Abstract
Cell proliferation and terminal differentiation are mutually exclusive in most cell lineages. The b-zip transcription factor CCAAT/enhancer-binding protein α (C/EBPα) induces proliferation arrest and differentiation in many cell types, suggesting that both activities are linked. Here we show that C/EBPα-mediated proliferation arrest and differentiation pathways can be separated by the E7 oncoprotein of the “high-risk” human papilloma virus 16. The E7 oncoprotein overrides C/EBPα-mediated cell cycle withdrawal without compromising the transactivation activity of C/EBPα or its ability to participate in differentiation. Uncoupling of both pathways depends on the casein kinase II site of the oncoprotein but not on its ability to neutralize pocket proteins or the cyclin-dependent kinase inhibitor protein p21. Our results suggest a bifurcation of C/EBPα-mediated proliferation arrest and differentiation pathways.
CCAAT/enhancer-binding protein (C/EBP) transcription factors are intimately linked to cellular differentiation and proliferation control in a variety of tissues, including adipose, liver, intestine, the hematopoietic system, and skin (1–7). The function of C/EBPs depends on tissues and cell types varying from regulation of genes involved in energy metabolism, memory formation, or host defense against pathogens (8–19).
Expression of C/EBPα abrogates proliferation in a number of normal, transformed, or tumorigenic cells, suggesting a tumor-suppressing activity of C/EBPα (7, 20, 21). The proliferation-inhibitory function of C/EBPα depends neither on p53 nor on retinoblastoma protein (Rb) (20), yet is dominant over SV40 large T antigen or c-myc (20, 21) and resides in the complex amino-terminal transactivation domain, which also mediates differentiation control (4, 22, 23). Several lines of evidence suggest that the proliferation inhibitory and differentiation-inducing activities of C/EBPα are functionally linked. For example, in several cell types, such as liver cells, gut cells, or adipocytes, endogenous C/EBPα expression inversely correlates to cell multiplication (1, 3, 24). Furthermore, in the preadipocyte cell line 3T3-L1, expression of C/EBPα antisense RNA blocks terminal differentiation and simultaneously prevents cell cycle exit (25), whereas ectopic expression of C/EBPα in fibroblasts induces both growth arrest and differentiation (7, 25, 26). It is, however, not known whether C/EBPα uses different pathways to mediate both differentiation and inhibition of proliferation. Here we show that C/EBPα-mediated proliferation- and differentiation-control pathways bifurcate and that both pathways can be separated from each other.
MATERIALS AND METHODS
Cells and Tissue Culture.
Cells were incubated in a humidified atmosphere with 5% CO2 at 37°C. BALB/c cells, p21−/− mouse embryonal fibroblasts (MEFs), and the BOSC 23 virus-packaging cell line (27) were propagated in DMEM (Gibco); 3T3-L1 cells were propagated in a mixture of DMEM and F-12 Ham’s Medium (1:1); and all were supplemented with 10% FCS. QT6 fibroblasts (28) were grown as described (29). BALB/c or p21−/− fibroblasts were stably transfected with the chimeric C/EBPαER construct (7) or with empty vector (pMV7 or pBabePuro, vector control). C/EBPαER-expressing cells received phenol red-free DMEM with charcoal-stripped FCS. The C/EBPαER fusion protein was activated by β-estradiol (final concentration, 0.5 μM), and after medium changes, the hormone was replenished. Control cultures received solvent only. Induction of adipogenic differentiation on confluent cultures was induced exactly as described in ref. 30. The differentiation medium was exchanged every second day.
Vector Construction.
A rat C/EBPαER EcoRI fragment was fused to the hormone-binding domain of the human estrogen receptor (7) and cloned into pMV7 (31) or pBabePuro (32). Human papilloma virus (HPV)-1, -6, and -16 E7 coding regions (33) were cloned as EcoRI-SalI fragments in the pBabePuro vector with or without a hemagglutinin (HA) tag. A PCR fragment of the HPV-16 E6 coding region was cloned as a BamHI-EcoRI fragment into the pBabePuro vector.
Transfections.
For stable expression, 5 × 104 cells were transfected by a calcium phosphate method using 5 μg DNA. Twelve hours later, cells were washed, and, after another 24 h, antibiotics were added (0.8 mg/ml of G-418 or 2.5 μg/ml of puromycin). Resistant colonies were isolated (BALB/c C/EBPαER) or pooled (150–200 colonies in the case of E6 and E7 or empty vector transfections). For transient expression, 8 × 106 cells were transfected as described (29, 34) by using MSV-C/EBPα, HPV-16 E7, and E7 mutants in pBabePuro.
Retroviral Methods.
The ecotropic-packaging cell line, BOSC 23, was transiently transfected, and infectious virus was harvested (27). Target cells (5 × 105) were infected as described (27), and selection for antibiotic resistance was started 53 h later.
Growth Curves.
Cells (1 × 105) were seeded in phenol red-free DMEM, and either β-estradiol (0.5 μM) or solvent was added. Aliquots were taken every 24 h, stained with trypan blue (Sigma), and counted by microscopic inspection by using a Neubauer counting chamber.
Western Blot Analysis.
Western blotting was performed as described (35). Rabbit polyclonal antiserum (anti-C/EBPα, 1:3,000) in combination with a horseradish peroxidase-coupled anti-rabbit antibody (Dianova, Hamburg, Germany, 1:5,000) or a mAb raised against the HA tag (Babco, Richmond, CA, 1:1,000) in combination with a horseradish peroxidase-coupled anti-mouse antibody (Promega, 1:5,000) was used. Immunoreactivity was detected by chemiluminescence.
Northern Blot Analysis.
The poly(A)+ fraction from total RNA (36) was isolated (29), and Northern blots were performed as described (35). The restriction fragments used as probes for no. 461 and chicken glyceraldehyde-3-phosphate dehydrogenase have been described (37, 38). E6 and E7 probes correspond to the oncogene inserts as described above. A mouse probe for aP2/422 was generated by reverse transcription–PCR.
Oil-Red-O and Hematoxylin Staining.
Cells were washed with PBS, fixed with 4% paraformaldehyde, stained with Oil-Red-O, counterstained with Harris hematoxylin staining solution, and analyzed by bright-field microscopy.
RESULTS
C/EBPα is expressed in skin cells and appears to be involved in the differentiation of keratinocytes (6, 39, 40), the natural host of HPVs (41, 42). To complete the papilloma viral life cycle, both DNA replication and cell differentiation are required (43). Thus, papillomaviruses have adopted strategies to uncouple differentiation from cellular proliferation pathways, and the papilloma viral oncogenes supposedly are involved in separating these pathways.
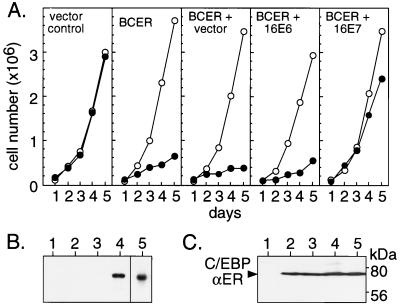
To examine whether C/EBPα-induced inhibition of proliferation can be abrogated by papilloma viral oncogenes, we constructed a BALB/c fibroblast cell line, BCER, which harbors a conditional C/EBPα-estrogen receptor chimera that previously has been shown to arrest proliferation in an estrogen-dependent fashion (7). Fig. 1A shows that estrogen treatment inhibited growth of BCER cells whereas solvent-treated BCER cells or hormone-treated parental cells remained unaffected. Next, we stably expressed the HPV-16-derived E6 or E7 oncogenes in BCER cells. Comparison of growth curves from pools of oncogene-expressing and control cultures (pools of approximately 200 colonies from each transfection to compensate for clonal variations) showed that E7 restored cell multiplication of hormone-treated BCER cells, whereas E6 did not (Fig. 1A). Release of growth inhibition by E7 was not due to loss of C/EBPαER expression as revealed by Western blotting (Fig. 1C).
Figure 1.
Abrogation of C/EBPα-induced proliferation arrest by the E7 oncogene. (A) C/EBPαER-expressing cells (BCER) were stably transfected with empty vector (+ vector) and E6 and E7 (+ 16E6 and + 16E7) expression vectors, respectively. Pools of approximately 200 independent colonies were examined for growth kinetics in the presence (●) or absence (○) of estrogen (0.5 μM). (B) RNA analysis of E6 and E7 oncogene expression. Polyadenylated RNA from vector control cells (lane 1), BCER cells (lane 2), or pools of each transfection (empty vector, lane 3; 16E7, lane 4; 16E6, lane 5) were examined by Northern blotting. (C) Protein analysis of C/EBPαER-fusion protein expression in various BCER pools. Cell lysates from vector control cells (lane 1), BCER cells (lane 2), or pools as in A and B (empty vector, lane 3; 16E7, lane 4; 16E6, lane 5) were examined by immunoblotting.
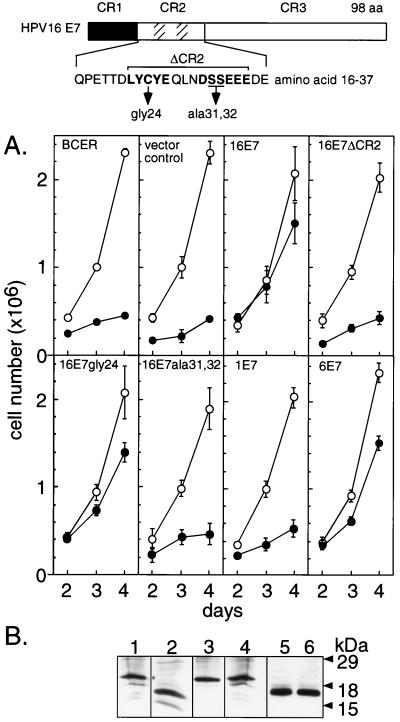
Next, we set out to determine pathways involved in C/EBPα-mediated proliferation arrest by using several HPV-16 E7 mutants. As shown in Fig. 2, we generated BCER cells that stably expressed a set of E7 mutants and determined cell multiplication from triplicate pools of transfected cells (approximately 200 puromycin-resistant colonies per pool) in the presence and absence of estrogen. When the conserved region (CR) 2 of HPV-16 E7 was removed (16E7ΔCR2; amino acids 21–35 deleted), the ability of the oncogene to restore growth was lost entirely (Fig. 2A). This region harbors two functional motifs (Fig. 2): a LXCXE motif, which binds to the pocket proteins Rb, p107, and p130 (44–48) and is involved in suppression of p21-mediated growth arrest (49), and a casein kinase II (CKII) phosphorylation site. Both motifs determine the transformation potential of E7 and its ability to collaborate with the ras oncogene (46, 48, 50). To distinguish between contributions of the LXCXE and the CKII motifs, respectively, we examined point mutants that previously have been shown to abrogate the function of either motif. Surprisingly, the point mutation in 16E7gly24 (Cys-24 to Gly), which abrogates pocket protein binding, did not affect the ability of E7 to promote growth of estrogen-treated BCER cells (Fig. 2A). Similar results were obtained with two additional pocket protein-binding-site mutants 16E7ala22 (Leu-22 to Ala) and 16E7gln26 (Glu-26 to Gln; data not shown). However, the 16E7ala31/32 mutant that lacks the CKII phosphorylation site failed to restore proliferation (Fig. 2A), suggesting that this protein region is required for interference with C/EBPα-induced proliferation inhibition.
Figure 2.
Effect of E7 mutants on C/EBPα-induced proliferation arrest. Schematic representation of E7 wt and mutant constructs used. CRs are indicated at the top. Within CR2 the pocket protein-binding motif and the CKII phosphorylation motif are indicated by hatched areas. The amino acid sequence of the wt HPV-16 E7 CR2 is shown below that. The pocket protein-binding and CKII phosphorylation motifs are indicated by bold letters, and point mutations are shown below them. (A) BCER cells were stably transfected in triplicates with pBabePuro constructs as indicated in the individual plots. Growth curves were determined (means of triplicates and SDs) from pools (approximately 200 colonies per pool) of transfected cells grown in the presence (●) or absence (○) of estrogen (0.5 μM). (B) Immunoblot analysis of HA epitope-tagged E7 constructs. BCER cells were infected with recombinant retroviruses encoding 16E7 (lane 1), 16E7ΔCR2 (lane 2), 16E7gly24 (lane 3), E7ala13/32 (lane 4), 1E7 (lane 5), and 6E7 (lane 6). Nuclear extracts were examined by Western blotting.
The importance of the E7 CKII site for the abrogation of C/EBPα-mediated proliferation block was corroborated by analysis of E7 proteins from other HPV types. HPV-1 is structurally different in the region comprising the CKII site but binds to pocket proteins with affinity similar to HPV-16 E7 (33). In contrast, E7 proteins of HPV-6 and -11 have CKII-like sites, but weakly associate with pocket proteins (47). As shown in Fig. 2A, HPV-1 E7 failed to abrogate C/EBPαER-induced proliferation arrest whereas HPV-6 E7 released the C/EBPα-induced proliferation inhibition. Results similar to HPV-6 E7 were obtained with HPV-11 E7 (data not shown). These results confirm the importance of the 16E7-type CKII site in restoring proliferation of BCER cells after C/EBPα activation.
Although RNA expression levels of wild-type (wt) and mutant E7 transcripts in stably transfected BCER pools were similar (data not shown), we consistently failed to detect E7 proteins by using commercially available antisera. To exclude that differences in oncoprotein levels accounted for the observed E7 effects, we generated epitope-tagged oncoproteins encoded by high-titered recombinant retroviruses (27) and infected BCER cells. Tens of thousands of individually infected cells per dish were obtained as judged by selection for puromycin resistance. Growth kinetics from all infected cultures were virtually indistinguishable from previous results by using pools of stably transfected BCER cells (data not shown). Moreover, immunoblot analysis showed that expression levels of various E7 proteins did not account for different growth kinetics (Fig. 2B). Taken together, our data show that the CKII site is required for abrogation of C/EBPα-mediated growth arrest, but not the high-affinity pocket protein-binding function in E7 CR2.
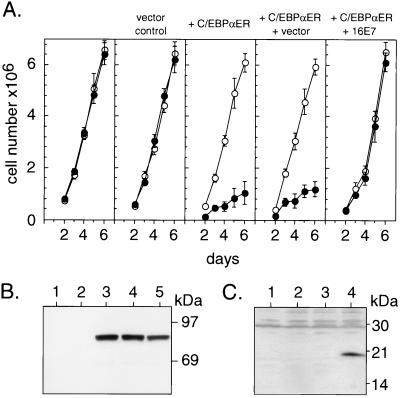
C/EBPα-mediated growth arrest may occur by stabilization of the cyclin kinase inhibitor protein p21 (51, 52). Furthermore, E7 was shown to neutralize p21 activity (49, 53). Neutralization of p21, however, depends on regions in E7 (49, 53) that were not involved in abrogating the antiproliferative function of C/EBPα. To determine unequivocally the contribution of p21 in both C/EBPα-mediated proliferation arrest and E7-rescued cell multiplication, we examined the activity of the C/EBPαER chimera in p21-deficient murine fibroblasts in the absence or presence of E7. As shown in Fig. 3A, C/EBPα still interferes with cell cycle progression in the absence of p21. Moreover, HPV-16 E7 also defeats C/EBPα-induced proliferation arrest in p21-deficient cells (Fig. 3A). This indicates that in fibroblasts, p21 is not the critical target of C/EBPα-mediated growth inhibition and is not involved in the release of proliferation arrest by E7.
Figure 3.
Induction of growth arrest by C/EBPα and its abrogation in the absence of p21. (A) p21−/− MEFs were stably transfected with empty vector (vector control) or with the chimeric C/EBPαER construct (+ C/EPBαER). A C/EPBαER-expressing clone was infected with recombinant retroviruses encoding vector (+ vector) or HA-tagged HPV-16 E7 (+ 16E7) (infection rate, >90%). Cells were examined for growth kinetics (means of triplicates and SDs) in the presence (●) and absence (○) of estrogen (0.5 μM). (B) Immunoblot analysis of C/EBPαER-fusion protein expression in p21−/− MEF. Lysates from p21−/− parental cells (lane 1), vector control (lane 2), C/EBPαER-transfected cells (lane 3), or C/EBPαER-expressing cells infected with recombinant retroviruses encoding vector (lane 4) or HA-tagged HPV-16 E7 (lane 5) were examined. (C) Immunoblot analysis of HPV-16 E7 expression in p21−/− MEFs. Nuclear extracts from p21−/− MEF transfected with empty vector (lane 1) or C/EBPαER (lane 2) and from C/EBPαER-expressing cells infected with recombinant retroviruses encoding the empty vector (lane 3) or HA-tagged HPV-16 E7 (lane 4) were examined.
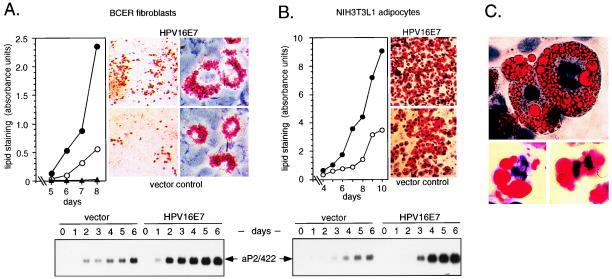
Both the antiproliferative activity and the differentiation-inducing capacity of C/EBPα depend on the presence of its transactivation domain (4, 22). To determine whether E7 also interferes with C/EBPα-mediated differentiation, we examined inducible adipogenesis in BCER cells after hormone activation of the C/EBPαER chimera in the presence or absence of E7. We stimulated fat cell differentiation with an adipogenic hormone mixture as described for a 3T3L1 preadipocyte cell line expressing a comparable C/EBPαER chimera (7). Strikingly, the E7 oncogene in BCER cells did not inhibit fat cell differentiation; in contrast, lipid accumulation even was enhanced compared with vector control cells (Fig. 4A). We observed earlier onset, increased size, and larger numbers of lipid-accumulating fat cell clones in HPV-16 E7-expressing BCER cells. This result was supported further by increased expression of the fat cell-specific gene aP2/422 (Fig. 4A). At this level of analysis the CKII E7 mutant-expressing BCER cells were indistinguishable from control or parental cells (data not shown). These observations suggested that the E7 oncoprotein separates C/EBPα-mediated proliferation and differentiation pathways, which both depend on the complex transactivation domain (4, 22, 23). To rule out the possibility of an artifact caused by the chimeric C/EBPαER protein, we chose the preadipocytic 3T3-L1 cell line, in which expression of endogenous C/EBP proteins and differentiation into fat is well characterized (2, 30, 54–56). In response to C/EBPα expression, 3T3L1 cell multiplication ceases and differentiation commences (7, 25, 26). We established 3T3-L1 cells stably expressing wt E7, the CKII phosphorylation-defective mutant, or empty vector, respectively, pooled approximately 150 puromycin-resistant colonies, and induced adipogenic differentiation according to the standard protocol (30, 56). Samples were retrieved at different time points and stained for lipid accumulation as shown in Fig. 4B. Similar to BCER cells, adipogenic differentiation was enhanced by wt HPV-16 E7. Enhancement of fat cell differentiation by E7 in 3T3L1 cells was also revealed by increased expression of the adipocyte-specific gene aP2/422 (Fig. 4B). Similar to what has been observed in BCER cells, CKII E7 mutant-expressing L1 cells were indistinguishable from control or parental cells (data not shown).
Figure 4.
Uncoupling of differentiation and cell division in BCER and 3T3-L1 cells. (A) Quantitative evaluation of differentiating BCER cells (Left). Vector control BCER cells (○) and E7-expressing BCER cells (●) in the presence of estrogen (0.5 μM) or vector control BCER cells in the absence of estrogen (▴) were subjected to the standard differentiation protocol, and samples (days 5–8) were stained with Oil-Red-O. Dishes were scanned and the absorbance was plotted to determine adipocyte differentiation. Photographs (at ×40 and ×400 original magnification) were taken at day 8 to visualize characteristic fat accumulation. (Lower) Northern analysis of differentiating BCER cells. Five micrograms of total RNA per lane was examined by using a probe for the adipocyte-specific aP2/422 gene. (B) Quantitative evaluation of differentiating 3T3L1 cells. Vector control 3T3-L1 cells (○) or E7-expressing 3T3-L1 cells (●) were subjected to the standard differentiation protocol, and samples (days 4–10) were stained and evaluated as in A (Left). Photographs (Right) were taken from differentiating cultures at day 10 (×100). (Lower) RNA analysis of differentiating 3T3L1 cells. Five micrograms of total RNA per lane was examined as in A. (C) Differentiation of 3T3-L1 cells expressing HPV-16 E7 was induced by the standard protocol. Cells were stained with hematoxylin and Oil-Red-O after 7 (Upper) or 10 days (Lower) and examined microscopically for mitotic figures (×1,000).
A striking difference between HPV-16 E7 and control L1 cells was revealed when differentiating cultures were examined on the single-cell level. As shown in Fig. 4C and Table 1, mitotic figures were detected with a high frequency in lipid laden L1 cells that express HPV-16 E7. Mitotic figures characteristic for all phases of cell division could be identified throughout adipocyte differentiation, indicating that E7 stimulates mitosis even in terminally differentiated fat cells. No mitotic figures were found in vector control cells or in the parental cell line. In comparison with wt E7, the 16E7ala31/32 mutant was compromised severely in its ability to induce mitosis in mature L1 cells, although differentiated mitotic cells still could be found to a low percentage (Table 1). The possibility that E7 alters the expression levels of C/EBPα or its isoform p42:p30 ratio was ruled out by Western blotting by using C/EBPα-specific antibodies (data not shown). We conclude from these experiments that E7 uncouples the connection between C/EBPα-induced differentiation and proliferation arrest pathways and that the structure encompassing the CKII site in E7 plays an important role in disconnecting both pathways from each other.
Table 1.
Cell division in differentiated adipocytes
| Cell line | Frequency of mitotic figures; cell counts (%)
|
|
|---|---|---|
| Experiment I | Experiment II | |
| 3T3L1 parental | 1/3,635 (<0.03) | 0/3,279 (<0.03) |
| 3T3L1 E7HPV-16 | 63/4,361 (1.4) | 51/3,692 (1.38) |
| 3T3L1 E7Ala31,32 | 12/5,467 (0.22) | 10/4,792 (0.21) |
3T3L1 cultures, as indicated in the left column, were differentiated for 10 days, and several thousand differentiated cells, as indicated, were microscopically inspected for mitotic figures. Enumeration shows absolute numbers of mitotic cells out of absolute numbers of all differentiated cells evaluated. Percentages of differentiated mitotic cells are given in parentheses for easier comparison. Two independent experiments are shown.
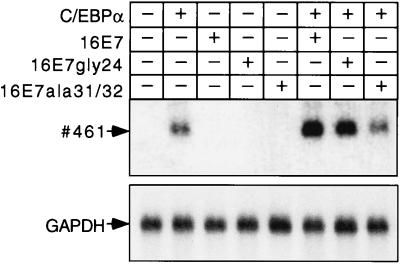
To determine whether E7 enhances the transactivation function of C/EBPα independently from adipogenic hormone cocktails or complex differentiation processes that involve the activation of other transcription factors such as C/EBPβ or PPARγ2 (2, 57), we turned to a different system. Previously, we have shown that endogenous, chromatin-embedded, myelomonocytic C/EBP target genes can be activated in QT6 fibroblasts after transient effector transfection (29, 34). Thus, QT6 fibroblasts were transiently transfected with C/EBPα and E7 expression plasmids, and the activation of the myelomonocytic C/EBP target gene 461 (29, 38) was analyzed after 18 h (Fig. 5). Expression of 461 depended on C/EBPα but not on the HPV-16 E7 oncogene (Fig. 5). However, in conjunction with C/EBPα, 461 expression was enhanced by wt 16E7 or 16E7gly24 but not by the 16E7ala31/32 mutant. Similar results were obtained with the myelomonocytic gene mim-1 (58), another direct target gene of C/EBP (34, 59) (data not shown). These results show that the HPV-16 E7 oncoprotein does not interfere negatively with, but enhances, C/EBPα-mediated transactivation and that enhancement of transactivation depends on the E7 CKII site. Furthermore, collaboration between C/EBPα and E7 occurs in various cell types, is not restricted to adipocytes, and does not require adipogenic hormones.
Figure 5.
Activation of resident C/EBPα target genes is enhanced by the E7 oncogene. Expression plasmids, as indicated, were transfected in QT6 fibroblasts. After 18 h, cells were lysed and polyadenylated mRNA was examined by Northern blot analysis with 461 and glyceraldehyde-3-phosphate dehydrogenase probes.
DISCUSSION
Here we show that C/EBPα-mediated proliferation arrest and differentiation pathways can be separated from each other. Fibroblasts and adipocytes that become proliferation-arrested by C/EBPα maintain or resume cell division once they also express HPV-16 E7. Intriguingly, in both BALB/c cells harboring a conditional C/EBPα version (BCER cells) or 3T3-L1 cells expressing endogenous C/EBPα, HPV-16 E7 simultaneously enhanced adipogenic differentiation. Both sets of data were reconciled by the observation that in E7-expressing cells proliferation was maintained during differentiation. These results were unexpected because cell growth and differentiation usually are mutually exclusive and, to the best of our knowledge, unprecedented for an oncogene. That HPV-16 E7 reinitiates or maintains cell division during C/EBPα-induced differentiation is highly reminiscent of the oncoprotein’s ability to induce DNA replication during epithelial cell differentiation (60). Our results, therefore, may be relevant for HPV pathology because C/EBP proteins have been detected in keratinocytes (6, 39), which are natural host cells of HPV multiplication and tumor formation (41, 42).
Activation of C/EBP target genes in various cell types is enhanced and differentiation proceeds significantly faster and in more cells once E7 is expressed. It is tempting, therefore, to speculate that E7 has a dual function in viral genome regulation and differentiation control and that the oncoprotein E7 takes advantage of the C/EBP pathway as a potent activator of differentiation genes. The proliferation-suppressing activity of C/EBPα, however, is undesired during viral genome replication, and, thus, E7 inhibits this particular C/EBPα activity. This would allow completion of the viral life cycle, which requires differentiation-dependent transcription of the late genes and replication of the viral genome (43). It would be interesting to know whether the capacity of HPV-16 E7 to override C/EBPα-mediated cell cycle arrest (which was not observed with E7 from the nonmalignant HPV1 subtype) might reflect the potential of HPV-16 to induce malignant conversion and whether E7 also affects the activity of other C/EBP proteins such as C/EBPβ or C/EBPδ.
The mechanism by which the HPV-16 E7 oncoprotein uncouples C/EBPα-mediated growth arrest and differentiation remains to be revealed; however, it is clear that the domain containing the CKII phosphorylation site and the negative charges of this region are required. This conclusion is based on the observations that both the phosphorylation-deficient HPV-16 E7ala31/32 mutant as well as the HPV-1 E7 failed to abrogate growth arrest. Although HPV-1 E7 harbors two negatively charged amino acid residues at equivalent positions, it contains adjacent proline residues that might alter the overall structure of this region. An interesting possibility is that the E7 protein directly interacts with C/EBPα and modifies its transactivation potential as has been shown for E7 and “bZIP” proteins of the AP-1 family (61). Preliminary data using phosphorylated and unphosphorylated E7 proteins employing the yeast two-hybrid system and biochemical-binding assays, however, would indicate that this is not the case (C.M., unpublished observations).
Although the significance of the CKII phosphorylation site for the transformation potential of HPV-16 E7 has been demonstrated, its role is still a matter of debate (50, 62, 63). Phosphorylation of the CKII site has been shown to enhance the binding of E7 to TATA box-binding protein (TBP), the core component of the basal transcription factor complex TFIID (63). Mutation of the CKII site reduced E7-mediated transactivation from the adenovirus E2 promoter (48, 64). Our data show that the CKII site of E7 is involved in the stimulation of C/EBP-dependent target gene expression and the enhancement of 3T3-L1 adipocyte differentiation. Whether these effects are mediated by association of E7 with TBP and whether and how C/EBPα participates remain to be clarified.
What are the proliferation-inhibitory pathways that C/EBPα utilizes? It has been shown that C/EBPα binds to Rb, an activity that is required for the differentiation function of C/EBPα (65, 66), and that C/EBPα enhances expression of the cdk inhibitor p21 (51, 52). The E7 oncoprotein is able to interfere with both the Rb and the p21 proliferation-inhibitory pathways (44–47, 49, 53). Unexpectedly, neither the pocket protein-binding nor p21-inactivation mutants of HPV-16 E7 (49) were compromised in their ability to overrule C/EBPα-induced proliferation arrest. Furthermore, C/EBPα was able to induce proliferation arrest in p21-deficient cells (this work) or in cells with inactivated Rb (20). Therefore, it is likely that C/EBPα uses a novel pathway to mediate proliferation arrest and that this proliferation-inhibitory pathway is targeted by the CKII site of the E7 oncoprotein.
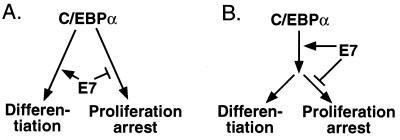
Our data reveal that both functions of the transactivation domain of C/EBPα, proliferation arrest and differentiation, can be separated from each other. It remains to be shown, however, at what stage this occurs and how uncoupling is achieved. It is conceivable that the antimitotic and the differentiation functions reside in different parts of the highly modular transactivation domain of C/EBPα (see model in Fig. 6A) and that both functions are differentially modulated by E7. Alternatively, E7 could enhance the transactivation potential of C/EBPα but block the activity of a downstream C/EBPα target that mediates its antiproliferative effect (Fig. 6B). Because mutation of the CKII site of E7 impairs both the transactivation-enhancing as well as the proliferation-stimulating function of the oncogene, we favor the first model (Fig. 6A). It is tempting to speculate that the transactivation domain of C/EBPα allows complex formation with different cofactors. Such cofactors could mediate activation of tissue-specific genes and induction of proliferation arrest. E7 could join such a differentiation-inducing complex, stabilize it, or enhance its activity and simultaneously prevent or inhibit docking of proliferation-suppressing cofactors. Alternatively, E7 could inactivate a putative proliferation-suppressing cofactor, thereby allowing more C/EBPα to engage in differentiation complex formation. E7 merely would change an equilibrium of different C/EBPα complexes with divergent functions toward the differentiation side. How that happens exactly awaits the isolation of C/EBPα mutants that selectively have lost their proliferation-inhibitory function or their ability to induce differentiation.
Figure 6.
Bifurcation of C/EBPα-mediated proliferation arrest and differentiation. (A) The antiproliferative and differentiation functions of C/EBPα reside in different parts of its modular transactivation domain and are modulated differentially by E7. (B) E7 enhances the transactivation potential of C/EBPα but blocks the activity of a downstream C/EBPα target that mediates its antiproliferative effect.
Acknowledgments
We thank Cor Calkhoven, M. Eilers, Thomas Graf, and S. A. Ness for discussions, suggestions, and critical reading of the manuscript. We thank S. L. McKnight and R. M. Umek for providing the C/EBPαER and MSVC/EBPα constructs and the C/EBPα-specific antibody, H. Land and B. Amati for providing the pBabePuro vector, and H. Land and Sally Newman for providing the p21−/− MEFs. The work was supported by a grant to A.L. (LE770/2-2) from the Deutsche Forschungsgemeinschaft “cell cycle regulation” priority program.
ABBREVIATIONS
- C/EBPα
CCAAT/enhancer binding protein α
- CR
conserved region
- CKII
casein kinase II
- HPV
human papilloma virus
- HA
hemagglutinin
- Rb
retinoblastoma protein
- MEF
mouse embryonal fibroblast
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 2.Cao Z, Umek R M, McKnight S L. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekaran C, Gordon J I. Proc Natl Acad Sci USA. 1993;90:8871–8875. doi: 10.1073/pnas.90.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freytag S O, Paielli D L, Gilbert J D. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 5.Scott L M, Civin C I, Rorth P, Friedman A D. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 6.Swart G W M, van Groningen J J M, van Ruissen F, Bergers M, Schalkwijk J. Biol Chem. 1997;378:373–379. doi: 10.1515/bchm.1997.378.5.373. [DOI] [PubMed] [Google Scholar]
- 7.Umek R M, Friedman A D, McKnight S L. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 8.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akira S, Kishimoto T. Immunol Rev. 1992;127:25–50. doi: 10.1111/j.1600-065x.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 10.Alberini C M, Ghirardi M, Metz R, Kandel E R. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 11.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 12.Friedman A D, Landschulz W H, McKnight S L. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 13.Juan T S, Wilson D R, Wilde M D, Darlington G J. Proc Natl Acad Sci USA. 1993;90:2584–2588. doi: 10.1073/pnas.90.7.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestner K H, Christy R J, Lane M D. Proc Natl Acad Sci USA. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKnight S L, Lane M D, Gluecksohn W S. Genes Dev. 1989;3:2021–2024. doi: 10.1101/gad.3.12b.2021. [DOI] [PubMed] [Google Scholar]
- 16.Ramji D P, Vitelli A, Tronche F, Cortese R, Ciliberto G. Nucleic Acids Res. 1993;21:289–294. doi: 10.1093/nar/21.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, et al. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka T, Akira S, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 20.Hendricks-Taylor L R, Darlington G J. Nucleic Acids Res. 1995;23:4726–4733. doi: 10.1093/nar/23.22.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freytag S O, Geddes T J. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- 22.Lin F T, MacDougald O A, Diehl A M, Lane M D. Proc Natl Acad Sci USA. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nerlov C, Ziff E B. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 24.Mischoulon D, Rana B, Bucher N L, Farmer S R. Mol Cell Biol. 1992;12:2553–2560. doi: 10.1128/mcb.12.6.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin F-T, Lane M D. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 26.Lin F T, Lane M D. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moscovici C, Moscovici M G, Jiminez H, Lai M M C, Hayman M J, Vogt P K. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 29.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 30.Yeh W C, Cao Z, Classon M, McKnight S L. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 31.Kirschmeier P T, Housey G M, Johnson D M, Perkins A S, Weinstein B. DNA. 1988;7:219–225. doi: 10.1089/dna.1988.7.219. [DOI] [PubMed] [Google Scholar]
- 32.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciccolini F, Di Pasquale G, Carlotti F, Crawford L, Tommasino M. Oncogene. 1994;9:2633–2638. [PubMed] [Google Scholar]
- 34.Ness S A, Kowenz L E, Casini T, Graf T, Leutz A. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 35.Müller C, Kowenz-Leutz E, Grieser-Ade S, Graf T, Leutz A. EMBO J. 1995;14:6127–6135. doi: 10.1002/j.1460-2075.1995.tb00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 37.Dugaiczyk A, Haron J A, Stone E M, Dennison O E, Rothblum K N, Schwartz R J. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- 38.Nakano T, Graf T. Oncogene. 1992;7:527–534. [PubMed] [Google Scholar]
- 39.McKnight S L. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 771–795. [Google Scholar]
- 40.Maytin E V, Habener J F. J Invest Dermatol. 1998;110:238–246. doi: 10.1046/j.1523-1747.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- 41.zur Hausen H, de Villiers E M. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 42.zur Hausen H. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 43.Howley P M. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 947–978. [Google Scholar]
- 44.Davies R, Hicks R, Crook T, Morris J, Vousden K. J Virol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyson N, Howley P M, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 46.Dyson N, Guida P, Munger K, Harlow E. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Münger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phelps W C, Münger K, Yee C L, Barnes J A, Howley P M. J Virol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones D L, Alani R M, Münger K. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Firzlaff J M, Lüscher B, Eisenman R N. Proc Natl Acad Sci USA. 1991;88:5187–5191. doi: 10.1073/pnas.88.12.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 52.Timchenko N A, Harris T E, Wilde M, Bilyeu T A, Burgess-Beusse B L, Finegold M J, Darlington G J. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green H, Kehinde O. Cell. 1974;1:113–116. [Google Scholar]
- 55.Green H, Kehinde O. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 56.Student A K, Hsu R Y, Lane M D. J Biol Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 57.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 58.Ness S A, Marknell Å, Graf T. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 59.Burk O, Mink S, Ringwald M, Klempnauer K H. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng S, Schmidt G D, Murant T, Broker T R, Chow L T. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 61.Antinore M J, Birrer M J, Patel D, Nader L, McCance D J. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- 62.Firzlaff J M, Galloway D A, Eisenman R N, Lüscher B. New Biol. 1989;1:44–53. [PubMed] [Google Scholar]
- 63.Massimi P, Pim D, Storey A, Banks L. Oncogene. 1996;12:2325–2330. [PubMed] [Google Scholar]
- 64.Edmonds C, Vousden K H. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P-L, Riley D J, Chen Y, Lee W-H. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]