Abstract
The coronavirus responsible for the severe acute respiratory syndrome (SARS-CoV) contains a small envelope protein, E, with putative involvement in host cell apoptosis and virus morphogenesis. It has been suggested that E protein can form a membrane destabilizing transmembrane (TM) hairpin, or homooligomerize to form a regular TM α-helical bundle. We have shown previously that the topology of the α-helical putative TM domain of E protein (ETM), flanked by two lysine residues at C and N termini to improve solubility, is consistent with a regular TM α-helix, with orientational parameters in lipid bilayers that are consistent with a homopentameric model. Herein, we show that this peptide, reconstituted in lipid bilayers, shows sodium conductance. Channel activity is inhibited by the anti-influenza drug amantadine, which was found to bind our preparation with moderate affinity. Results obtained from single or double mutants indicate that the organization of the transmembrane pore is consistent with our previously reported pentameric α-helical bundle model.
Keywords: structure/function studies, viral protein topologies, structural proteins, protein structure prediction
The causative agent of severe acute respiratory syndrome (SARS), SARS-CoV, is a member of the family Coronaviridae (Rota et al. 2003), which causes common colds in humans and is responsible for serious diseases in other animal species. The lipid bilayer that surrounds the virions typically embeds three proteins: spike (S), membrane (M), and envelope (E) protein. These proteins are studied for their important roles in receptor binding and virion budding.
The E protein of the SARS-CoV virus is a small, 76-amino acid, integral membrane protein with one putative transmembrane α-helical (TMH) hydrophobic domain, 20–30 amino acids long, flanked by a short N-terminal region of <10 amino acids and a longer C-terminal tail, both more hydrophilic. This protein has been found to be critical for viral budding in mouse hepatitis virus (MHV) (Fischer et al. 1998). Also, in many other coronaviruses, coexpression of M and E proteins, which probably interact via their cytoplasmic domains in pre-Golgi compartments (Lim and Liu 2001), is necessary for morphogenesis (Bos et al. 1996; Corse and Machamer 2001).
Further functional roles suggested for E protein are in promoting apoptosis (An et al. 1999; Chen et al. 2001) and alteration of membrane permeability when expressed in Escherichia coli and mammalian cells (Liao et al. 2004). A somewhat related phenomenon is its cation-selective ion channel activity (Wilson et al. 2004) observed in vitro, which has been reported for a synthetic, full-length, SARS-CoV E protein, and also for its N-terminal (40 residues) fragment that encompasses the α-helical transmembrane domain (Wilson et al. 2004). The channel activity of E protein from two other coronaviruses has been shown to be blocked by hexamethylene amiloride (Wilson et al. 2006), and this effect has been correlated with inhibition of replication of these viruses. This points to a possible pharmacological target during coronavirus infection, although the precise function of these putative channels is not well known.
These results suggest that the E protein in coronaviruses, specifically that of SARS-CoV, belong to the viroporin class of proteins (Pinto et al. 1992; Ewart et al. 1996; van Kuppeveld et al. 1997; Bodelon et al. 2002; Fischer and Sansom 2002; Gonzalez and Carrasco 2003). One of the members of this class, M2 from influenza A virus, binds the drug amantadine with high affinity (Hay et al. 1985). Amantadine has long been used for the treatment of influenza A (Fleming 2001). This drug has also been used to abolish ion conductance of hepatitis C virus p7 protein (HCV), which was also suggested to be a viroporin (Griffin et al. 2003). Indeed, amantadine, in combination with other drugs, has shown some degree of success in HCV clinical trials (Torre et al. 2001; Zilly et al. 2002). By analogy, amantadine treatment may be beneficial against SARS-CoV infection.
The channel properties suggested for SARS-CoV E protein may be caused by oligomerization of its transmembrane domain (ETM) to form an α-helical bundle. Indeed, we have shown recently that the synthetic transmembrane domain of SARS-CoV E, flanked by two lysine residues (K2-ETM-K2) to improve solubility, insert in lipid bilayers as a regular α-helix (Torres et al. 2006). Multiple infrared dichroic data were consistent with one of the homopentameric models we obtained computationally using evolutionary conservation data (Torres et al. 2005). We proposed that this α-helical bundle was responsible for the channel activity observed. This prediction, however, was in contrast with a short α-helical hairpin topology, suggested using a slightly longer ETM, which did not contain flanking lysine residues (Arbely et al. 2004; Khattari et al. 2006).
Herein, we have performed experiments to determine if peptide K2-ETM-K2 and several of its mutants show channel properties consistent with those observed for the full-length and 40-residue N-terminal peptide of SARS-CoV E (Wilson et al. 2004). Also, we wanted to test if amantadine can block this channel activity, and measure its binding affinity. Our results suggest that our modified peptide, K2-ETM-K2, explains the channel properties of ETM, and can be used as a model to study the channel formed by SARS-CoV E.
Results and Discussion
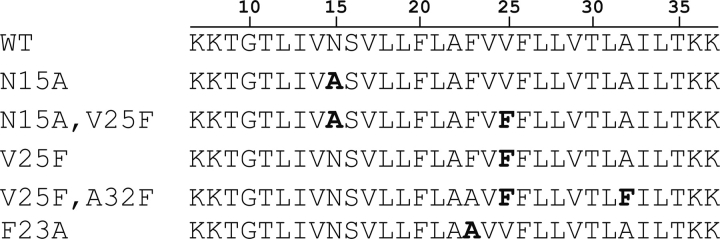
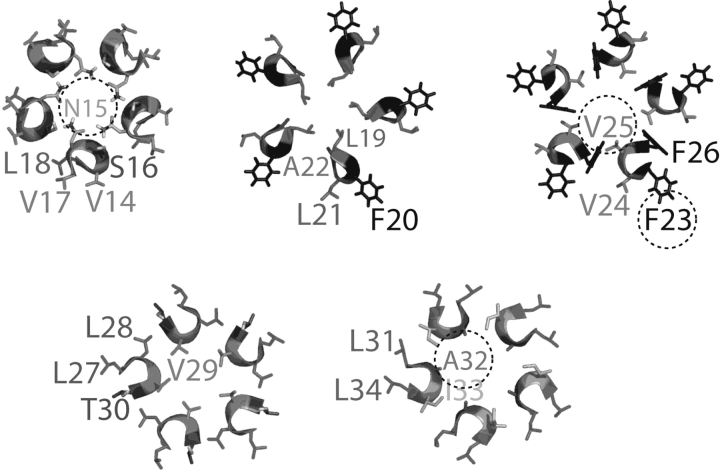
The peptides synthesized for K2-ETM-K2 and its mutants are shown in ▶. The positions of these mutated residues in the context of the α-helical pentameric bundle model we reported previously (Torres et al. 2006), are shown in ▶. This latter figure shows that residues N15, V25, and A32 face the lumen of the pore, whereas F23 is oriented toward lipidic environment.
Figure 1.
Synthetic peptides corresponding to the transmembrane α-helical domain of SARS-CoV E (ETM) protein (residues 9–35) and several of its mutants, flanked by two lysine residues at the C and N termini. The mutations introduced are indicated in bold. The numbering (top) corresponds to the SARS-CoV E protein sequence.
Figure 2.
Slices through the ETM pentameric α-helical bundle (Torres et al. 2005, 2006). The residues mutated in this work are indicated inside the circles.
SDS electrophoreses for all the peptides shown in ▶ (not shown) showed a similar pattern of mobility, consistent with dimers, trimers, and pentamers. No significant amount of monomers (∼3.3 kDa) were detected, even in the case of double mutations. In the latter case, it is likely that native monomer–monomer interactions are affected; therefore, the bands found are likely to represent nonnative interactions induced by SDS. These results were identical to those we published for the wild-type (labeled WT in ▶) (Torres et al. 2005, 2006).
Conductance experiments
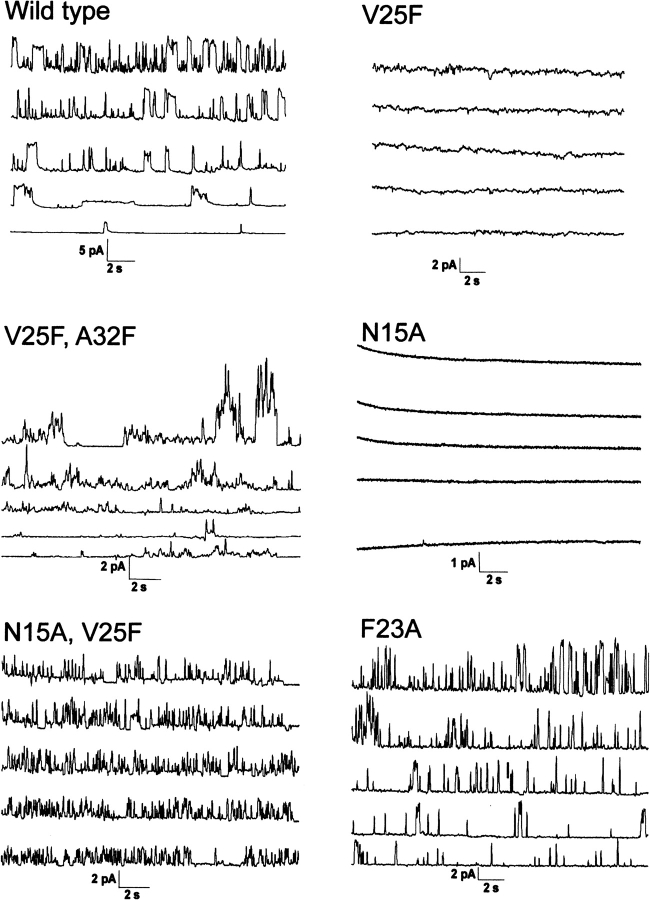
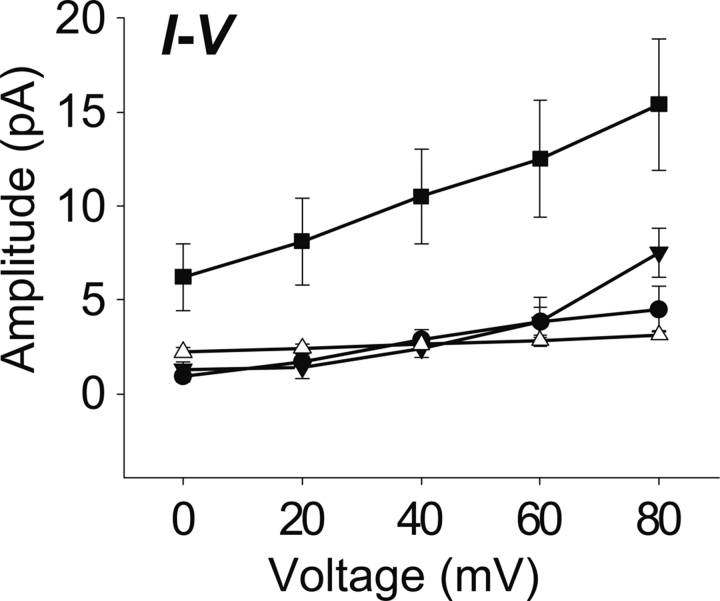
▶ shows the conductance results for our peptide K2-ETM-K2 and its mutants. The I/V plots corresponding to these experiments, which were used to estimate the conductance, are shown in ▶. The wild-type (WT in ▶) conductance (∼45 pS) is consistent with that observed in previous reports for the full-length and N-terminal fragment (Wilson et al. 2004). This demonstrates that the stretch of amino acids used here is responsible for channel activity. Also, we point out that this conductance is observed in the presence of the two lysine residues at N and C termini, which were introduced to increase solubility and facilitate purification of the synthetic peptide.
Figure 3.
ETM ion channel activity in planar lipid bilayers for peptides shown in ▶. Openings are deviations from the baseline (short dashed line). The trans chamber was earthed and the cis chamber was held at various potentials between 0 (bottom) and 80 mV (top), in 20-mV intervals.
Figure 4.
Current–voltage plots (I–V plots) are shown in the lower right panel. Single-channel conductances (slopes of the linear fit obtained from SigmaPlot, version 9.2) were 44.5 pS for WT (•), 114 pS for F23A (▪), 77 pS for V25F–A32F (▾), 10 pS for N15A–V25F (△). The data represents the average from 4 to 10 recordings.
The single-mutants, N15A and V25F, did not show any conductance at any of the voltages tested (see ▶), consistent with the orientation of these residues toward the center of the pore (see ▶) (Torres et al. 2006). The single-mutation F23A showed conductance (114 pS), consistent with an orientation of this residue toward the lipidic environment (see ▶).
The double-mutants, V25F–A32F and N15A–V25F, showed some conductance, 77 pS and 10 pS, respectively, despite the fact that they contain the mutations V25F and N15A. These latter mutations on their own, as shown above, completely abolish conductance. The high conductance observed in this mutant could be due to nonspecific destabilization of the membrane.
To test this hypothesis, we compared the ion selectivity of WT and double-mutant V25F–A32F (▶). This figure (panel C) shows an asymmetrical current–voltage relationship for the wild-type sequence (WT), which indicates that it is more permeable to sodium ions than to calcium ions. In contrast, the double mutant shows a symmetrical current–voltage relationship which indicates no cation specificity.
Figure 5.
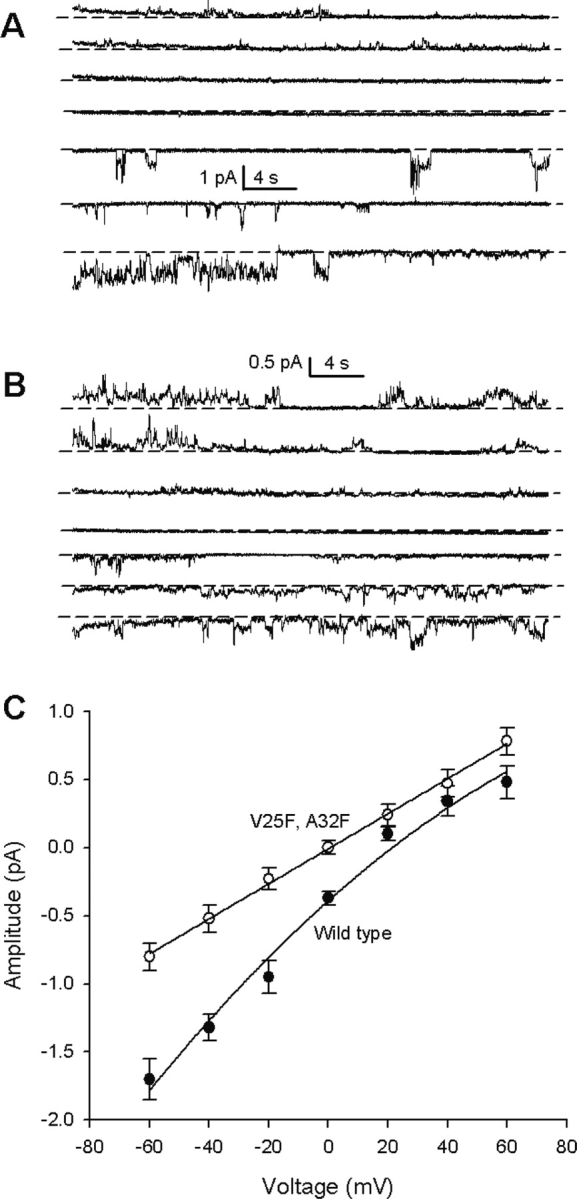
Comparison of ion selectivity (see Materials and Methods), with single-channel currents, between wild-type sequence (A) and V25F–A32F double mutant (B). Voltages are from −60 mV (bottom) to 60 mV (top), in steps of 20 mV. Short dashed lines indicate the closed state. The corresponding current–voltage relationship is shown in (C). A line is drawn only to guide the eye. Data for wild-type (•) and double-mutant V25F–A32F (○) are indicated. Vertical bars represent two standard deviations.
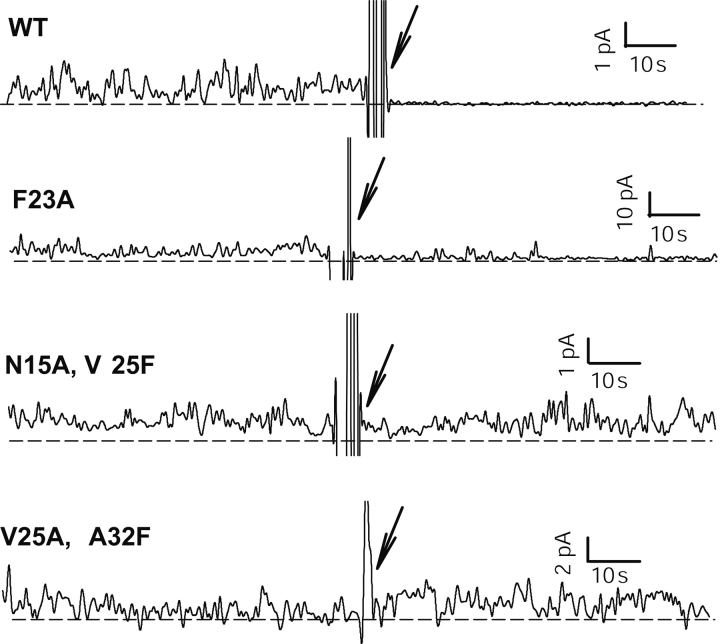
The effect of the addition of amantadine on conductance is shown in ▶. Amantadine blocks conductance in the native sequence (WT) and in the mutant F23A, suggesting a native, or close to native, organization of the monomers in these two samples. However, conductance in the double mutants (N15A–V25F and V25A–A32F) was not affected by the addition of amantadine, which may reflect a significant structural change. Two situations are possible: (1) Amantadine still binds but is not effective in inhibiting ion flow, or (2) amantadine does not bind at all. The latter, as stated above, would explain the high conductance of the double mutants relative to the single mutants by a destabilization of the membrane.
Figure 6.
Effect of amantadine addition (100 μM, see arrow) on the conductance of the peptides shown in ▶, except N15A and V25F, which did not show conductance (▶).
Surface plasmon resonance
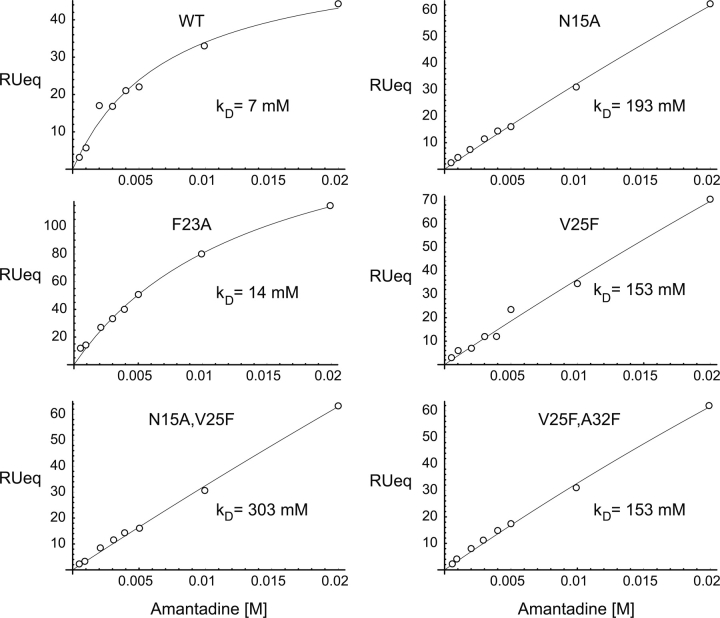
To determine the affinity of amantadine for the pore formed by ETM and its mutants in lipid bilayers, we used surface plasmon resonance (SPR). ▶ depicts the amantadine dose–response curves obtained under equilibrium conditions for the sequences shown in ▶. The data were fitted using a model that assumes a one-to-one binding of amantadine and channel.
Figure 7.
Representative amantadine SPR dose–response curves. The data is shown in white circles, and were fitted (solid line) using Formula 1. The determined standard deviation of the k D did not exceed 20% of the average value in each case.
▶ shows that amantadine binds with relatively high affinity to native ETM (∼10−3 M). This affinity is comparable to that observed for influenza A M2, ∼10−3–10−4 M, depending on the strain (Astrahan et al. 2004). As predicted from the model in ▶, significant binding was only observed for F23A (k D ∼14 mM), whereas only what appeared to be nonspecific aggregation was observed for N15A, V25F, and A32F, suggesting an alteration of the native oligomeric structure in these samples. This is also consistent with our observation that conductance cannot be inhibited by amantadine in V25F–A23F or N15AV25F (▶), despite the fact that these samples show high conductance. Therefore, we conclude that the latter is due to membrane permeabilization in this case.
The observation that mutation at N15 disrupts ion channel conductance and amantadine binding is expected, attending the conservation of a polar residue (S, Q, or N) at this position (Torres et al. 2005) in all coronavirus envelope proteins. However, the results obtained with the other mutations could not have been anticipated in the absence of our model. In summary, our results not only confirm that the ion conductance properties of SARS-CoV E reside in the transmembrane domain, we also show that a peptide flanked by two lysine residues (K2-ETM-K2) (Torres et al. 2006) for which a homopentameric α-helical bundle model was proposed, is a good model to study its channel properties. Further, we report that this peptide incorporated in lipid bilayers binds amantadine with relatively high affinity, at least comparable to influenza A M2. This result may have therapeutic implications in the treatment of SARS infection.
Materials and Methods
Peptide synthesis
Peptides corresponding to the transmembrane domain of SARS-CoV E, from residues 9–35, with two lysine residues flanking both N and C termini, were obtained as described previously (Torres et al. 2006) from W.M. Keck Biotechnology Resource Center. The peptides were cleaved from the resin with trifluoroacetic acid (TFA) and lyophilized. The lyophilized peptides were dissolved in trifluoroethanol (TFE), TFA, and acetonitrile (1:1:4, v/v/v; final peptide concentration ca. 5 mg/mL) and injected to a 20-mL Jupiter 5 C4–300 column (Phenomenex) equilibrated with H2O. Peptide elution was achieved with a linear gradient to a final solvent composition of 10% H2O, 90% acetonitrile, using a Waters 600 HPLC system. All solvents contained 0.1% (v/v) TFA. The resulting fractions were pooled and lyophilized. Peptide purity was confirmed by electrospray mass spectrometry (ESI), and the sample did not have deletions or truncations, with a purity of ∼95%.
Peptide reconstitution in lipid bilayers
Approximately 1 mg of lyophilized peptide and 10 mg of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (Avanti polar lipids) were dissolved in 500 μL hexafluoro-2-propanol (HFIP). The solution was mixed at 37°C for 1 h and HFIP was completely evaporated. The protein–lipid mixture was dissolved in filtered and degassed phosphate-buffered saline (PBS), pH 6.6 (10 mM Na2HPO4 · NaH2PO4, 120 mM NaCl, and 2.7 mM KCl) to a final lipid concentration of 3 mM. The solution was mixed for 20 min, resulting in a suspension of peptide reconstituted in liposomes. Proper ETM reconstitution in lipid bilayers was confirmed by Fourier Transform InfraRed (FTIR) spectroscopy (Torres et al. 2000), with an amide I band centered at 1655 cm−1 (not shown) consistent with α-helix. The liposomes were resized to a uniform diameter of 100 nm by extrusion through an Avanti Mini Extruder with a 100 nm polycarbonate membrane.
Surface plasmon resonance
Binding measurements were performed on a BIAcore 3000 system, with L1 chip as substrate, which allows liposome binding (Cooper et al. 2000). The L1 chip was cleaned prior to each experiment with 40 mM Triton X-100 (20 μL at a flow rate of 20 μL/min). The liposomes were immobilized on one of the flow cells of the chip (80 μL at a flow rate of 2 μL/min). Excess unbound liposomes was removed with 10 mM NaOH in PBS (50 μL at a flow rate of 100 μL/min) followed by PBS flow for 4 h at a rate of 5 μL/min. Amantadine was injected (60 μL at a flow rate of 20 μL/min) in the concentration range of 0.5–20 mM on the membrane immobilized on the chip. At the end of each injection, after equilibrium, amantadine was allowed to dissociate for 1 min under PBS. Subsequently, the surface was regenerated with PBS (30 μL/min flow rate for 15 min).
Each experiment included the immobilization of empty liposomes (i.e., those without peptide) in one of the flow cells, which was used as a reference. The mean RU (resonance units) of the last 10–15 sec of amantadine injection on the empty liposomes' cell were subtracted from the mean RU, at the same time period, in the cell containing the reconstituted ETM. Due to the fact that the on- and off-rate constants of amantadine binding to ETM were rapid, all thermodynamic analyses were based on equilibrium, steady-state measurements (see below).
The binding results were represented as dose–response curves, in which the final RUeq is plotted against amantadine concentration. The dose–response curves were fitted according to Equation 1, where the association constant is given by 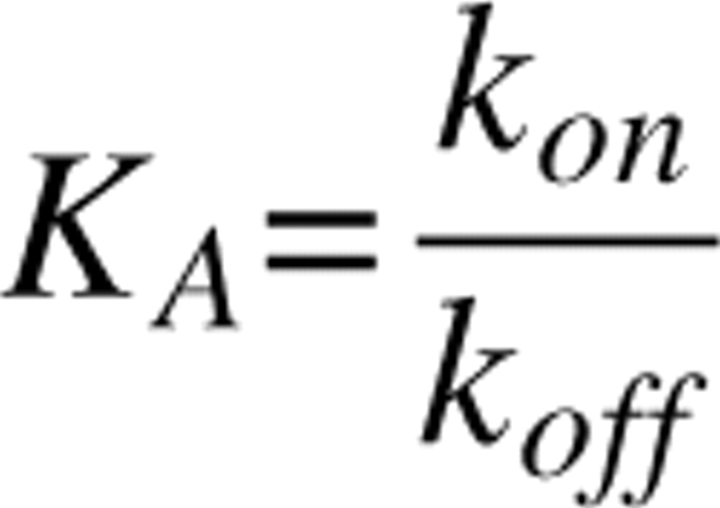
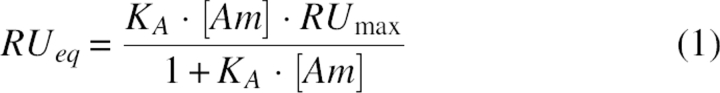 |
and [Am] is the concentration of amantadine injected, RUmax is the RU when all the ETM channels are saturated with amantadine, and RUeq is the RU at equilibrium for a given concentration of amantadine (Astrahan et al. 2004). From K A, K D can be obtained (K D = K A −1). The dissociation constant (K D) was derived using the Biaevaluation 3.0 program (Biacore). The goodness of fit was estimated as χ2 values, where a good fit is estimated as χ2 < 10. These values were well below 10 in all cases (ETM, 1.98; V25F, 0.799; V25F–A32F, 1.31; N15A, 1.69; N15A–V25F, 1.3; F23A, 0.621).
The calculated RUMAX was normalized by multiplication by a constant factor (Astrahan et al. 2004) and the fitting curve program was then run once again to produce calibrated χ2, to compare the different ETMs χ2. The model used to analyze the surface plasmon resonance (SPR) data assumed a 1:1 binding stoichiometry between amantadine and ETM (Astrahan et al. 2004).
Ion conductance
Synthetic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylserine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylcholine (POPC), and dimyristoyl-phosphotidylcholine (DMPC) were purchased from Avanti Polar Lipids. Decane (≥99%) and (N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonicacid]) were obtained from Sigma-Aldrich, chloroform from Sigma-Aldrich, and ethanol (≥99%) and sodium chloride (NaCl) were obtained from Fluka. Ion channel formation of ETM was measured by (Wilson et al. 2004) forming planar lipid bilayers across a 250-μm aperture in the wall of a Delrin cup separating the aqueous solutions in the cis and trans chambers. The working volume was of 1 mL in each chamber. A lipid mixture of 5:3:2, POPE:POPS:POPC (50 mg/mL) was dissolved in chloroform and dried under N2 gas. The lipid cake was then resuspended in decane. A small amount of lipid mixture was painted across the aperture of the Delrin cup. About 14–20 μg of SARS-CoV ETM protein was added to the cis chamber under continuous stirring, until ion channel activity was detected, typically after 15–30 min. The chambers contained 500 mM NaCl, 5 mM HEPES, pH 7.2 (cis), and 5 mM NaCl and 5 mM of HEPES, pH 7.2 (trans). Recordings were performed in asymmetric ionic conditions using a different buffer. The cis and trans solutions were connected via an Ag/AgCl electrode and 1 M NaCl, 2% agar bridge. Currents were amplified using a Warner Instrument amplifier and digitized at 5 kHz and filtered at 10 kHz using pCLAMP 9.2 software (Axon Instruments, Inc.). The trans chamber was set as reference and the cis chamber was held at different potentials ranging from 0 to 80 mV, in 20 mV increments. Stirring was stopped after detection of channel activity. The ion selectivity experiment was performed with 500 mM NaCl, 5 mM HEPES (pH 7.2) in the cis chamber, and 500 mM CaCl2, 5 mM HEPES (pH 7.2) in the trans chamber.
Acknowledgments
J.T. thanks Singapore's Ministry of Education, Grant ARC 7/05, and the Biomedical Research Council (BMRC) of Singapore, Grant (04/1/22/19/361), for financial support. We also thank Susana Geifman Shochat for useful discussions in the analysis of the SPR data.
Footnotes
Reprint requests to: Jaume Torres, School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551; e-mail: jtorres@ntu.edu.sg; fax: 65-6791-3856.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062730007.
References
- An S., Chen, C.J., Yu, X., Leibowitz, J.L., and Makino, S. 1999. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 73 7853–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E., Khattari, Z., Brotons, G., Akkawi, M., Salditt, T., and Arkin, I.T. 2004. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 341 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrahan P., Kass, I., Cooper, M.A., and Arkin, I.T. 2004. A novel method of resistance for influenza against a channel-blocking antiviral drug. Proteins 55 251–257. [DOI] [PubMed] [Google Scholar]
- Bodelon G., Labrada, L., Martinez-Costas, J., and Benavente, J. 2002. Modification of late membrane permeability in avian reovirus-infected cells: Viroporin activity of the S1-encoded nonstructural p10 protein. J. Biol. Chem. 277 17789–17796. [DOI] [PubMed] [Google Scholar]
- Bos E.C., Luytjes, W., van der Meulen, H.V., Koerten, H.K., and Spaan, W.J. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., An, S., and Makino, S. 2001. Induction of apoptosis in murine coronavirus-infected 17Cl-1 cells. Adv. Exp. Med. Biol. 494 615–620. [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Hansson, A., Lofas, S., and Williams, D.H. 2000. A vesicle capture sensor chip for kinetic analysis of interactions with membrane-bound receptors. Anal Biochem 277 196–205. [DOI] [PubMed] [Google Scholar]
- Corse E. and Machamer, C.E. 2001. Infectious bronchitis virus envelope protein targeting: Implications for virus assembly. Adv. Exp. Med. Biol. 494 571–576. [DOI] [PubMed] [Google Scholar]
- Ewart G.D., Sutherland, T., Gage, P.W., and Cox, G.B. 1996. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 70 7108–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen, C.F., Masters, P.S., and Samsonoff, W.A. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72 7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W.B. and Sansom, M.S. 2002. Viral ion channels: Structure and function. Biochim. Biophys. Acta 1561 27–45. [DOI] [PubMed] [Google Scholar]
- Fleming D.M. 2001. Managing influenza: Amantadine, rimantadine and beyond. Int. J. Clin. Pract. 55 189–195. [PubMed] [Google Scholar]
- Gonzalez M.E. and Carrasco, L. 2003. Viroporins. FEBS Lett. 552 28–34. [DOI] [PubMed] [Google Scholar]
- Griffin S.D., Beales, L.P., Clarke, D.S., Worsfold, O., Evans, S.D., Jaeger, J., Harris, M.P., and Rowlands, D.J. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 535 34–38. [DOI] [PubMed] [Google Scholar]
- Hay A.J., Wolstenholme, A.J., Skehel, J.J., and Smith, M.H. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4 3021–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattari Z., Brotons, G., Akkawi, M., Arbely, E., Arkin, I.T., and Salditt, T. 2006. SARS coronavirus E protein in phospholipid bilayers: An X-ray study. Biophys. J. 90 2038–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lescar, J., Tam, J.P., and Liu, D.X. 2004. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 325 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P. and Liu, D.X. 2001. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276 17515–17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L.H., Holsinger, L.J., and Lamb, R.A. 1992. Influenza virus M2 protein has ion channel activity. Cell 69 517–528. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste, M.S., Monroe, S.S., Nix, W.A., Campagnoli, R., Icenogle, J.P., Penaranda, S., Bankamp, B., Maher, K., Chen, M.H., et al. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300 1394–1399. [DOI] [PubMed] [Google Scholar]
- Torre F., Giusto, R., Grasso, A., Brizzolara, R., Campo, N., Sinelli, N., Balestra, V., and Picciotto, A. 2001. Clearance kinetics of hepatitis C virus under different antiviral therapies. J. Med. Virol. 64 455–459. [DOI] [PubMed] [Google Scholar]
- Torres J., Adams, P.D., and Arkin, I.T. 2000. Use of a new label, 13C=18O, in the determination of a structural model of phospholamban in a lipid bilayer. Spatial restraints resolve the ambiguity arising from interpretations of mutagenesis data. J. Mol. Biol. 300 677–685. [DOI] [PubMed] [Google Scholar]
- Torres J., Wang, J., Parthasarathy, K., and Liu, D.X. 2005. The transmembrane oligomers of coronavirus protein E. Biophys. J. 88 1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Parthasarathy, K., Lin, X., Saravanan, R., Kukol, A., and Liu, D.X. 2006. Model of a putative pore: The pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 91 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppeveld F.J., Hoenderop, J.G., Smeets, R.L., Willems, P.H., Dijkman, H.B., Galama, J.M., and Melchers, W.J. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16 3519–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay, C., Gage, P., and Ewart, G. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Gage, P., and Ewart, G. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilly M., Lingenauber, C., Desch, S., Vath, T., Klinker, H., and Langmann, P. 2002. Triple antiviral re-therapy for chronic hepatitis C with interferon-alpha, ribavirin and amantadine in nonresponders to interferon-alpha and ribavirin. Eur. J. Med. Res. 7 149–154. [PubMed] [Google Scholar]