Abstract
The second osmotic virial coefficients of seven proteins—ovalbumin, ribonuclease A, bovine serum albumin, α-lactalbumin, myoglobin, cytochrome c, and catalase—were measured in salt solutions. Comparison of the interaction trends in terms of the dimensionless second virial coefficient b2 shows that, at low salt concentrations, protein–protein interactions can be either attractive or repulsive, possibly due to the anisotropy of the protein charge distribution. At high salt concentrations, the behavior depends on the salt: In sodium chloride, protein interactions generally show little salt dependence up to very high salt concentrations, whereas in ammonium sulfate, proteins show a sharp drop in b2 with increasing salt concentration beyond a particular threshold. The experimental phase behavior of the proteins corroborates these observations in that precipitation always follows the drop in b2. When the proteins crystallize, they do so at slightly lower salt concentrations than seen for precipitation. The b2 measurements were extended to other salts for ovalbumin and catalase. The trends follow the Hofmeister series, and the effect of the salt can be interpreted as a water-mediated effect between the protein and salt molecules. The b2 trends quantify protein–protein interactions and provide some understanding of the corresponding phase behavior. The results explain both why ammonium sulfate is among the best crystallization agents, as well as some of the difficulties that can be encountered in protein crystallization.
Keywords: protein interactions, protein crystallization, osmotic second virial coefficient, self-interaction chromatography
The relation between the molecular structure of a protein and the interaction potential between two such molecules is the foundation of a thermodynamic approach to understanding and predicting the behavior of protein solutions. Ultimately, such a relation enables calculation of the phase behavior and thus understanding of the formation of different phases, such as crystals, dense liquid phases, gels, and aggregates. In practice, the interactions between proteins in solutions are modulated by additives such as salts, hydrophilic polymers, or small organic molecules that may be present in formulations or added to induce crystallization. The focus of the present work is limited to exploring the role of different salts in tuning protein interactions.
The interactions between proteins include contributions from at least electrostatic and van der Waals forces, hydration effects, hydrogen bonding, salt bridging, and ion binding. This inherent complexity of the potential of mean force (PMF) between protein molecules adds to the difficulty of understanding the effects of salts on the PMF. Experimentally, it is important to obtain an unambiguous measurement of protein–protein interactions under a variety of solution conditions. Among the different approaches that have been explored to quantify protein–protein interactions, measurement of protein solubilities was probably the first thermodynamic variable explored as a function of the concentrations of different additives. Unfortunately, protein solubility depends on the solid phase formed (Arakawa and Timasheff 1985), and consequently its theoretical interpretation is not straightforward. The second osmotic virial coefficient (B2), on the other hand, depends only on the PMF, and for this reason it has become the variable of choice for investigating the effects of different additives.
B2 is defined by the virial equation of state, which describes the nonideality of the osmotic pressure, and it is related to the PMF U(r,Ω1,Ω2) via (McQuarrie 2000)
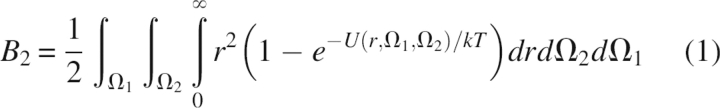 |
Here, r is the protein center-to-center separation distance, and Ω1 and Ω2 are the solid angles that describe the orientations of the two protein molecules. This expression indicates that, in general, positive (repulsive) values of U give rise to positive B2 values, while negative (attractive) values of U produce negative B2 values. Measurements at very low salt concentrations may be dominated by an additional Donnan contribution (Asthagiri et al. 2005), but this is negligible at the higher salt concentrations that are of primary interest here.
B2 can be measured using any of several different experimental techniques, including membrane osmometry (Vilker et al. 1981; Haynes et al. 1992; Wu and Prausnitz 1999), static light scattering (SLS) (Curtis et al. 1998; Velev et al. 1998; Guo et al. 1999; Piazza and Pierno 2000), ultracentrifugation (Behlke and Ristau 1999), small-angle X-ray scattering (SAXS) (Bonneté et al. 1999; Vivarès and Bonneté 2002), small-angle neutron scattering (SANS) (Gripon et al. 1996, 1997; Velev et al. 1998), size-exclusion chromatography (Bloustine et al. 2003), and self-interaction chromatography (SIC) (Tessier et al. 2002a). The latter technique is the most efficient in terms of time and protein consumption (Patro and Przybycien 1996; Tessier et al. 2002a), and for this reason it was used in the present investigation. With SIC, the protein of interest is covalently immobilized on the surface of chromatographic particles, and B2 is calculated from the measured retention time of the same protein through the SIC column.
As is apparent from Equation 1, B2 has units of volume, and its value therefore depends on the size of the protein. The B2 values published in the literature are often expressed in units of mol.mL/g2, for which the notation B22 is adopted. However, instead of B22, which depends on the molecular weight, it is more appropriate to compare B2 values for different proteins in terms of the dimensionless second osmotic virial coefficient (b2), for which B2 is normalized by the excluded volume contribution of a sphere of equal volume (B 2 HS), which is simply four times the molecular volume. This yields (Bonneté and Vivarès 2002)
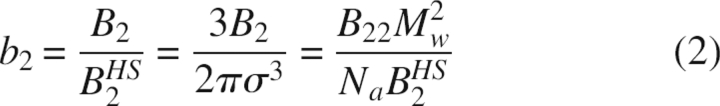 |
Here, σ is the protein equivalent diameter, Mw the molecular weight, and Na Avogadro's number. The results obtained for different proteins are presented here as b2, but these values can easily be converted to B22 using the molecular weights (▶).
Table 1.
Isoelectric point, molecular weight, global charge at pH 7, and equivalent diameter of the proteins studied
Both osmotic pressure and B2 measurements have long been used in studies of the thermodynamic properties of protein solutions (Kupke 1960; Adams et al. 1978), but it is only in about the last decade that a renewed interest in these approaches has been stimulated by interest in developing rational methods to crystallize proteins. George and Wilson (1994) made an important contribution in recognizing a correlation between slightly negative values of B22 and solution conditions favorable for protein crystallization. Their results may seem intuitive in retrospect because of the association of negative B2 values with attractive interactions, but their observations led to the quantitative concept of a “crystallization slot,” a window of B22 values between −1 and −8 mol.mL/g2 for which proteins are likely to crystallize (George and Wilson 1994; George et al. 1997). This concept greatly contributed to the development of a rational approach to crystallize proteins because it permits quantification of the interactions between proteins.
Using this methodology, the effects of different salts (Curtis et al. 1998, 2002; Piazza 1999; Tessier et al. 2002b), surfactants (Jia et al. 2005; Velev et al. 2005), polymers (Kulkarni et al. 2000; Vivarès and Bonneté 2002), alcohols (Farnum and Zukoski 1999; Liu et al. 2004; Berger et al. 2005), and sugar additives (Chi et al. 2003; Valente et al. 2005) on protein–protein interactions have been investigated. However, B2 measurements remain difficult, and many reports present limited data that offer only a snapshot of the effects studied. Many of those studies are also limited to frequently studied proteins such as lysozyme, and they are consequently not representative of the interaction trends that can be observed for other proteins. For this reason, particular emphasis is given here to comparison of the b2 trends for different proteins. The two salts that were investigated in depth, ammonium sulfate and sodium chloride, represent two qualitatively different forms of behavior, and they are consequently good candidates to study the effects of salt on protein interactions. Ammonium sulfate is a good salting-out agent and is among the most successful additives in driving protein crystallization (Gilliland and Davies 1984; Gilliland 1988). On the other hand, sodium chloride is a poor salting-out agent that has had only limited success in protein crystallization (McPherson 2001). These differences have often been described in terms of the Hofmeister series, which describes the effect of salts on a wide variety of phenomena (Collins and Washabaugh 1985; Cacace et al. 1997). The choice of ammonium sulfate also enables comparison of the b2 measurements with the crystallization behavior previously reported with ammonium sulfate for some of the proteins investigated, such as ovalbumin (Miller et al. 1983; Stein et al. 1991; Judge et al. 1995), ribonuclease A (Kunitz 1939), α-lactalbumin (Chrysina et al. 2000), horse heart myoglobin (Kendrew 1950; Lawrie 1951), and catalase (Sumner and Dounce 1937).
In the present work, the b2 measurements are intended to provide a global perspective on the effects of the salt on protein–protein interactions. For this reason, b2 has been measured for ovalbumin, ribonuclease A, bovine serum albumin, α-lactalbumin, myoglobin, cytochrome c, and catalase at pH 7 in different salts. The seven proteins that were studied were selected to have different sizes and isoelectric points in order to give a broad and representative picture of the interactions between proteins (▶). However, the conditions under which those proteins crystallize were not a criterion of selection, and, as always, the results can be biased by the selection of proteins.
Results
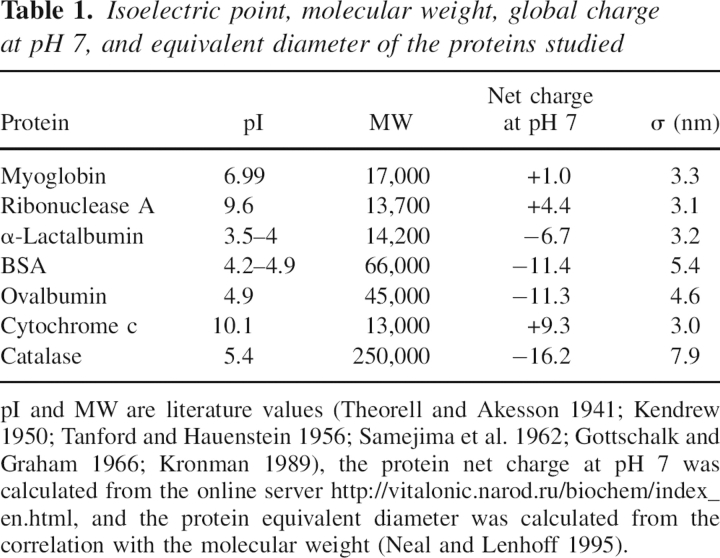
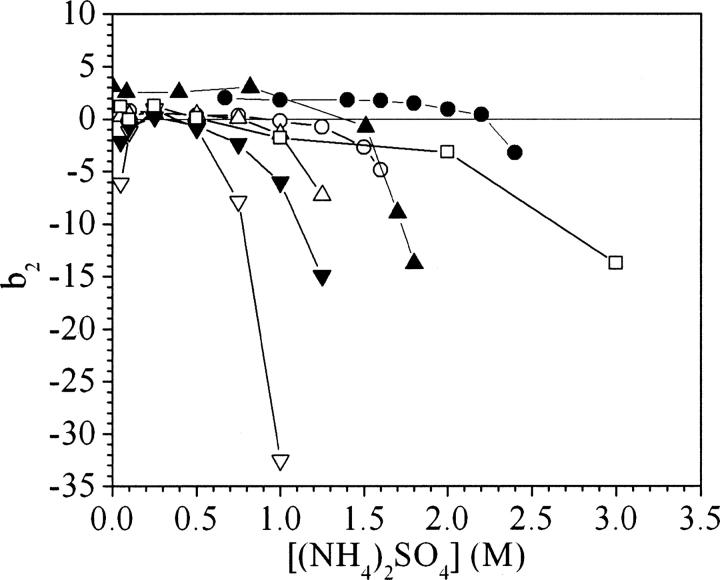
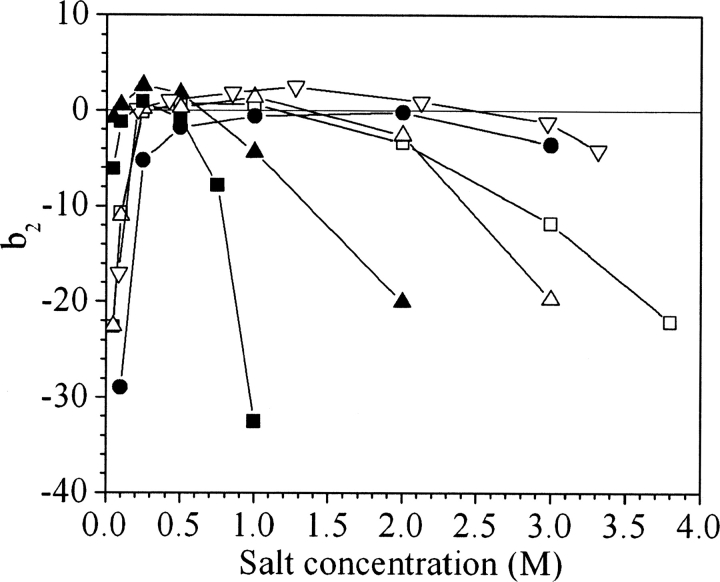
▶ and ▶ present the b2 values for ovalbumin, ribonuclease A, cytochrome c, catalase, α-lactalbumin, myoglobin, and BSA for various concentrations of ammonium sulfate and sodium chloride at pH 7. The error in the measurements based on the reproducibility of Vr values is smaller than the size of the symbols. However, uncertainties in quantities used in SIC calculations such as protein immobilization density, protein radius, and V0 may introduce additional errors in b2 on the order of 0.5–1, but these errors are systematic and would not change the trends observed, which represent the principal focus of this work.
Figure 1.
Values of b2 for ovalbumin (○), ribonuclease A (△), cytochrome c (□), catalase (▽), and α-lactalbumin (▾) in ammonium sulfate at pH 7, myoglobin (•) at pH 6 (Tessier et al. 2002b), and bovine serum albumin (▴) at pH 6.2 (Tessier et al. 2002b).
Figure 2.
Values of b2 for ovalbumin (○), ribonuclease A (△), cytochrome c (□), catalase (▽), α-lactalbumin (▾), myoglobin (•), and bovine serum albumin (▴) in sodium chloride at pH 7.
The experimental trends can be divided into two classes based on the salt concentration. Below ∼0.25–0.5 M, electrostatic interactions must play an important role in the PMF, whereas at higher salt concentrations they are almost completely screened, so other contributions govern protein interactions. ▶ and ▶ illustrate how protein interactions can be either repulsive or attractive in the low-salt region (<0.25–0.5 M). For myoglobin and cytochrome c, positive b2 values indicate that protein interactions are repulsive, whereas for catalase and α-lactalbumin, the negative b2 values reflect attractive interactions. The same trends are observed at low salt concentration for both sodium chloride and ammonium sulfate.
However, in the high-salt region the trends with sodium chloride and ammonium sulfate are drastically different. For sodium chloride, b2 is constant even up to 4 M salt, with the exception of catalase, for which it becomes slightly negative. In ammonium sulfate, b2 is initially constant, but with increasing salt concentration it drops, indicating the onset of attractive interactions. The threshold concentration corresponding to the b2 decrease is different for the seven proteins studied. Among these, catalase shows the earliest decrease, around 1 M ammonium sulfate, whereas for myoglobin and cytochrome c, the b2 decline occurs well above 2 M ammonium sulfate. For the other proteins, the b2 decreases occur between 1 and 2 M. The trends in b2 reflect the effects of the salt on protein–protein interactions, and in this respect ammonium sulfate and sodium chloride behave very differently. Whereas ammonium sulfate is observed systematically to cause b2 to decrease to negative values, sodium chloride is observed to have very little or no effect on protein interactions.
The connection between b2 and phase behavior has practical implications for protein crystallization, and for this reason batch crystallization experiments were performed on myoglobin and catalase in ammonium sulfate at pH 7. Myoglobin was observed to crystallize into clusters of thin needles (▶) within 1–2 wk for pH values of 6–8 and ammonium sulfate concentrations of 2–2.4 M. At higher salt concentrations, however, myoglobin forms only aggregates. In comparison, myoglobin remains soluble in sodium chloride at up to 4 M concentration. The crystallization experiments were restricted to a range of pH values of 6–8 because myoglobin irreversibly denatures below pH 5, probably due to the loss of the heme group (Yang and Phillips 1996; Chi and Asher 1998). The upper limit was determined by the acid–base properties of the ammonium ion (pKa = 9.2), which causes the release of ammonia above pH 8.
Figure 3.
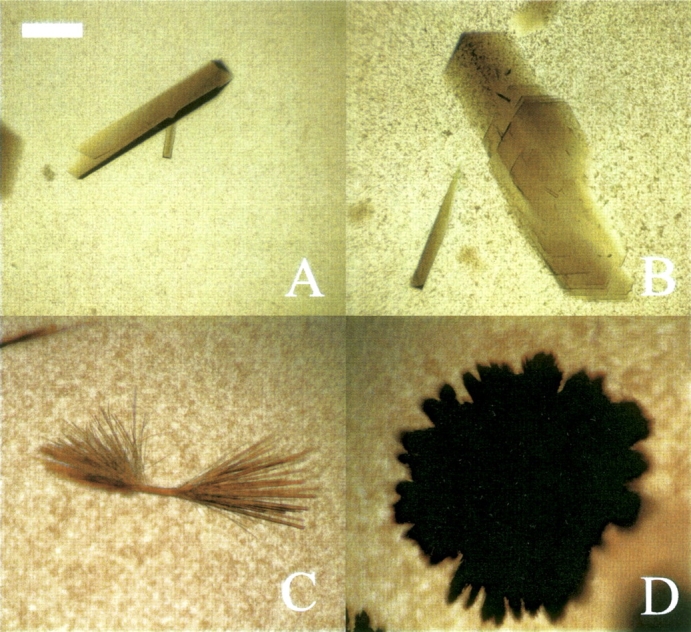
(A,B) Crystals of catalase formed in ammonium sulfate at pH 7, 5 mM bis-tris. (C,D) Crystals of myoglobin formed in ammonium sulfate at pH 7, 5 mM bis-tris. The scale bar represents 50 μm.
The second protein to be studied by batch crystallization was catalase, for which crystals were obtained for pH values of 4–8 and ammonium sulfate concentrations of 0.25–1 M. Depending on the salt concentration, either needles, plates, clusters of sheets, or single crystals were obtained, but, as observed for myoglobin, only aggregates were obtained for salt concentrations above those inducing protein crystallization. Catalase was also observed to form hexagonal crystals at 4°C in 5 mM bis-tris, pH 7 in a region where b2 is highly negative at room temperature.
For both proteins, the b2 decrease in concentrated ammonium sulfate solutions coincides with protein crystallization and is followed by the formation of aggregates. In comparison, high sodium chloride concentrations have very little or no effect on protein interactions and phase behavior for the seven different proteins investigated.
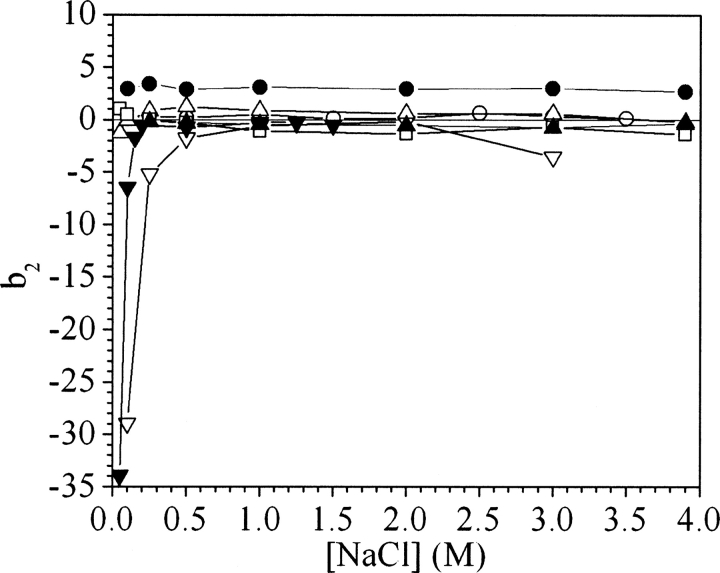
The comparison between ammonium sulfate and sodium chloride was extended to different salts in order to generalize the present observations. Values of b2 were measured at pH 7 for ovalbumin and catalase in potassium thiocyanate, potassium chloride, sodium formate, sodium acetate, sodium malonate, and potassium phosphate (▶, ▶).
Figure 4.
Values of b2 for ovalbumin in sodium chloride (•), potassium chloride (▽), potassium thiocyanate (○), sodium acetate (□), sodium formate (△), ammonium sulfate (▪), and potassium phosphate (▾), all at pH 7.
Figure 5.
Values of b2 for catalase in sodium chloride (•), potassium chloride (▽), sodium formate (△), sodium acetate (□), sodium malonate (▴), and ammonium sulfate (▪), all at pH 7.
At low salt concentrations, the trends are the same as previously observed for ammonium sulfate and sodium chloride, and catalase interactions are clearly attractive.
At high salt concentrations, the different salts follow the same order in inducing attractive interactions for ovalbumin and catalase, but catalase was found to be much more sensitive to the salts than was ovalbumin. The concentration at which the b2 decline begins follows the order potassium phosphate < ammonium sulfate < sodium malonate < sodium formate ∼ sodium acetate < sodium chloride ∼ potassium chloride < potassium thiocyanate. This series corresponds closely to the order observed in the Hofmeister series (Hofmeister 1888).
According to ▶, potassium phosphate and ammonium sulfate are the only two effective salting-out agents for ovalbumin, whereas in sodium formate, sodium acetate, and potassium chloride, b2 does not vary with salt concentration, similar to the results for sodium chloride. In potassium thiocyanate, the b2 values are flat but show a slight increase above 2 M salt. Unlike the case for ovalbumin, all the salts cause b2 to drop for catalase (▶), and sodium malonate, sodium acetate, and sodium formate have an effect intermediate to those of ammonium sulfate and sodium chloride.
Discussion
The seven proteins studied here provide a global perspective on the effects of salt on protein interactions that allow different interaction patterns to be identified. These include the existence of attractive interactions at low salt concentrations, the flat b2 profiles in sodium chloride, and the b2 drops in concentrated ammonium sulfate solutions (▶, ▶). However, b2 does not provide any information about the physical origins of the changes in interactions observed experimentally. Protein–protein interactions are complex and, despite recent progress in computational methods, there are no molecularly based models able to describe the effects of the salt on protein–protein interactions in their full complexity. As a result, it can be difficult to identify with certainty the physical origins of the trends observed experimentally, but what is generally known about the physical chemistry of protein interactions allows the changes in the b2 trends to be attributed to one or another particular aspect of protein interactions.
Lysozyme has been the subject of numerous investigations, and, in particular, its b2 values in sodium chloride have been used as standard measurements to assess the accuracy of different experimental techniques (Rosenbaum and Zukoski 1996; Gripon et al. 1997; Velev et al. 1998; Bonneté et al. 1999; Moon et al. 2000; Piazza and Pierno 2000; Curtis et al. 2002; Tessier et al. 2002b; Bloustine et al. 2003; Teske et al. 2004). The b2 values for lysozyme are positive at low salt concentrations and decrease progressively with increasing salt concentration. The commonly accepted approach considers lysozyme interactions as a balance between repulsive electrostatic and attractive van der Waals interactions (Vilker et al. 1981; Haynes et al. 1992; Coen et al. 1995), even if this does not explain all the trends observed experimentally (Retailleau et al. 1997; Piazza and Pierno 2000). In this picture, which emanates from the Derjaguin–Landau–Verwey–Overbeek (DLVO) theory of colloidal interactions (Israelachvili 1992; Verwey and Overbeek 1999), the change in interactions is attributed to the screening effect of the salt on electrostatic interactions between proteins. A few proteins, e.g., ribonuclease A, α-chymotrypsinogen, subtilisin properase, and purafect subtilisin, show the same b2 trends as lysozyme as a function of increasing salt concentration over a restricted range of solution conditions (Velev et al. 1998; Pjura et al. 2000; Pan and Glatz 2003). However, even among those proteins, ribonuclease A and α-chymotrypsinogen show inconsistencies with the DLVO picture of protein stability under certain solution conditions, and in this perspective lysozyme seems to exhibit a relatively unusual form of behavior. These observations are confirmed by the present results, which show that none of the seven proteins studied behaves like lysozyme.
In particular, ▶ and ▶ show two characteristic trends that are noteworthy in their difference to lysozyme. The first distinctive feature is the existence of attractive interactions at low salt concentrations. This trend, which was observed previously under certain solution conditions for ribonuclease A, α-chymotrypsinogen, and β-lactoglobulin A (Velev et al. 1998; Pjura et al. 2000; Piazza et al. 2002; Tessier et al. 2003), can be interpreted as resulting from attractive electrostatic interactions due in a complex way to the anisotropy of the protein charge distribution. Several aspects of the b2 trends support this interpretation. The attraction is screened at lower salt concentrations in ammonium sulfate than in sodium chloride and it almost disappears above 0.25 M salt concentration, two characteristic signatures of an electrostatic phenomenon as predicted by the Debye–Hückel theory (Israelachvili 1992; Hiemenz and Rajagopalan 1997). The anisotropy of the charge distribution for some proteins was postulated in early experimental work on proteins (Cohn and Edsall 1943; Kirkwood 1967), and it became obvious once the structures of proteins were determined by X-ray diffraction and their electrostatic potential could be calculated using now widely available computational techniques (Honig and Nicholls 1995). Using a Poisson-Boltzmann approach, ribonuclease A, α-chymotrypsinogen, and β-lactoglobulin have been investigated computationally (McClurg and Zukoski 1998; Neal et al. 1998; Fogolari et al. 2000), and the results show for all three proteins that pairwise electrostatic interactions can be attractive in individual configurations.
The second distinctive feature in ▶ and ▶ is the stability of most protein solutions in sodium chloride even at high salt concentrations, resulting in a flat b2 profile, as noted previously (Tessier and Lenhoff 2003). Of the seven proteins studied, only catalase shows more attractive interactions with increasing sodium chloride concentration. This trend is not unexpected, as sodium chloride is known to be a weak salting-out agent, but it strongly contradicts the DLVO picture of protein stability, and shows that proteins can be stable in solution even when electrostatic interactions are screened.
In some cases, ion binding has been suggested to be the cause of repulsive interactions between proteins and as a result to increase protein solubility. The b2 values for ovalbumin in MgCl2 and lysozyme in MgBr2 are increasingly repulsive with salt concentration (Guo et al. 1999; Grigsby et al. 2000; Tessier et al. 2002a). The solubility of xylose isomerase shows a minimum and then increases with magnesium sulfate concentration (Vuolanto et al. 2003). To explain this behavior, it was postulated that magnesium ions bind to and stabilize those proteins in solution (Grigsby et al. 2000). Preferential interaction measurements showing that lysozyme interacts preferentially with magnesium ions (Arakawa and Timasheff 1982, 1984; Arakawa et al. 1990), and the ability of hydrated magnesium ions to form stronger hydrogen bonds than water itself (Brown 2002), are consistent with this hypothesis.
However, not all ions have a strong propensity to bind to proteins (Collins 1995, 1997), and ion binding cannot explain, for example, protein solubility in concentrated electrolyte solutions when the salt is known to be preferentially excluded from the surface of the protein. This occurs, for example, in sulfate and phosphate salts (Arakawa and Timasheff 1982, 1984), in which proteins can nonetheless be soluble up to relatively high salt concentrations before salting out. Preferential interaction measurements show that both sulfate and phosphate ions are preferentially excluded from the surface of proteins, and this is consistent with the analysis of crystal structures, which reveals only a few ions binding proteins with great specificity (Quiocho et al. 1987, 1989; Chakrabarti 1993), even when proteins crystallize in concentrated salt solutions. For example, ovalbumin (1OVA) and myoglobin (1WLA), which crystallize in ∼2 M and 2.2 M ammonium sulfate, respectively, have only one sulfate ion each in their PDB files, which is insufficient to suggest that their high solubilities are due to salt binding.
The hydration of proteins is another phenomenon that has often been invoked to explain protein stability (Cohn and Edsall 1943; Edsall and McKenzie 1983; Rupley and Careri 1991). Since the development of computational methods able to calculate the hydration free energy, mainly in the context of the protein docking problem (Sitkoff et al. 1994; Janin 1999; Orozco and Luque 2000; Wong and McCammon 2003), its contributions have become more widely appreciated. When a contact forms between two proteins, the water molecules must be removed from the interface, and this gives rise to hydration effects. On average, the surfaces of proteins are 57% nonpolar, 24% polar, and 19% charged (Miller et al. 1987). Protein hydration is consequently the sum of contributions from residues of different types. The dehydration of nonpolar groups generally favors contact formation, but the dehydration of the polar and charged groups has an enthalpic cost that must be compensated (Janin 1999; Elcock et al. 2001; Paliwal et al. 2005). This contribution can therefore contribute to protein solubility in concentrated electrolyte solutions even if electrostatic interactions are fully screened.
From the point of view of protein crystallization, the sharp drop in b2 observed in ammonium sulfate solutions with increasing salt concentration is important, as it begins the region corresponding to slightly negative values of b2, where crystallization is possible. Protein salting out in concentrated electrolyte solutions is one of the oldest known characteristics of protein phase behavior, and it has commonly been attributed to a water-mediated effect between protein and salt molecules (Hofmeister 1888; Cohn and Edsall 1943). This explanation, which is consistent with the previous arguments about the protein solubility in concentrated salt solutions, is based on experimental observations. First, effective salting-out agents are strongly hydrated; e.g., small ions with a high charge density are better salting-out agents than are larger ions with a lower charge density (Collins 1997, 2004). Second, preferential hydration measurements suggest that strongly hydrated salts such as sulfate and phosphate are preferentially excluded from the surface of proteins (Arakawa and Timasheff 1982, 1984). These two arguments are mutually consistent, but they are both global arguments, and, as noted earlier, there is no molecularly based model able to describe the effect of the salt on protein interactions.
Because the drop in b2 is associated experimentally with protein salting out, it implies that the b2 drop at high salt concentrations is due to a water-mediated effect, and consequently so is the relative efficiency of the different salts in changing the value of b2 for ovalbumin and catalase (▶, ▶). For ovalbumin, the agreement between the present b2 measurements and the Hofmeister series is unsurprising, as the Hofmeister series was originally established based on the salting out of egg white (Hofmeister 1888; Kunz et al. 2004), which is 50%–60% ovalbumin and has a pH close to 7.4.
The interpretation of the b2 drop in ammonium sulfate in terms of a water-mediated effect explains qualitatively the relative efficiency of different salts on proteins such as ovalbumin and catalase. However, the effect of the salt can be much more complex. Ion binding, for example, follows the inverse Hofmeister series (Klotz and Urquhart 1949; Scatchard and Black 1949). As a result, weak salting-out agents have a stronger tendency to bind proteins (Collins 1995, 1997), which can also affect protein interactions. For example, the b2 trend observed for ovalbumin with increasing potassium thiocyanate concentration (▶) is consistent with thiocyanate ions binding to ovalbumin, as previously observed for other proteins (Scatchard et al. 1950, 1959).
From a practical point of view, protein crystallization is accomplished by starting from a soluble protein solution and progressively reducing protein solubility. In terms of interactions, it corresponds to finding solution conditions for which protein–protein interactions are weakly attractive. The experimental investigation of ovalbumin and catalase crystallization in ammonium sulfate shows that these conditions correspond to the drop in b2 values in concentrated salt solutions. The seven proteins investigated here all show a sharp drop in the values of b2 at different salt concentrations. Although attractive interactions are necessary to drive protein crystallization, the steepness of the drop in b2 values can be a disadvantage, as it indicates that there is only a narrow range of salt concentrations for which b2 is slightly negative. Therefore, finding the exact domain of salt concentrations favorable to the formation of crystals can be one of the difficulties encountered when proteins are crystallized by empirical screening.
The trends for catalase show that moderate salting-out agents have a wider range of concentrations corresponding to slightly negative values of b2 (▶). Therefore malonate, formate, and acetate salts, which have a behavior intermediate between ammonium sulfate and sodium chloride, can be interesting alternatives to induce protein crystallization. However, ovalbumin, which is more soluble, simply does not salt out with a weaker precipitant than ammonium sulfate or potassium phosphate. These elements seem to explain the efficiency of sodium malonate previously suggested based on empirical screening (McPherson 2001). The b2 values obtained for catalase show that the effect of sodium malonate is only slightly weaker than that of ammonium sulfate. The key to its success seems to be a good balance between the range of concentrations over which it induces protein crystallization and its strength as a salting-out agent.
The results in ▶ also suggest that the solubility of the salt can be an issue when proteins require extremely high salt concentrations before salting out. For example, sodium sulfate, which is soluble up to only 1.3 M at 20°C, cannot salt-out ovalbumin or BSA, whereas ammonium sulfate, which is soluble up to 4.03 M at 20°C, does (Söhnel and Novotny 1985). The difference in solubility between sulfate salts that is observed experimentally is consequently one of the features that explain their efficiency as crystallization agents. Thus, it is not surprising that ammonium sulfate, which is highly soluble and a strong salting-out agent, is among the most successful additives for protein crystallization (McPherson 2001).
The b2 trends also explain why the formation of aggregates is commonly interpreted as a positive indicator of the proximity of solution conditions favorable to crystal growth in crystallization screens (Gilliland and Davies 1984). The results show that the appearance of precipitates generally follows the b2 decrease, and that crystallization by salting-out occurs in the presence of aggregates or at salt concentrations slightly below where aggregates form. This is clearly illustrated by the phase behavior experiments with myoglobin and catalase. As a consequence, the formation of aggregates indicates the domain of salt concentrations corresponding to slightly negative b2 values, providing basically the same information as obtained from b2 measurements. In terms of protein consumption, even the most economical methods to measure b2 cannot match simple crystallization screens. However, because the range of the crystallization slot also depends on the slope of the b2 curve, b2 measurements also provide information about the range of salt concentrations over which there are slightly negative b2 values, which cannot be obtained otherwise.
Conclusions
The present measurements are among the most extensive sets of b2 values that describe the effects of salt on protein–protein interactions. Three trends are noteworthy. First, at low salt concentrations, the interactions can be either attractive or repulsive. This behavior has an electrostatic origin, and it is probably due to the anisotropy of the charge distribution on the protein and its correlation with high-complementarity nonelectrostatic interactions. Second, most proteins have a flat b2 profile with increasing sodium chloride concentration, which explains the high solubility of most proteins in highly concentrated solutions of sodium chloride. Third, the b2 drop in solutions of ammonium sulfate and other strongly hydrated salts occurs over a narrow range of concentrations, and the effects of different salts follow the Hofmeister series with few exceptions. Those trends coincide with protein salting-out, which according to the classical interpretation corresponds to a water-mediated effect between protein and salt molecules.
From these three features, it is possible to define different classes of behavior for the proteins studied, but all differ from that of lysozyme, which appears to be anomalous. From a practical point of view, the drop in b2 values coincides with the formation of amorphous aggregates, and when a protein crystallizes, it typically does so at slightly lower salt concentrations than that at which aggregates appear. The sharp b2 drop in ammonium sulfate results in a narrow window of salt concentrations favorable to the formation of crystals, and this explains some of the difficulties in crystallizing proteins using empirical screening procedures.
Materials and Methods
Proteins and solutions
Myoglobin from horse skeletal muscle (M-0630); bovine serum albumin (A-7638); ovalbumin from chicken eggs, crystallized and lyophilized (A-2512); catalase from bovine liver (C-40); ribonuclease A from bovine pancreas, salt-fractionated and chromatographically purified (R-5503); α-lactalbumin, calcium depleted, from bovine milk (L-6010); and cytochrome c from bovine heart (C-2037) were obtained from Sigma. The proteins were used without further purification with the exception of α-lactalbumin, which was dialyzed overnight against a 1 mM CaCl2 solution using a Slide-A-Lyzer 10,000 MW cut-off (66410) (Pierce Biotechnology).
The chemicals used for protein immobilization, namely N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) (E-6383), N-hydroxysuccinimide (NHS) (H-7377), glutaraldehyde (G-5882), and ethanolamine (E-9508), were purchased from Sigma. The different salts and buffers, ammonium sulfate (A-2939), potassium thiocyanate (P-2713), bis-tris (B-7535), calcium chloride (C-3881), sodium malonate (M-4795), sodium formate (S-2140), citric acid (C-1909), and MES (M-8250) were also purchased from Sigma. Sodium chloride (S-271), sodium hydroxide (S-318), sodium phosphate dibasic (S-374), potassium phosphate monobasic (P-285), potassium chloride (P-217), sodium acetate (S-205), and hydrochloric acid (A144–212) were purchased from Fisher Scientific. Tris (819,620) was purchased from INC.
All solutions were prepared with deionized water that was further purified using a Milli-Q Plus deionization system (Millipore). The pH was adjusted with concentrated solutions of sodium hydroxide and hydrochloric acid.
Protein concentrations were measured using a Lambda 4B spectrophotometer (Perkin-Elmer). The absorbance was measured at 280 nm using E 1cm 1% = 7.35 for ovalbumin, 7.14 for ribonuclease A, 6.6 for bovine serum albumin, 20.9 for α-lactalbumin, 17.0 for myoglobin, 23.9 for cytochrome c, and 13.5 for catalase (Sober 1970).
Immobilization
Myoglobin, ribonuclease A, bovine serum albumin, and cytochrome c were immobilized using glutaraldehyde following a protocol similar to that described previously (Tessier et al. 2002b). Ovalbumin, catalase, and α-lactalbumin were immobilized using EDC/NHS (Carraway and Koshland 1972; Bauminger and Wilchek 1980; Grabarek and Gergely 1990; Sehgal and Vijay 1994). Between 60 and 100 mg of protein were dissolved in 10 mL of 5 mM MES, pH 6.5 buffer containing 0.1 M NaCl. Two milliliters of Toyopearl AF-Amino 650 (8002) chromatographic particles (Tosoh Bioscience) were washed with 1 L of deionized water on a glass frit with a Supor 200 membrane disc filter (60301) (Pall) and resuspended in an 18-mL glass vial with the protein solution. One hundred fifty milligrams of EDC and 10 mg of NHS were introduced and set up to react overnight on a rotary mixer. Once the reaction was complete, the particles were extensively washed with deionized water and stored in the immobilization buffer. The immobilization density was determined using the Micro BCA protein assay (23235) (Pierce Biotechnology).
Chromatographic procedures and b2 calculation
The particles with immobilized protein were packed into a 3 mm × 50 mm microbore glass chromatography column (993301) (Colbert Associates) using a Pharmacia FPLC system at a flow rate of 5 mL/min. The retention time was measured using an ÄKTA Purifier equipped with an autosampler (GE Healthcare). The injection volumes were 20 μL at protein concentrations of 1–5 mg/mL, and all the measurements were made in triplicate. The buffer flow rates were 0.1 mL/min for the measurements and 0.5 mL/min during equilibration.
Values of b2 were calculated from the retention volume. The retention factor was calculated from (Tessier et al. 2002a)
 |
where Vr is the retention volume and V0 the dead volume, which was determined using a column without immobilized protein (Tessier et al. 2002a). The excluded volume contribution to the second osmotic virial coefficient was obtained as four times the protein volume, which implicitly assumes the protein to be a sphere. The protein volume was calculated from its correlation with the molecular weight (Neal and Lenhoff 1995). The value of b2 was then found from (Tessier et al. 2002a)
 |
where ρs is the immobilization density and ϕ is the phase ratio, defined as ϕ = As/V0. The protein-accessible surface area As was obtained from the literature (DePhillips and Lenhoff 2000).
Batch crystallization experiments
Myoglobin and catalase were dissolved in deionized water to obtain concentrated protein solutions of ∼40 mg/mL. The buffers were sodium citrate at pH 3, sodium acetate at pH 4 and 5, MES at pH 6, bis-tris at pH 7, and Tris at pH 8. Batch experiments were prepared on a 96-well plate (21–377–203) (Fisher Scientific). The salt concentrations were investigated in the ranges 0–3.2 M ammonium sulfate from pH 6–8 for myoglobin and 0–2 M ammonium sulfate from pH 3–8 for catalase. Each well was covered with microbatch paraffin oil (HR3–421) (Hampton Research). The protein concentration in all the experiments was 10 mg/mL. The evolution of the phase behavior was recorded every day during the first week and subsequently every 2 wk up to the point that no further change was observed.
Acknowledgments
We thank the National Science Foundation for financial support (grant number BES-0519191). A.M.S.-O. was supported under a Chemistry–Biology Interface Training Grant from the National Institutes of Health.
Footnotes
Reprint requests to: Abraham M. Lenhoff, Center for Molecular and Engineering Thermodynamics, Department of Chemical Engineering, University of Delaware, Newark, DE 19716, USA; e-mail: lenhoff@udel.edu; fax: (302) 831-4466; or Eric W. Kaler, Center for Molecular and Engineering Thermodynamics, Department of Chemical Engineering, University of Delaware, Newark, DE 19716, USA; e-mail: kaler@udel.edu; fax: (302) 831-6751.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072957907.
References
- Adams E.T., Wan, P.J., and Crawford, E.F. 1978. Membrane and vapor pressure osmometry. Methods Enzymol. 48 69–154. [DOI] [PubMed] [Google Scholar]
- Arakawa T. and Timasheff, S.N. 1982. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry 21 6545–6552. [DOI] [PubMed] [Google Scholar]
- Arakawa T. and Timasheff, S.N. 1984. Mechanism of protein salting in and salting out by divalent cation salts: Balance between hydration and salt binding. Biochemistry 23 5912–5923. [DOI] [PubMed] [Google Scholar]
- Arakawa T. and Timasheff, S.N. 1985. Theory of protein solubility. Methods Enzymol. 114 49–77. [DOI] [PubMed] [Google Scholar]
- Arakawa T., Bhat, R., and Timasheff, S.N. 1990. Preferential interactions determine protein solubility in 3 component solutions: The MgCl2 system. Biochemistry 29 1914–1923. [DOI] [PubMed] [Google Scholar]
- Asthagiri D., Paliwal, A., Abras, D., Lenhoff, A.M., and Paulaitis, M.E. 2005. A consistent experimental and modeling approach to light-scattering studies of protein–protein interactions in solution. Biophys. J. 88 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauminger S. and Wilchek, M. 1980. The use of carbodiimides in the preparation of immunizing conjugates. Methods Enzymol. 70 151–159. [DOI] [PubMed] [Google Scholar]
- Behlke J. and Ristau, O. 1999. Analysis of the thermodynamic nonideality of proteins by sedimentation equilibrium experiments. Biophys. Chem. 76 13–23. [DOI] [PubMed] [Google Scholar]
- Berger B.W., Blamey, C.J., Naik, U.P., Bahnson, B.J., and Lenhoff, A.M. 2005. Roles of additives and precipitants in crystallization of calcium- and integrin-binding protein. Cryst. Growth Des. 5 1499–1507. [Google Scholar]
- Bloustine J., Berejnov, V., and Fraden, S. 2003. Measurements of protein–protein interactions by size exclusion chromatography. Biophys. J. 85 2619–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneté F. and Vivarès, D. 2002. Interest of the normalized second virial coefficient and interaction potentials for crystallizing large macromolecules. Acta Crystallogr. D 58 1571–1575. [DOI] [PubMed] [Google Scholar]
- Bonneté F., Finet, S., and Tardieu, A. 1999. Second virial coefficient: Variations with lysozyme crystallization conditions. J. Cryst. Growth 196 403–414. [Google Scholar]
- Brown D. 2002. The chemical bond in inorganic chemistry: The bond valence model. Oxford University Press, New York.
- Cacace M.G., Landau, E.M., and Ramsden, J.J. 1997. The Hofmeister series: Salt and solvent effects on interfacial phenomena. Q. Rev. Biophys. 30 241–277. [DOI] [PubMed] [Google Scholar]
- Carraway K.L. and Koshland, D.E. 1972. Carbodiimide modification of proteins. Methods Enzymol. 25 616–650. [DOI] [PubMed] [Google Scholar]
- Chakrabarti P. 1993. Anion binding sites in protein structures. J. Mol. Biol. 234 463–482. [DOI] [PubMed] [Google Scholar]
- Chi Z.H. and Asher, S.A. 1998. UV resonance Raman determination of protein acid denaturation: Selective unfolding of helical segments of horse myoglobin. Biochemistry 37 2865–2872. [DOI] [PubMed] [Google Scholar]
- Chi E.Y., Krishnan, S., Kendrick, B.S., Chang, B.S., Carpenter, J.F., and Randolph, T.W. 2003. Roles of conformational stability and colloidal stability in the aggregation of recombinant human granulocyte colony-stimulating factor. Protein Sci. 12 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysina E.D., Brew, K., and Acharya, K.R. 2000. Crystal structures of apo- and holo-bovine α-lactalbumin at 2.2 Å resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 275 37021–37029. [DOI] [PubMed] [Google Scholar]
- Coen C.J., Blanch, H.W., and Prausnitz, J.M. 1995. Salting-out of aqueous proteins: Phase-equilibria and intermolecular potentials. AIChE J. 41 996–1004. [Google Scholar]
- Cohn E.J. and Edsall, J.T. 1943. Proteins, amino acids and peptides as ions and dipolar ions. Reinhold Publishing Corporation, New York.
- Collins K.D. 1995. Sticky ions in biological systems. Proc. Natl. Acad. Sci. 92 5553–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.D. 1997. Charge density-dependent strength of hydration and biological structure. Biophys. J. 72 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.D. 2004. Ions from the Hofmeister series and osmolytes: Effects on proteins in solution and in the crystallization process. Methods 34 300–311. [DOI] [PubMed] [Google Scholar]
- Collins K.D. and Washabaugh, M.W. 1985. The Hofmeister effect and the behavior of water at interfaces. Q. Rev. Biophys. 18 323–422. [DOI] [PubMed] [Google Scholar]
- Curtis R.A., Montaser, A., Prausnitz, J.M., and Blanch, H.W. 1998. Protein–protein and protein–salt interactions in aqueous protein solutions containing concentrated electrolytes. Biotechnol. Bioeng. 58 451. [DOI] [PubMed] [Google Scholar]
- Curtis R.A., Montaser, A., Prausnitz, J.M., and Blanch, H.W. 2002. Protein–protein interactions in concentrated electrolyte solutions. Hofmeister-series effects. Biotechnol. Bioeng. 57 11–21. [DOI] [PubMed] [Google Scholar]
- DePhillips P. and Lenhoff, A.M. 2000. Pore size distributions of cation-exchange adsorbents determined by inverse size-exclusion chromatography. J. Chromatogr. A. 883 39–54. [DOI] [PubMed] [Google Scholar]
- Edsall J.T. and McKenzie, H.A. 1983. Water and proteins. II. The location and dynamics of water in protein systems and its relation to their stability and properties. Adv. Biophys. 16 53–183. [DOI] [PubMed] [Google Scholar]
- Elcock A.H., Sept, D., and McCammon, J.A. 2001. Computer simulation of protein–protein interactions. J. Phys. Chem. B 105 1504–1518. [Google Scholar]
- Farnum M. and Zukoski, C. 1999. Effect of glycerol on the interactions and solubility of bovine pancreatic trypsin inhibitor. Biophys. J. 76 2716–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogolari F., Ragona, L., Licciardi, S., Romagnoli, S., Michelutti, R., Ugolini, R., and Molinari, H. 2000. Electrostatic properties of bovine β-lactoglobulin. Proteins 39 317–330. [DOI] [PubMed] [Google Scholar]
- George A. and Wilson, W.W. 1994. Predicting protein crystallization from a dilute-solution property. Acta Crystallogr. D 50 361–365. [DOI] [PubMed] [Google Scholar]
- George A., Chiang, Y., Guo, B., Arabshahi, A., Cai, Z., and Wilson, W.W. 1997. Second virial coefficient as predictor in protein crystal growth. Methods Enzymol. 276 100–110. [DOI] [PubMed] [Google Scholar]
- Gilliland G.L. 1988. A biological macromolecule crystallization database: A basis for a crystallization strategy. J. Cryst. Growth 90 51–59. [Google Scholar]
- Gilliland G.L. and Davies, D.R. 1984. Protein crystallization: The growth of large-scale single crystals. Methods Enzymol. 104 370–381. [DOI] [PubMed] [Google Scholar]
- Gottschalk A. and Graham, B. 1966. The basic structure of glycoproteins. In The proteins (ed. H. Neurath), pp. 95–151. Academic Press, New York.
- Grabarek Z. and Gergely, J. 1990. Zero-length crosslinking procedure with the use of active esters. Anal. Biochem. 185 131–135. [DOI] [PubMed] [Google Scholar]
- Grigsby J.J., Blanch, H.W., and Prausnitz, J.M. 2000. Diffusivities of lysozyme in aqueous MgCl2 solutions from dynamic light-scattering data: Effect of protein and salt concentrations. J. Phys. Chem. B 104 3645–3650. [Google Scholar]
- Gripon C., Legrand, L., Rosenman, I., Vidal, O., Robert, M.-C., and Boué, F. 1996. Study of protein–protein interactions in undersaturated and supersaturated lysozyme solutions in heavy water as a function of temperature. C.R. Acad. Sci. Ser. IIb: Mec. Phys. Chim. Astron. 322 565–571. [Google Scholar]
- Gripon C., Legrand, L., Rosenman, I., Vidal, O., Robert, C., and Boué, F. 1997. Lysozyme–lysozyme interactions in under- and super-saturated solutions: A simple relation between the second virial coefficients in H2O and D2O. J. Cryst. Growth 178 575–584. [Google Scholar]
- Guo B., Kao, S., McDonald, H., Asanov, A., Combs, L.L., and Wilson, W.W. 1999. Correlation of second virial coefficients and solubilities useful in protein crystal growth. J. Cryst. Growth 196 424–433. [Google Scholar]
- Haynes C.A., Tamura, K., Korfer, H.R., Blanch, H.W., and Prausnitz, J.M. 1992. Thermodynamic properties of aqueous α-chymotrypsin solutions from membrane osmometry mesurements. J. Chem. Phys. 96 905–912. [Google Scholar]
- Hiemenz P.C. and Rajagopalan, R. 1997. Principles of colloid and surface chemistry, 3rd ed. Marcel Dekker, New York.
- Hofmeister F. 1888. Zur lehre von der wirkung der salze. Arch. Exp. Pathol. Pharmakol. 24 247–260. [Google Scholar]
- Honig B. and Nicholls, A. 1995. Classical electrostatics in biology and chemistry. Science 268 1144–1149. [DOI] [PubMed] [Google Scholar]
- Israelachvili J.N. 1992. Intermolecular and surface forces. Academic Press, San Diego, CA.
- Janin J. 1999. Hydration and protein–protein interaction. In Hydration processes in biology (eds. M.C. Bellissent-Funel et al.). IOS Press, Amsterdam.
- Jia Y.W., Narayanan, J., Liu, X.-Y., and Liu, Y. 2005. Investigation on the mechanism of crystallization of soluble protein in the presence of nonionic surfactant. Biophys. J. 89 4245–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge R.A., Johns, M.R., and White, E.T. 1995. Protein-purification by bulk crystallization: The recovery of ovalbumin. Biotechnol. Bioeng. 48 316–323. [DOI] [PubMed] [Google Scholar]
- Kendrew J.C. 1950. The crystal structure of horse met-myoglobin. I. General features: The arrangement of the polypeptides chains. Proc. R. Soc. Lond. A 201 62–89. [Google Scholar]
- Kirkwood J.G. 1967. Proteins. Gordon and Breach, New York.
- Klotz I.M. and Urquhart, J.M. 1949. The binding of organic ions by proteins. Buffer effects. J. Phys. Chem. 53 100–114. [PubMed] [Google Scholar]
- Kronman M.J. 1989. Metal-ion binding and the molecular conformational properties of α-lactalbumin. Crit. Rev. Biochem. Mol. Biol. 24 565–667. [DOI] [PubMed] [Google Scholar]
- Kulkarni A.M., Chatterjee, A.P., Schweizer, K.S., and Zukoski, C.F. 2000. Effects of polyethylene glycol on protein interactions. J. Chem. Phys. 113 9863–9873. [Google Scholar]
- Kunitz M. 1939. Isolation from beef pancreas of a crystalline protein possessing ribonuclease activity. Science 90 112–113. [DOI] [PubMed] [Google Scholar]
- Kunz W., Henle, J., and Ninham, B.W. 2004. “Zur lehre von der wirkung der salze” (About the science of the effect of salts): Franz Hofmeister's historical papers. Curr. Opin. Colloid Interface Sci. 9 19–37. [Google Scholar]
- Kupke D.W. 1960. Osmotic pressure. Adv. Protein Chem. 15 57–130. [DOI] [PubMed] [Google Scholar]
- Lawrie R.A. 1951. Crystalline forms of myoglobin from horse heart. Nature 167 802–804. [DOI] [PubMed] [Google Scholar]
- Liu W., Bratko, D., Prausnitz, J.M., and Blanch, H.W. 2004. Effect of alcohols on aqueous lysozyme–lysozyme interactions from static light-scattering measurements. Biophys. Chem. 107 289–298. [DOI] [PubMed] [Google Scholar]
- McClurg R.B. and Zukoski, C.F. 1998. The electrostatic interaction of rigid, globular proteins with arbitrary charge distributions. J. Colloid Interface Sci. 208 529–542. [DOI] [PubMed] [Google Scholar]
- McPherson A. 2001. A comparison of salts for the crystallization of macromolecules. Protein Sci. 10 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuarrie D.A. 2000. Statistical mechanics. University Science Books, Sausalito, CA.
- Miller M., Weinstein, J.N., and Wlodawer, A. 1983. Preliminary X-ray analysis of single-crystals of ovalbumin and plakalbumin. J. Biol. Chem. 258 5864–5866. [PubMed] [Google Scholar]
- Miller S., Janin, J., Lesk, A.M., and Chothia, C. 1987. Interior and surface of monomeric proteins. J. Mol. Biol. 196 641–656. [DOI] [PubMed] [Google Scholar]
- Moon Y.U., Anderson, C.O., Blanch, H.W., and Prausnitz, J.M. 2000. Osmotic pressures and second virial coefficients for aqueous saline solutions of lysozyme. Fluid Phase Equil. 168 229–239. [Google Scholar]
- Neal B.L. and Lenhoff, A.M. 1995. Excluded-volume contribution to the osmotic second virial coefficient for proteins. AIChE J. 41 1010–1014. [Google Scholar]
- Neal B.L., Asthagiri, D., and Lenhoff, A.M. 1998. Molecular origins of osmotic second virial coefficients of proteins. Biophys. J. 75 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco M. and Luque, J. 2000. Theoretical methods for the description of the solvent effect ion biomolecular systems. Chem. Rev. 100 4187–4225. [DOI] [PubMed] [Google Scholar]
- Paliwal A., Asthagiri, D., Abras, A., Lenhoff, A.M., and Paulaitis, M.E. 2005. Light-scattering studies of protein solutions: Role of hydration in weak protein–protein interactions. Biophys. J. 89 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. and Glatz, C.E. 2003. Solvent effects on the second virial coefficient of subtilisin and solubility. Cryst. Growth Des. 3 203–207. [Google Scholar]
- Patro S.Y. and Przybycien, T.M. 1996. Self-interaction chromatography: A tool for the study of protein–protein interactions in bioprocessing environments. Biotechnol. Bioeng. 52 193–203. [DOI] [PubMed] [Google Scholar]
- Piazza R. 1999. Interactions in protein solutions near crystallization: A colloid physics approach. J. Cryst. Growth 196 415–423. [Google Scholar]
- Piazza R. and Pierno, M. 2000. Protein interactions near crystallization: A microscopic approach to the Hofmeister series. J. Phys. Condens. Matter 12 A443–A449. [Google Scholar]
- Piazza R., Iacopini, S., and Galliano, M. 2002. BLGA protein solutions at high ionic strength: Vanishing attractive interactions and “frustrated” aggregation. Europhys. Lett. 59 149–154. [Google Scholar]
- Pjura P.E., Lenhoff, A.M., Leonard, S.A., and Gittis, A.G. 2000. Protein crystallization by design: Chymotrypsinogen without precipitants. J. Mol. Biol. 300 235–239. [DOI] [PubMed] [Google Scholar]
- Quiocho F.A., Sack, J.S., and Vyas, N.K. 1987. Stabilization of charges on isolated ionic groups sequestered in proteins by polarized peptide units. Nature 329 561–564. [DOI] [PubMed] [Google Scholar]
- Quiocho F.A., Wilson, D.S., and Vyas, N.K. 1989. Substrate specificity and affinity of a protein modulated by bound water molecules. Nature 340 404–407. [DOI] [PubMed] [Google Scholar]
- Retailleau P., Riès-Kautt, M., and Ducruix, A. 1997. No salting-in of lysozyme chloride observed at low ionic strength over a large range of pH. Biophys. J. 73 2156–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D.F. and Zukoski, C.F. 1996. Protein interactions and crystallization. J. Cryst. Growth 169 752–758. [Google Scholar]
- Rupley J.A. and Careri, G. 1991. Protein hydration and function. Adv. Protein Chem. 41 37–172. [DOI] [PubMed] [Google Scholar]
- Samejima T., Kamata, M., and Shibata, K. 1962. Dissociation of bovine liver catalase at Low pH. J. Biochem. 51 181–187. [DOI] [PubMed] [Google Scholar]
- Scatchard G. and Black, E.S. 1949. The effect of salts on the isoionic and isoelectric points of proteins. J. Phys. Chem. 53 88–100. [PubMed] [Google Scholar]
- Scatchard G., Scheinberg, I.H., and Armstrong, S.H. 1950. Physical chemistry of protein solutions. V. The combination of human serum albumin with thiocyanate ion. J. Am. Chem. Soc. 72 540–546. [Google Scholar]
- Scatchard G., Wu, Y.V., and Shen, A.L. 1959. Physical chemistry of protein solutions. X. The binding of small anions by serum albumin. J. Am. Chem. Soc. 81 6104–6109. [Google Scholar]
- Sehgal D. and Vijay, I.K. 1994. A method for the high-efficiency of water-soluble carbodiimide-mediated amidation. Anal. Biochem. 218 87–91. [DOI] [PubMed] [Google Scholar]
- Sitkoff D., Sharp, K.A., and Honig, B. 1994. Accurate calculation of hydration free energies using macroscopic solvent models. J. Chem. Phys. 98 1978–1988. [Google Scholar]
- Sober H.A. 1970. Handbook of biochemistry. Selected data for molecular biology, 2nd ed. The Chemical Rubber Company, Cleveland, OH.
- Söhnel O. and Novotny, P. 1985. Densities of aqueous solutions of inorganic substances. Elsevier Science, Amsterdam, Netherlands.
- Stein P.E., Leslie, A.G.W., Finch, J.T., and Carrell, R.W. 1991. Crystal-structure of uncleaved ovalbumin at 1.95 Å resolution. J. Mol. Biol. 221 941–959. [DOI] [PubMed] [Google Scholar]
- Sumner J.B. and Dounce, A.L. 1937. Crystalline catalase. J. Biol. Chem. 121 417–424. [Google Scholar]
- Tanford C. and Hauenstein, J.D. 1956. Hydrogen ion equilibria of ribonuclease. J. Am. Chem. Soc. 78 5287–5291. [Google Scholar]
- Teske C.A., Blanch, H.W., and Prausnitz, J.M. 2004. Measurement of lysozyme–lysozyme interactions with quantitative affinity chromatography. J. Phys. Chem. B 108 7437–7444. [Google Scholar]
- Tessier P.M. and Lenhoff, A.M. 2003. Measurements of protein self-association as a guide to crystallization. Curr. Opin. Biotechnol. 14 512–516. [DOI] [PubMed] [Google Scholar]
- Tessier P.M., Lenhoff, A.M., and Sandler, S.I. 2002a. Rapid measurement of protein osmotic second virial coefficients by self-interaction chromatography. Biophys. J. 82 1620–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier P.M., Vandrey, S.D., Berger, B.W., Pazhianur, R., Sandler, S.I., and Lenhoff, A.M. 2002b. Self-interaction chromatography: A novel screening method for rational protein crystallization. Acta Crystallogr. D 58 1531–1535. [DOI] [PubMed] [Google Scholar]
- Tessier P.M., Johnson, H.R., Pazhianur, R., Berger, B.W., Prentice, J.L., Bahnson, B.J., Sandler, S.I., and Lenhoff, A.M. 2003. Predictive crystallization of ribonuclease A via rapid screening of osmotic second virial coefficients. Proteins 50 303–311. [DOI] [PubMed] [Google Scholar]
- Theorell H. and Akesson, A. 1941. Studies on cytochrome c. III. Titration curves. J. Am. Chem. Soc. 63 1818–1820. [Google Scholar]
- Valente J.J., Verma, K.S., Manning, M.C., Wilson, W.W., and Henry, C.S. 2005. Second virial coefficient studies of cosolvent-induced protein self-interaction. Biophys. J. 89 4211–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velev O.D., Kaler, E.W., and Lenhoff, A.M. 1998. Protein interactions in solution characterized by light and neutron scattering: Comparison of lysozyme and chymotrypsinogen. Biophys. J. 75 2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velev O.D., Pan, Y.H., Kaler, E.W., and Lenhoff, A.M. 2005. Molecular effects of anionic surfactants on lysozyme precipitation and crystallization. Cryst. Growth Des. 5 351–359. [Google Scholar]
- Verwey E.J.W. and Overbeek, J.T.G. 1999. Theory of the stability of lyophobic colloids. Dover Publications, Mineola, NY. [DOI] [PubMed]
- Vilker V.L., Colton, C.K., and Smith, K.A. 1981. The osmotic-pressure of concentrated protein solutions: Effect of concentration and pH in saline solutions of bovine serum albumin. J. Colloid Interface Sci. 79 548–566. [Google Scholar]
- Vivarès D. and Bonneté, F. 2002. X-ray scattering studies of Aspergillus flavus urate oxidase: Towards a better understanding of PEG effects on the crystallization of large proteins. Acta Crystallogr. D 58 472–479. [DOI] [PubMed] [Google Scholar]
- Vuolanto A., Uotila, S., Leisola, M., and Visuri, K. 2003. Solubility and crystallization of xylose isomerase from Streptomyces rubiginosus . J. Cryst. Growth 257 403–411. [Google Scholar]
- Wong C.F. and McCammon, J.A. 2003. Protein simulation and drug design. Adv. Protein Chem. 66 87–121. [DOI] [PubMed] [Google Scholar]
- Wu J.Z. and Prausnitz, J.M. 1999. Osmotic pressures of aqueous bovine serum albumin solutions at high ionic strength. Fluid Phase Equil. 155 139–154. [Google Scholar]
- Yang F. and Phillips, G.N. 1996. Crystal structures of CO-, deoxy- and met- myoglobins at various pH values. J. Mol. Biol. 256 762–774. [DOI] [PubMed] [Google Scholar]