DC.
- Guimarães C.R.W., Repasky, M.P., Chandrasekhar, J., Tirado-Rives, J., and Jorgensen, W.L. 2003. Contributions of conformational compression and preferential transition state stabilization to the rate enhancement by chorismate mutase. J. Am. Chem. Soc. 125 6892–6899. [DOI] [PubMed] [Google Scholar]
- Guimarães C.R.W., Udier-Blagović, M., Tubert-Brohman, I., and Jorgensen, W.L. 2005. Effects of Arg90 neutralization on the enzyme-catalyzed rearrangement of chorismate to prephenate. J. Chem. Theory Comput. 1 617–625. [DOI] [PubMed] [Google Scholar]
- Guo H., Cui, Q., Lipscomb, W.N., and Karplus, M. 2001. Substrate conformational transitions in the active site of chorismate mutase: Their role in the catalytic mechanism. Proc. Natl. Acad. Sci. 98 9032–9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge A. and Thornton, J.M. 2005. Understanding nature's catalytic toolkit. Trends Biochem. Sci. 30 622–629. [DOI] [PubMed] [Google Scholar]
- Harris D., Loew, G., and Waskell, L. 2001. Calculation of the electronic structure and spectra of model cytochrome P450 compound I. J. Inorg. Biochem. 83 309–318. [DOI] [PubMed] [Google Scholar]
- Heine A., DeSantis, G., Luz, J.G., Mitchell, M., Wong, C.-H., and Wilson, I.A. 2001. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 294 369–374. [DOI] [PubMed] [Google Scholar]
- Himo F., Eriksson, L.A., Maseras, F., and Siegbahn, P.E.M. 2000. Catalytic mechanism of galactose oxidase: A theoretical study. J. Am. Chem. Soc. 122 8031–8036. [Google Scholar]
- Hofmann T., Hodges, R.S., and James, M.N.G. 1984. Effect of pH on the activities of penicillopepsin and Rhizopus pepsin and a proposal for the productive substrate binding mode in penicillopepsin. Biochemistry 23 635–643. [DOI] [PubMed] [Google Scholar]
- Houk K.N., Leach, A.G., Kim, S.P., and Zhang, X. 2003. Binding affinities of host–guest, protein–ligand, and protein–transition-state complexes. Angew. Chem. Int. Ed. Engl. 42 4872–4897. [DOI] [PubMed] [Google Scholar]
- Hur S. and Bruice, T.C. 2003a. Comparison of formation of reactive conformers (NACs) for the Claisen rearrangement of chorismate to prephenate in water and in the E. coli mutase: The efficiency of the enzyme catalysis. J. Am. Chem. Soc. 125 5964–5972. [DOI] [PubMed] [Google Scholar]
- Hur S. and Bruice, T.C. 2003b. Just a near attack conformer for catalysis (chorismate to prephenate rearrangements in water, antibody, enzymes, and their mutants). J. Am. Chem. Soc. 125 10540–10542. [DOI] [PubMed] [Google Scholar]
- Hur S. and Bruice, T.C. 2003c. The near attack conformation approach to the study of the chorismate to prephenate reaction. Proc. Natl. Acad. Sci. 100 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland L.J., Tomaszek Jr, T.A., Roberts, G.D., Carr, S.A., Magaard, V.W., Bryan, H.L., Fakhoury, S.A., Moore, M.L., Minnich, M.D., Culp, J.S., et al. 1991. Human immunodeficiency virus-1 protease. 1. Initial velocity studies and kinetic characterization of reaction intermediates by oxygen-18 isotope exchange. Biochemistry 30 8441–8453. [DOI] [PubMed] [Google Scholar]
- Ireton G.C., Black, M.E., and Stoddard, B.L. 2003. The 1.14 Å crystal structure of yeast cytosine deaminase: Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure 11 961–972. [DOI] [PubMed] [Google Scholar]
- James M.N.G. and Sielecki, A.R. 1986. Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 Å resolution. Nature 319 33–38. [DOI] [PubMed] [Google Scholar]
- Kast P. and Hilvert, D. 1996. Is chorismate mutase a protoypic entropy trap? Activation parameters for the Bacillus subtilis enzyme. Tetrahdron Lett. 37 2691–2694. [Google Scholar]
- Khanjin N.A., Snyder, J.P., and Menger, F.M. 1999. Mechanism of chorismate mutase: Contribution of conformational restriction to catalysis in the Claisen rearrangement. J. Am. Chem. Soc. 121 11831–11846. [Google Scholar]
- Kim H. and Lipscomb, W.N. 1990. Crystal structure of the complex of carboxypeptidase A with a strongly bound phosphonate in a new crystalline form: Comparison with structures of other complexes. Biochemistry 29 5546–5555. [DOI] [PubMed] [Google Scholar]
- Ko T.-P., Lin, J.-J., Hu, C.-Y., Hsu, Y.-H., Wang, A.H.-J., and Liaw, S.-H. 2003. Crystal structure of yeast cytosine deaminase. J. Biol. Chem. 278 19111–19117. [DOI] [PubMed] [Google Scholar]
- Lassila J.K., Privett, H.K., Allen, B.D., and Mayo, S.L. 2006. Combinatorial methods for small-molecule placement in computational enzyme design. Proc. Natl. Acad. Sci. 103 16710–16715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach A.G., Houk, K.N., and Reymond, J.-J. 2004. Theoretical investigation of the origins of catalysis of a retro-Diels–Alder reaction by antibody 10F11. J. Org. Chem. 69 3683–3692. [DOI] [PubMed] [Google Scholar]
- Lee A.Y., Karplus, P.A., Genem, B., and Clardy, J. 1995. Atomic-structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 117 3627–3628. [Google Scholar]
- Lee Y.S., Worthington, S.E., Krauss, M., and Brooks, B.R. 2002. Reaction mechanism of chorismate mutase studied by the combined potentials of quantum mechanics and molecular mechanics. J. Phys. Chem. B 106 12059–12065. [Google Scholar]
- Lin H. and Truhlar, D.G. 2006. QM/MM: What have we learned, where are we, and where do we go from here? Theor. Chem. Acc. 117 185–199. [Google Scholar]
- Lundberg M., Blomberg, M.R.A., and Siegbahn, P.E.M. 2003. Modeling water exchange on monomeric and dimeric Mn centers. Theor. Chem. Acc. 110 130–143. [Google Scholar]
- Manjasetty B.A., Powlowski, J., and Vrielink, A. 2003. Crystal structure of a bifunctional aldolase–dehydrogenase: Sequestering a reactive and volatile intermediate. Proc. Natl. Acad. Sci. 100 6992–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margareta R.A. and Siegbahn, P.E.M. 2006. Quantum chemistry applied to the mechanisms of transition metal containing enzymes—Cytochrome c oxidase, a particularly challenging case. J. Comput. Chem. 27 1373–1384. [DOI] [PubMed] [Google Scholar]
- Martí S., Andrés, J., Moliner, V., Silla, E., Tuñón, I., and Bertrán, J. 2003. A comparative study of Claisen and Cope rearrangements catalyzed by chorismate mutase. An insight into enzymatic efficiency: Transition state stabilization or substrate preorganization. J. Am. Chem. Soc. 126 311–319. [DOI] [PubMed] [Google Scholar]
- Mock W.L. 1997. Zinc proteinases. In Comprehensive biological catalysis: A mechanistic reference (ed. M. Sinnott), pp. 425–446. Academic Press Limited, London, UK.
- Na J. and Houk, K.N. 1996. Predicting antibody catalyst selectivity from optimum binding of catalytic groups to a hapten. J. Am. Chem. Soc. 118 9204–9205. [Google Scholar]
- Na J., Houk, K.N., and Hilvert, D. 1996. Transition state of the base-promoted ring-opening of isoxazoles. Theoretical prediction of catalytic functionalities and design of haptens for antibody production. J. Am. Chem. Soc. 118 6462–6471. [Google Scholar]
- Nachon F., Asojo, O.A., Borgstahl, G.E.O., Masson, P., and Lockridge, O. 2005. Role of water in aging of human butyrylcholinesterase inhibited by echothiophate: The crystal structure suggests two alternative mechanisms of aging. Biochemistry 44 1154–1162. [DOI] [PubMed] [Google Scholar]
- Nitiss K.C., Malik, M., He, X., White, S.W., and Nitiss, J.L. 2006. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl. Acad. Sci. 103 8953–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notenboom V., Birsan, C., Nitz, M., Rose, D.R., Warren, R.A.J., and Withers, S.G. 1998. Insights into transition state stabilization of the β-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants. Nat. Struct. Biol. 5 812–818. [DOI] [PubMed] [Google Scholar]
- Pauling L. 1946. Molecular architecture and biological reactions. Chem. Eng. News 24 1375–1377. [Google Scholar]
- Pauling L. 1948. The nature of forces between large molecules of biological interest. Nature 161 707–709. [DOI] [PubMed] [Google Scholar]
- Prabhakar R. and Siegbahn, P.E.M. 2003. A comparison of the mechanism for the reductive half-reaction between pea seedling and other copper amine oxidases (CAOs). J. Comput. Chem. 24 1599–1609. [DOI] [PubMed] [Google Scholar]
- Prabhakar R., Vreven, T., Morokuma, K., and Musaev, D.G. 2005. Elucidation of the mechanism of selenoprotein glutathione peroxidase (GPx)-catalyzed hydrogen peroxide reduction by two glutathione molecules: A density functional study. Biochemistry 44 11864–11871. [DOI] [PubMed] [Google Scholar]
- Ranaghan K.E., Ridder, L., Szefczyk, B., Sokalski, W.A., Hermann, J.C., and Mulholland, A.J. 2004. Transition state stabilization and substrate strain in enzyme catalysis: Ab initio QM/MM modelling of the chorismate mutase reaction. Org. Biomol. Chem. 2 968–980. [DOI] [PubMed] [Google Scholar]
- Ryu D.H., Zhou, G., and Corey, E.J. 2005. Nonparallelism between reaction rate and dienophile-catalyst affinity in catalytic enantioselective Diels–Alder reactions. Org. Lett. 7 1633–1636. [DOI] [PubMed] [Google Scholar]
- Senn H.M. and Thiel, W. 2007. QM/MM methods for biological systems. Top. Curr. Chem. 268 173–290. [Google Scholar]
- Sherwood P. 2000. Hybrid quantum mechanics/molecular mechanics approaches. In Modern methods and algorithms of quantum chemistry (ed. J. Grotendorst), pp. 257–277. John von Neumann Institute for Computing, Jülich, Germany.
- Siegbahn P.E.M. and Borowski, T. 2006. Modeling enzymatic reactions involving transition metals. Acc. Chem. Res. 39 729–738. [DOI] [PubMed] [Google Scholar]
- Siegbahn P.E.M. and Wirstam, M. 2001. Is the bis–oxo Cu2(III,III) state an intermediate in tyrosinase? J. Am. Chem. Soc. 123 11819–11820. [DOI] [PubMed] [Google Scholar]
- Siegbahn P.E.M., Westerberg, J., Svensson, M., and Crabtree, R.H. 1998. Nitrogen fixation by nitrogenases: A quantum mechanical chemical study. J. Phys. Chem. B 102 1615–1623. [Google Scholar]
- Siegbahn P.E.M., Blomberg, M.R.A., and Pavlov, M.W.N. 2001. The mechanism of the Ni-Fe hydrogenases: A quantum chemical perspective. J. Biol. Inorg. Chem. 6 460–466. [DOI] [PubMed] [Google Scholar]
- Simonson T. and Brooks III, C.L. 1996. Charge screening and the dielectric constant of proteins: Insights from molecular dynamics. J. Am. Chem. Soc. 118 8452–8458. [Google Scholar]
- Skopec C.E., Himmel, M.E., Matthews, J.F., and Brady, J.W. 2003. Energetics for displacing a single chain from the surface of microcrystalline cellulose into the active site of Acidothermus cellulolyticus Cel5A. Protein Eng. 16 1005–1015. [DOI] [PubMed] [Google Scholar]
- Štrajbl M., Shurki, A., Kato, M., and Warshel, A. 2003. Apparent NAC effect in chorismate mutase reflects electronic transition state stabilization. J. Am. Chem. Soc. 125 10228–10237. [DOI] [PubMed] [Google Scholar]
- Tang J. and Wong, R.N.S. 1987. Evolution in the structure and function of aspartic proteases. J. Cell. Biochem. 33 53–63. [DOI] [PubMed] [Google Scholar]
- Tantillo D.J., Chen, J., and Houk, K.N. 1998. Theozymes and compuzymes: Theoretical models for biological catalysis. Curr. Opin. Chem. Biol. 2 743–750. [DOI] [PubMed] [Google Scholar]
- Ujaque G., Tantillo, D.J., Hu, Y., Houk, K.N., Hotta, K., and Hilvert, D. 2002. Catalysis on the coastline: Theozyme, molecular dynamcis, and free energy perturbation analysis of antibody 21D8 catalysis of the decarboxylation of 5-nitro-3-carboxybenzisoxazole. J. Comput. Chem. 24 98–110. [DOI] [PubMed] [Google Scholar]
- Varrot A. and Davies, G.J. 2003. Direct experimental observation of the hydrogen-bonding network of a glycosidase along its reaction coordinate revealed by atomic resolution analyses of endoglucanase Cel5A. Acta Crystallogr. D Biol. Crystallogr. 59 447–452. [DOI] [PubMed] [Google Scholar]
- Venturini A., López-Ortiz, F., Alvarez, J.M., and González, J. 1998. Theoretical proposal of a catalytic mechanism for the HIV-1 protease involving an enzyme-bound tetrahedral intermediate. J. Am. Chem. Soc. 120 1110–1111. [Google Scholar]
- Warshel A. and Levitt, M. 1976. Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 103 227–249. [DOI] [PubMed] [Google Scholar]
- Wiest O. and Houk, K.N. 1994. On the transition state of the chorismate-prephenate rearrangement. J. Org. Chem. 59 7582–7584. [Google Scholar]
- Wirstam M., Blomberg, M.R.A., and Siegbahn, P.E.M. 1999. Reaction mechanism of compound I formation in heme peroxidases: A density functional theory study. J. Am. Chem. Soc. 121 10178–10185. [Google Scholar]
- Woodcock H.L., Hodošček, M., Sherwood, P., Lee, Y.S., Schaefer, H.F.I., and Brooks, B.R. 2003. Exploring the quantum mechanical/molecular mechanical replica path method: A pathway optimization of the chorismate to prephenate Claisen rearrangement catalyzed by chorismate mutase. Theor. Chem. Acc. 109 140–148. [Google Scholar]
- Yang S.W., Burgin, A.B.J., Huizenga, B.N., Robertson, C.A., Yao, K.C., and Nash, H.A. 1996. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. 93 11534–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini A., Jiang, L., Cheng, G., Althoff, E.A., Röthllisberger, D., Wollacott, A.M., Meiler, J., and Baker, D. 2006. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 15 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan C.-G., Zheng, F., and Landry, D.W. 2003. Fundamental reaction mechanism for cocaine hydrolysis in human butyrylcholinesterase. J. Am. Chem. Soc. 125 2462–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. and Houk, K.N. 2005. Why enzymes are proficient catalysts: Beyond the Pauling paradigm. Acc. Chem. Res. 38 379–385. [DOI] [PubMed] [Google Scholar]
- Zhang X., Deng, Q., Yoo, S.H., and Houk, K.N. 2002. Origins and predictions of stereoselective antibody catalysis: Theoretical analysis of Diels–Alder catalysis by 39A11 and its germ-line antibody. J. Org. Chem. 67 9043–9053. [DOI] [PubMed] [Google Scholar]