Abstract
USP33/VDU1 is a deubiquitinating enzyme that binds to the von Hippel–Lindau tumor suppressor protein. It also regulates thyroid hormone activation by deubiquitinating type 2 iodothyronine deiodinase. USP33/VDU1 contains a ZF UBP domain, a protein module found in many proteins in the ubiquitin–proteasome system. Several ZF UBP domains have been shown to bind ubiquitin, and a structure of a complex of the ZF UBP domain of isoT/USP5 and ubiquitin is available. In the present work, the solution structure of the ZF UBP domain of USP33/VDU1 has been determined by NMR spectroscopy. The structure differs from that of the USP5 domain, which contains only one of the three Zn ions present in the USP33/VDU1 structure. The USP33/VDU1 ZnF UBP domain does not bind to ubiquitin.
Keywords: VHL, deubiquitination, HDAC6, IMP
The attachment of poly-ubiquitin chains to target proteins is essential for regulated protein turnover via the ubiquitin–proteasome system (Hershko and Ciechanover 1998). Poly- or mono-ubiquitination is also important for processes such as signal transduction, transcription, DNA repair, and protein trafficking. Like most post-translational modifications, ubiquitination is a reversible process, and the human genome codes for hundreds of deubiquitinating enzymes (Nijman et al. 2005). Human ubiquitin carboxyl-terminal hydrolase 33 (USP33) is a member of the ubiquitin-specific proteases family of deubiquitinating enzymes. USP33 and a closely related deubiquitinase, USP20, have been reported to interact with pVHL, a protein mutated in von Hippel–Lindau (VHL) disease, and they are sometimes referred to as VHL-interacting deubiquitinating enzymes 1 and 2 (VDU1 and VDU2) (Li et al. 2002a,b). pVHL is part of a ubiquitin ligase complex that, when oxygen is available, ubiquitinates the alpha-subunit of hypoxia-inducible factor (HIF-1α) and targets it for proteasomal degradation (Kaelin Jr. 2007). Both USP33/VDU1 and USP20/VDU2 are ubiquitinated by pVHL, and VDU2 deubiquitinates and stabilizes HIF-1α (Li et al. 2005). USP33/VDU1 and USP20/VDU2 also regulate the supply of active thyroid hormone by deubiquitining type 2 iodothyronine deiodinase (Curcio-Morelli et al. 2003). The N-terminal region of USP33/VDU1 contains a ZnF UBP domain (Seigneurin-Berny et al. 2001; Hook et al. 2002). This protein module is also found in other deubiquitinating enzymes as well as in HDAC6 (Boyault et al. 2006), a microtubule-associated deacetylase involved in the disposal of misfolded proteins, and BRAP2/IMP, an E3 ubiquitin ligase that regulates mitogenic signaling (Matheny et al. 2004). The ZnF UBP domains of USP5, yeast BRAP2/IMP, and HDAC6 have been shown to bind to ubiquitin with high affinity. The structure of a complex between ubiquitin and the ZnF UBP domain of USP5 has been determined (Reyes-Turcu et al. 2006). The domain primarily interacts with the C-terminal diglycine motif of ubiquitin. The majority of ZnF UBP domains, including that of USP33, are predicted to contain three Zn ions. The ZnF UBP domain of HDAC6, which has a pattern of Zn ligands similar to that of the USP33 domain, has been shown to bind three Zn ions. The USP5 domain is atypical in that it only contains one Zn ion. In this article we report the solution structure and ubiquitin binding properties of the ZnF UBP domain from USP33.
Results and Discussion
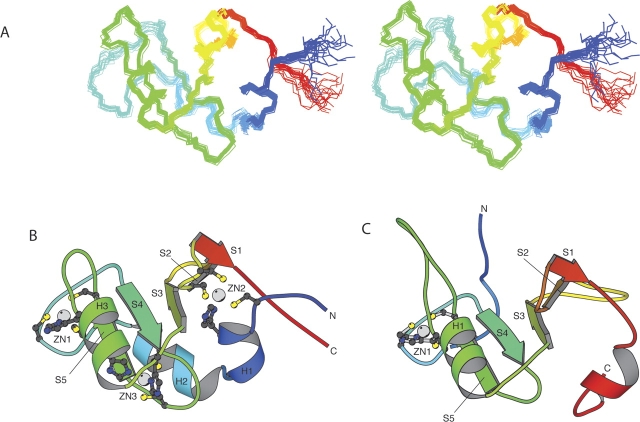
Nuclear magnetic resonance (NMR) spectroscopic methods were used to determine the solution structure of the ZnF UBP domain of human USP33. The structural statistics for the 20 lowest energy structures are summarized in ▶. An overlay of the 20 best structures is shown in ▶. The domain has an ααββαβββ secondary structure with all the β-strands antiparallel and in the order β2-β1-β3-β4-β5 (▶). The second and third helices pack on to opposite sides of the β-sheet. There are three bound Zn ions. One of these ions (Zn1) is ligated by two cystine residues in the loop between α2 and β1 and by a cystine and a histidine in the loop between β2 and α3. This site is equivalent to the single Zn ion-binding site in the USP5 domain. A second zinc ion (Zn2) is bound by a cystine and a histidine at the N terminus of the domain and by two cystine residues in turn between the final two strands of the sheet. The histidine in this binding site is at the N terminus of a small helix. The third Zn ion (Zn3) is bound by two histidines in α3 and by two cystines in the turn between β1 and β2.
Table 1.
Summary of conformational constraints and statistics for the 20 accepted structures of Znf-UBP domain
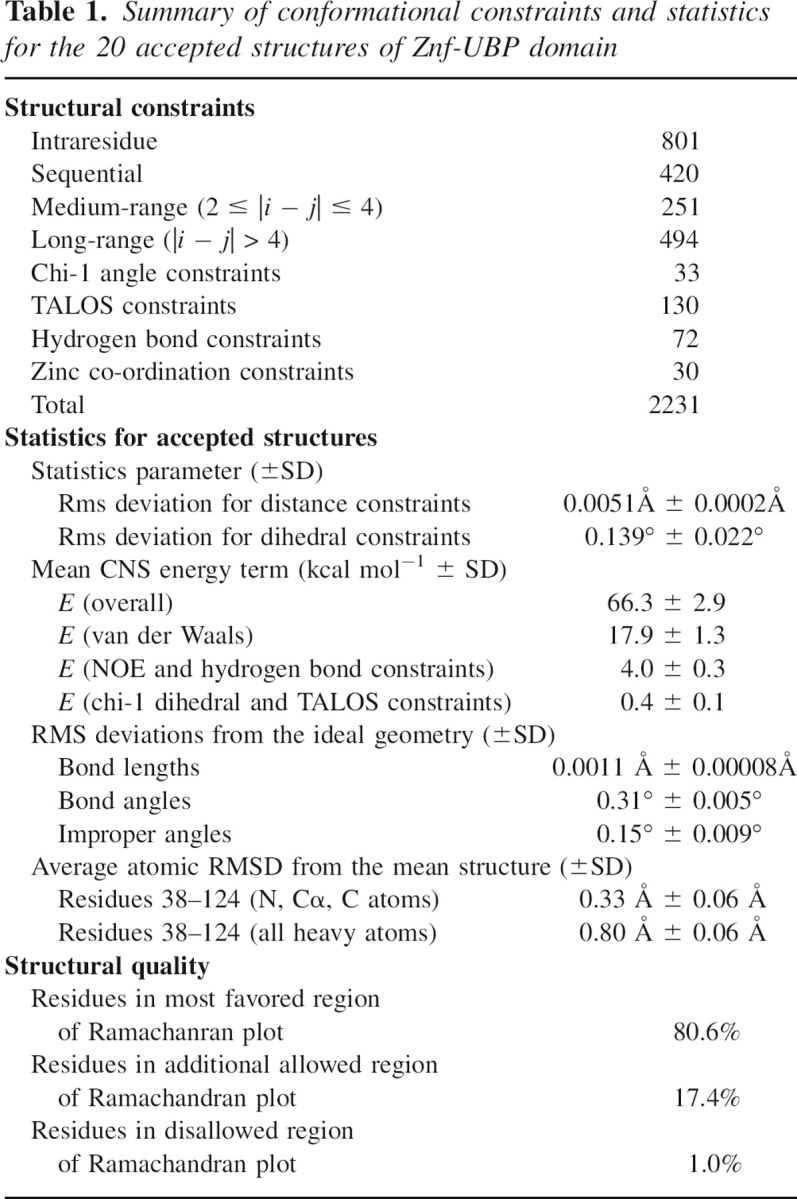
Figure 1.
Solution structure of human USP33 ZnF UBP domain. (A) An overlay of the backbone atoms of the 20 lowest energy structures. (B) Ribbon representation of the lowest energy structure. (C) The ZnF UBP domain of USP5 in the same orientation. (Prepared using the program MOLSCRIPT; Kraulis 1991.)
Although the USP5 ZnF UBP domain contains only one bound Zn ion, the central parts of the structures are very similar (▶). The only differences are that the loops between β2 and α3 and β3 and β4 are larger than in the USP33 domain, and the loop between β1 and β2 is shorter. When these regions are excluded, the central 51 residues of the domains can be superimposed with a root mean square deviation of 1.0 Å. The N- and C-terminal regions, however, differ significantly. The USP5 domain lacks the first two helices, probably because it does not have the Zn2 site, which in the USP33 structure anchors these helices to the rest of the domain. Most ZnF UBP domains are predicted to contain this site and are likely to have a structure similar to that of USP33. The region between the Zn2 and Zn1 coordination sites can apparently tolerate large insertions; the UBP22 domain for example has an additional 20 amino acids in this region compared to the USP33 domain. At the C terminus the USP33 domain structure ends after the last strand of the sheet and does not have the additional helix present in the USP5 domain. These differences have a significant effect on the relative orientations of the termini of ZnF UBP domains and could be important in providing alternative ways of positioning the domain with respect to the rest of the protein in which it is situated.
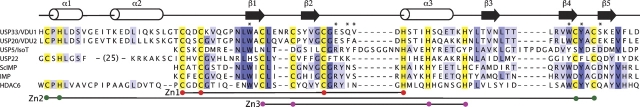
In contrast to what has been observed for some other ZnF UBP domains, we could detect no binding to ubiquitin by the USP33 domain. The USP5 domain binds to ubiquitin using residues located in a deep pocket and a loop (Reyes-Turcu et al. 2006). The equivalent loop in the USP33 domain is six residues shorter (▶). The differences in the length of this loop are unlikely to explain the lack of ubiquitin binding by the USP33 domain, as the ZnF UBP domains of HDAC6 and yeast IMP that do bind to ubiquitin have loops of similar length. Most of the residues in the pocket are conserved or are replaced with similar residues (▶). The only exception is Arg 221, which is replaced by a glutamate in USP33. The guanidino group of Arg 221 forms two hydrogen bonds with the main-chain carbonyl oxygen of ubiquitin Gly 75. The replacement of this residue with one of an opposite charge that cannot form these interacts is likely to account for the lack of binding by the USP33 domain. The USP20/VDU2 ZnF UBP domain also has a glutamate at this position and probably does not to bind to ubiquitin.
Figure 2.
Multiple sequence alignment of selected ZF UBP domains. The most highly conserved residues are boxed. Residues that contact ubiquitin in USP5 are marked with asterisks. The alignment was prepared using the program Jalview (Clamp et al. 2004).
USP5 hydrolyzes the isopeptide bonds between adjacent ubiquitins in unanchored poly-ubiquitin chains. The binding of an unmodified ubiquitin C terminus to the ZnF UBP domain of USP5 increases its catalytic activity, thereby preventing it from acting on poly-ubiquitin chains until they have been released from proteins by other deubiquitinating enzymes (Reyes-Turcu et al. 2006). USP33/VDU1 in contrast acts on poly-ubiquitin chains that are attached to proteins and therefore does not require this form of regulation. The ZnF UBP domain of yeast Ubp8, which deubiquitinates histone H2B, has a cystine at the position equivalent to Arg 221 in USP5, and this domain also probably does not bind to ubiquitin. In Ubp8 the ZnF UBP domain is required for interaction with the SAGA transcription regulatory histone acetylation complex (Ingvarsdottir et al. 2005). It is possible that the ZnF UBP domain of USP33 has similarly evolved a new regulatory function.
Materials and Methods
The DNA coding for residues 32–130 of human USP33 was amplified by PCR from a human cDNA library (Clontech) and cloned into the pRSETA (Invitrogen) expression vector. The resulting plasmid was transformed into Escherichia coli C41(DE3) cells. Cells were grown at 37°C to mid-log phase and induced with 1 mM IPTG. The temperature was then reduced to 25°C and the cells were grown for a further 16 h. Cells were lysed by sonication, and the ZnF UBP domain was purified by ion exchange chromatography using a Source Q column (Amersham Pharmacia) and gel filtration using a Superdex 75 HR column (Amersham Pharmacia). NMR spectra were recorded at 25°C on Bruker DRX 600 and AVANCE 800 MHz spectrometers. Samples contained 1 mM protein in 10 mM MES (pH 6.5), 50 mM NaCl. Resonance assignments were obtained using standard methods (Wüthrich 1986; Bax 1994). Distance constraints were derived from NOESY spectra recorded with a mixing time of 100 ms. Hydrogen bond constraints were included for a number of backbone NH groups whose signals were observed in a 2D 1H-15N-HSQC recorded in D2O at 298°K (pH 5.0). For hydrogen bond partners, two distance constraints were used where the distance (D)H-O(A) corresponded to 1.5–2.5 Å and (D)N-O(A) to 2.5–3.5 Å. Backbone torsional angle constraints were obtained from an analysis of C', N, Cα Hα, and Cβ chemical shifts using the program TALOS (Cornilescu et al. 1999). Structures were calculated using the standard torsion angle dynamics-simulated annealing protocol in the program CNS (Brunger et al. 1998). Ubiquitin binding was studied by recording a 2D 1H-15N-HSQC spectrum of 15N-labeled ZF UBP domain after the addition of equimolar amounts of unlabeled ubiquitin.
Footnotes
Reprint requests to: Mark Bycroft, MRC Centre for Protein Engineering, Hill Road, Cambridge CB2 2QH, UK; e-mail: mb10031@cus.cam.ac.uk; fax: 01223-336362.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072967807.
References
- Bax A. 1994. Multidimensional nuclear-magnetic-resonance methods for protein studies. Curr. Opin. Struct. Biol. 4 738–744. [Google Scholar]
- Boyault C., Gilquin, B., Zhang, Y., Rybin, V., Garman, E., Meyer-Klaucke, W., Matthias, P., Muller, C.W., and Khochbin, S. 2006. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 25 3357–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff, J., Searle, S.M., and Barton, G.J. 2004. The Jalview Java alignment editor. Bioinformatics 20 426–427. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C., Zavacki, A.M., Christofollete, M., Gereben, B., de Freitas, B.C., Harney, J.W., Li, Z., Wu, G., and Bianco, A.C. 2003. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel–Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J. Clin. Invest. 112 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover, A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67 425–479. [DOI] [PubMed] [Google Scholar]
- Hook S.S., Orian, A., Cowley, S.M., and Eisenman, R.N. 2002. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. 99 13425–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K., Krogan, N.J., Emre, N.C., Wyce, A., Thompson, N.J., Emili, A., Hughes, T.R., Greenblatt, J.F., and Berger, S.L. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G. 2007. The von Hippel–Lindau tumor suppressor protein and clear cell renal carcinoma. Clin. Cancer Res. 13 680s–684s. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. 1991. Molscript—A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Li Z., Na, X., Wang, D., Schoen, S.R., Messing, E.M., and Wu, G. 2002a. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel–Lindau tumor suppressor protein. J. Biol. Chem. 277 4656–4662. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang, D., Na, X., Schoen, S.R., Messing, E.M., and Wu, G. 2002b. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel–Lindau tumor suppressor. Biochem. Biophys. Res. Commun. 294 700–709. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang, D., Messing, E.M., and Wu, G. 2005. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 6 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny S.A., Chen, C., Kortum, R.L., Razidlo, G.L., Lewis, R.E., and White, M.A. 2004. Ras regulates assembly of mitogenic signalling complexes through the effector protein IMP. Nature 427 256–260. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas, M.P., Velds, A., Brummelkamp, T.R., Dirac, A.M., Sixma, T.K., and Bernards, R. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123 773–786. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Horton, J.R., Mullally, J.E., Heroux, A., Cheng, X., and Wilkinson, K.D. 2006. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124 1197–1208. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Verdel, A., Curtet, S., Lemercier, C., Garin, J., Rousseaux, S., and Khochbin, S. 2001. Identification of components of the murine histone deacetylase 6 complex: Link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21 8035–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K. 1986. NMR of proteins and nucleic acid. John Wiley, New York.