Abstract
ERM (Ezrin–Radixin–Moesin) proteins function as plasma membrane–actin cytoskeleton linkers and participate in the formation of specialized domains of the plasma membrane. We have investigated ezrin function in tubulogenesis of a kidney-derived epithelial cell line, LLC-PK1. Here we show that cells overproducing a mutant form of ezrin in which Tyr-353 was changed to a phenylalanine (Y353F) undergo apoptosis when assayed for tubulogenesis. While investigating the mechanism responsible for this apoptosis, we found that ezrin interacts with p85, the regulatory subunit of phosphatidylinositol 3-kinase (PI 3-kinase). Two distinct sites of ezrin are involved in this interaction, the amino-terminal domain containing the first 309 aa and the phosphorylated Tyr-353 residue, which binds to the carboxyl-terminal SH2 domain of p85. Cells producing Y353F ezrin are defective in activation of the protein kinase Akt, a downstream target of PI 3-kinase that protects cells against apoptosis. Furthermore, the apoptotic phenotype of these cells is rescued by production of a constitutively activated form of PI 3-kinase. Taken together, these results establish a novel function for ezrin in determining survival of epithelial cells by activating the PI 3-kinase/Akt pathway.
Ezrin belongs to a family of proteins known as ERM (Ezrin–Radixin–Moesin), that function as cross-linkers between the actin cytoskeleton and the plasma membrane (1). ERM proteins are involved in microvilli formation and breakdown (2–5) and are also necessary for cell–cell and cell–substrate adhesions (2, 6–8). The conserved amino-terminal globular domain of ERM proteins interacts with several transmembrane adhesion molecules, such as CD44, CD43, ICAM-1, -2, and -3, and with phosphatidylinositol (4,5)-bisphosphate at the plasma membrane (7, 9–14). Ezrin can also interact with the membrane through the peripheral protein EBP50, a PDZ-domain-containing protein (15, 16). F actin-binding sites have been mapped both to the carboxyl-terminal end (17) and to the amino-terminal domain of the ERM proteins (18).
Most of these binding sites appear to be masked in full-length molecules, suggesting that ERM proteins are present in an inactive, closed conformation in the cytoplasm (19, 20). The opening of cytoplasmic ERM proteins and their oligomerization and association with both membrane components and actin filaments are under the control of an activation step whose molecular nature is not yet understood (20–24). Cytoplasm-to-plasma membrane translocation of ERM proteins appears to be under the control of the small GTPase rho (25–27). Rho-dependent formation of actin stress fibers, focal contacts (28), and microvilli-like structures require ERM molecules (29). ERM proteins are phosphorylated at a carboxyl-terminal threonine by Rho kinase, a downstream target of Rho, and this phosphorylation interferes with their head-to-tail interaction (30). In vivo, the phosphorylation of moesin T558 by Rho kinase plays a crucial role in the formation of microvilli-like structures (29). In addition, binding of GDP-dissociation inhibitor rhoGDI to a cryptic site in the amino-terminal domain of ERM activates rho by releasing rhoGDI inhibition (31). ERM molecules are also regulated by tyrosine phosphorylation in response to growth factors. Epidermal growth factor, platelet-derived growth factor, and hepatocyte growth factor (HGF) stimulate tyrosine phosphorylation of ezrin (3, 32, 33). In ezrin, two tyrosine residues, corresponding to amino acids 145 and 353, are phosphorylated by the epidermal growth factor receptor (34).
In vivo, ezrin is preferentially produced in epithelial cells, where it localizes to their apical surface (35). To analyze ezrin function in epithelial cell morphogenesis, we took advantage of the ability of LLC-PK1 cells to form tubules in a three-dimensional collagen type I culture on HGF stimulation. We previously showed that overproduction of wild-type ezrin in LLC-PK1 cells potentiates their tubulogenesis, whereas overproduction of a mutated ezrin, in which both Tyr-145 and Tyr-353 were changed to phenylalanine, prevented tubule formation (3). Here we report that a point mutation in ezrin, Y353 to F, induces cell apoptosis by impairing the activation of the serine/threonine protein kinase Akt, a downstream effector of phosphatidylinositol 3-kinase (PI 3-kinase) (36). Therefore, in this morphogenetic assay, ezrin signals survival of LLC-PK1 cells.
MATERIALS AND METHODS
Cells, Drugs, and Antibodies.
LLC-PK1 (CCL 101; American Type Culture Collection) cells were grown in DMEM (GIBCO/BRL) supplemented with 10% FCS and maintained at 37°C in 10% CO2. As a source of HGF, we used the dia-filtered, human fibroblast MRC5-conditioned medium or recombinant human HGF (Becton-Dickinson Labware). LY294002 (Biomol, Plymouth Meeting, PA), Z-VAD-CH2F, and BocD-CH2F (Enzyme Systems Products, Livermore, CA) were dissolved in DMSO. P5D4 mAb (37) and rabbit polyclonal anti-ezrin antibody (19) were previously described. Affinity-purified rabbit polyclonal antibody raised against p85α subunit of PI 3-kinase was kindly provided by M. Thelen (Theodor-Kocher-Institut Der Universität, Bern, Switzerland). Rabbit polyclonal anti-Akt and rabbit phosphospecific Akt (Ser-473) antibodies were purchased from New England Biolabs. Mouse monoclonal anti bcl-2 antibody (clone 124) was purchased from Dako. The anti CD2 mAb Ox34 is a kind gift of D. Cantrell (Imperial Cancer Research Fund, London, England).
Stably Transfected Cell Lines.
Clones of LLC-PK1 cells producing Y353F ezrin were established as described (3). For phenotypic reversion, one Y353F clone was retransfected with plasmid RSV-tk-hygromycin or RSV-tk-hygromycin–hBcl-2 (provided by D. Vaux, Walter and Eliza Hall Institute, Victoria, Australia), or with pEF-BOS-rCD2p110-myc (provided by D. Cantrell) in combination with RSV-tk-hygromycin. Transfected cells were selected with 0.2 mg/ml hygromycin B in continuous presence of 0.7 mg/ml G418 and cloned as above.
Three-dimensional cultures.
Collagen type I gels were prepared as follows: 1 part DMEM 10× (GIBCO/BRL), 1 part NaHCO3 (37 g/liter), 1 part FCS were mixed with 3.5 parts of a suspension of 3 × 105 cells per ml and 3.5 parts of type I collagen at 5 mg/ml (Becton Dickinson Labware) at room temperature. The gel was covered with DMEM containing 10% FCS and 100 units/ml HGF (final concentration). Colonies originating from isolated cells were cultured for 7 days with one medium change unless otherwise indicated. Photographs were taken with an epifluorescence microscope (Leica) equipped with Nomarski interference optics. To assess apoptosis, the cells in the gels were fixed with methanol for 30 min at −20°C and stained with Hoechst 33258 (10 μg/ml) in PBS. In experiments testing the effects of LY294002, cultures were assayed after 24 h. Increasing concentrations of LY294002 in DMSO or only DMSO were added to the culture medium.
Immunoprecipitation.
Confluent monolayers of LLC-PK1 clones in 10 cm dish were lysed with 1 ml of RIPA buffer (50 mM Hepes, pH 7.4/10 mM EDTA/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS) containing 1 mM sodium orthovanadate and protease inhibitors (200 μg/ml Pefabloc/15 μg/ml benzamidine/1 μg/ml pepstatin/1 μg/ml antipain). Lysates were clarified by using a 10-min centrifugation at 20,000 × g at 4°C and were then added to 10 μl of protein A Sepharose (Amersham Pharmacia) previously preincubated for 30 min at 4°C with 1 μg of affinity-purified anti-p85 antibodies or 1 μg of nonspecific rabbit IgG (Sigma). Samples were electrophoresed in 6.5% polyacrylamide gels, blotted onto nitrocellulose, and probed with specific primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch). Immunoblots were developed by chemiluminescence detection (Boehringer Mannheim).
Purification of Bacterially Produced Proteins.
Recombinant proteins glutathione S-transferase (GST), GST–ezrin1–309, and GST–ezrin310–585 (the plasmid was a kind gift of P. Mangeat, Montpellier University) were purified as described (22). pGEX-2T plasmids containing the cDNA encoding the N-SH2 and C-SH2 domains of p85α (a kind gift of S. Fischer, Institut Cochin de Genetique Moleculaire, Paris) were purified by using the same procedure.
GST Pull-Down Experiment.
Human placenta was obtained from a consenting patient. Small fragments were homogenized in RIPA buffer, and the extracts were clarified by centrifuging at 30,000 × g for 30 min and then at 20,000 × g for 10 min. Cell lysate (9 mg) was diluted in 1 ml of RIPA buffer and incubated with 600 pmol of GST, GST–ezrin1–309 or GST–ezrin310–585 and 10 μl of glutathione beads (Amersham Pharmacia). After 1 h of incubation at 4°C, beads were washed four times with 1 ml of RIPA buffer and resuspended in SDS loading buffer.
Affinity-Binding Assay with Phosphorylated Peptides.
Synthetic peptides comprising the ezrin amino acids 348–358 were synthesized in the phosphorylated or nonphosphorylated forms. The peptide sequence is biotin-RQIKIWFQNRRMKWKKLRLQDY(P)EEKTK. For affinity assays, 25 μl of streptavidin Ultralink beads (Pierce) were preincubated with 300 μg of biotinylated peptides for 1 h at 4°C in buffer A (50 mM Hepes, pH 7.4/2 mM EDTA/1% Triton X-100/100 mM NaCl/50 μM ammonium molybdate/1 mM ZnCl2). LLC-PK1 cells (7 × 106) were lysed in 1 ml of cold buffer A supplemented with protease inhibitors. Extracts were clarified by using centrifugation and incubated with the beads. Binding of p85 SH2 domains was performed in 1 ml of buffer A with 40 μg of peptide bound to 10 μl of streptavidin beads and 1 μg of GST-p85 SH2 domain fusion proteins.
Akt Activation Assay.
Overnight serum-starved cells were recovered by using EDTA/trypsin treatment, washed once in DMEM containing 0.15 g/liter BSA, and resuspended at a concentration of 3 × 106 cells per ml in DMEM containing 0.15 g/liter BSA and 50 μg/ml soybean trypsin inhibitor. This cell suspension was embedded in collagen type I as described above, except that serum was replaced by water. Gelling solution (500 μl per well of a 48-well plate) was covered with 500 μl of DMEM containing 0.15 g/l BSA and 100 μM LY294002 or DMSO alone. After 2 h, the gels were washed once in 10 ml of PBS at 4°C. Cells were lysed in 1 ml of RIPA buffer. The collagen gels were pelleted by centrifuging at 3,300 × g for 2 min at 4°C. The supernatants were precipitated in 85% cold acetone. After 1 h at −20°C, proteins were pelleted by centrifuging at 3,300 × g for 2 min at 4°C and resuspended in 50 μl of SDS loading buffer. Ten-microliter samples were electrophoresed and transferred onto nitrocellulose membranes. Blots were probed with either polyclonal anti-Akt or anti-phosphospecific Akt (Ser-473) antibodies and revealed with alkaline phosphatase-conjugated anti-rabbit antibodies and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as substrates (Promega). The intensities of the bands were quantitated by using scanning densitometry on a Bio-Profil station (Vilbert-Lourmat, Marne La Vallée, France).
RESULTS
Production of Y353F Ezrin Confers an Apoptotic Phenotype to LLC-PK1 Cells in a Tubulogenesis Assay.
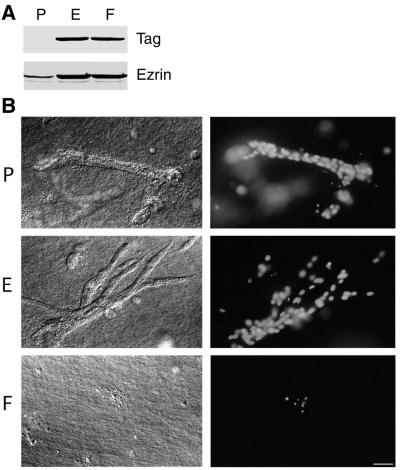
We transfected the kidney epithelial cell line LLC-PK1 with an ezrin cDNA construct in which Tyr-353 codon was replaced by an F codon. Stable clones (F cells) were isolated. F cells were compared with clones transfected with either the empty plasmid or with wild-type ezrin-expressing plasmid, P or E cells, respectively (3). Production of the exogenous proteins was detected by immunoblotting either with an anti-tag antibody or with an anti-ezrin antibody (Fig. 1A). The amount of exogenous tagged ezrin relative to the endogenous protein was estimated to be ≈10-fold higher in the clones further studied, by immunoblotting serially diluted extracts with the anti-ezrin antibody. All of the subsequent experiments were performed with at least two different clones of each type and gave identical results.
Figure 1.
Apoptosis of LLC-PK1 cells producing Y353F ezrin in a tubulogenesis assay. (A) Immunodetection of endogenous and overproduced tagged ezrin in stable LLC-PK1 clones. P, E, and F cells were transfected, respectively, with the empty vector, the same vector encoding VSV-G tagged wild-type ezrin, or VSV-G tagged Y353F ezrin. Total cellular lysates were analyzed with either P5D4 mAb (Tag) or an anti-ezrin serum. (B) Cells were cultured for 7 days in presence of HGF in a collagen type I gel. Cultures were examined with Nomarski optics (Left) and for Hoechst 33258 staining (Right). In a collagen type I gel, only apoptotic bodies, as highlighted by pyknotic nuclei, were observed in F cell cultures, whereas P and E cells were able to form tubules in presence of HGF. (Bar = 20 μm.)
These clones were tested for tubulogenesis in a three-dimensional collagen type I matrix in the presence of HGF. P and E cells were able to grow and differentiate into tubular structures. Overproduction of wild-type ezrin potentiated branching morphogenesis, as previously described (3). In sharp contrast, F cells were unable to differentiate into tubules. Hoechst staining of nuclei revealed that F cells contained condensed and fragmented chromatin. These features define a pyknotic nucleus, a hallmark of apoptosis. By 1 week, all of the F cells had died in the collagen matrix (Fig. 1B). This cell death was apoptosis, because it could be blocked by the caspase inhibitors Z-VAD-CH2F and BocD-CH2F (data not shown) or by expression of the antiapoptotic gene bcl-2 (see below).
This phenotype was specific to Y353F mutation because clones producing Y145F ezrin mutant survived (unpublished data). Interestingly, in standard culture conditions, no differences between F cells and control cells could be observed in proliferation and apoptosis rates. Y353F ezrin was correctly concentrated in the apical microvilli as assessed by using immunofluorescence. In addition, when cells were grown on collagen-coated dishes or on the top of a three-dimensional collagen gel or in a three-dimensional basement membrane matrix (Matrigel), apoptosis was not observed (data not shown).
Ezrin Interacts with PI 3-Kinase, an Enzyme Required for Cell Survival.
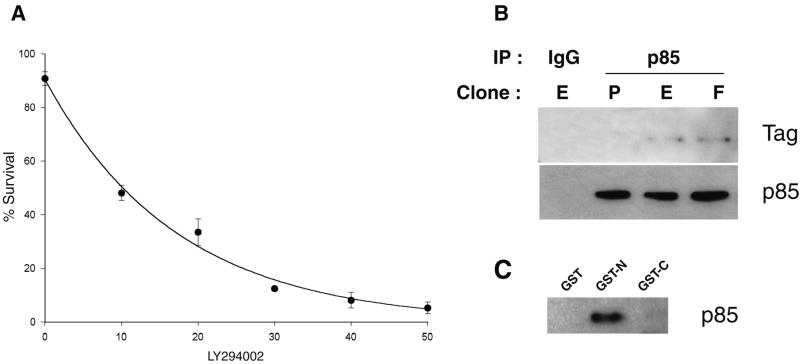
Because PI 3-kinase is a critical component in survival signaling, we tested the effect of the PI 3-kinase inhibitor, LY294002, in the tubulogenesis assay. LY294002 inhibited cell survival in a dose-dependent manner as estimated by scoring normal and pyknotic nuclei after Hoechst 33258 staining (Fig. 2A).
Figure 2.
PI 3-kinase, which is required for survival, interacts with ezrin. (A) LY294002 induces apoptosis of LLC-PK1 cells in the tubulogenesis assay. Mean ± SD of an experiment performed in triplicate is plotted. LY294002 concentration is given in μM. (B) Coimmunoprecipitation of either wild-type or Y353F ezrin with the p85 subunit of PI 3-kinase. Lysates from P, E, or F clones were immunoprecipitated with p85 affinity-purified antibodies or rabbit IgG. The immunoprecipitates were analyzed with P5D4 mAb (Tag) or polyclonal anti p85 antibodies. (C) p85 binding to the amino-terminal domain of ezrin. A lysate from placenta was incubated with immobilized GST, GST–ezrin1–309 (GST-N), or GST–ezrin310–585 (GST-C). After washing, bound material was analyzed with polyclonal anti-p85 antibodies.
Because both active PI 3-kinase and phosphorylated ezrin were required for cell survival, we tested whether PI 3-kinase and ezrin interacted. When p85, the regulatory subunit of PI 3-kinase, was immunoprecipitated from E and F clones, tagged ezrin was coprecipitated (Fig. 2B), suggesting that a complex containing ezrin and p85 exists in vivo. Interestingly, Y353F ezrin interacted with p85 as well as wild-type ezrin. To examine the regions in ezrin that mediate interaction with p85, we used the amino- and carboxyl-terminal domains of ezrin in fusion with GST to isolate interacting proteins from a placental lysate. p85 bound to the ezrin amino-terminal domain, but not to the carboxyl-terminal domain or to GST (Fig. 2C). This result suggests that a site in the amino-terminal domain interacts with p85, consistent with the coimmunoprecipitation result.
Activation of the PI 3-Kinase/Akt Pathway Requires Phosphorylation of Ezrin at Tyr-353.
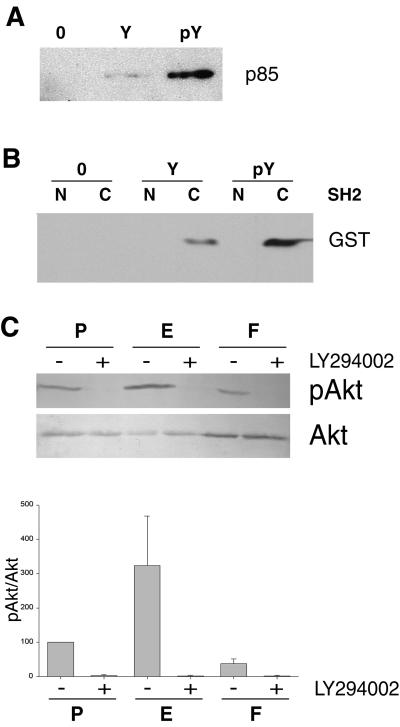
Because our morphogenesis assay indicated that the phosphorylated Tyr-353 was required for cell survival, we next investigated whether this residue was involved in an interaction with PI 3-kinase. An affinity assay was performed with an ezrin peptide encompassing the phosphorylated Tyr-353 and LLC-PK1 cell extracts. p85 bound to the phosphorylated peptide was easily detected whereas it was at the limit of detection when Tyr-353 of the peptide was not phosphorylated (Fig. 3A). To determine whether this interaction was direct and whether it involved the well characterized p85 SH2 domains, GST fusions of amino- and carboxyl-terminal SH2 domains were produced in Escherichia coli and purified. The phosphorylated peptide interacted with the carboxyl-terminal SH2 domain of the p85 subunit, but not with that of the amino terminus (Fig. 3B). This observation suggested that this second interaction involving phosphorylation of ezrin Tyr-353 and carboxyl-terminal SH2 domain of p85 was abrogated in the Y353F nonphosphorylatable form.
Figure 3.
Activation of the PI 3-kinase/Akt pathway requires ezrin phosphorylation at residue Tyr-353. (A) Binding of p85 to a phosphorylated peptide corresponding to amino acids 348–358 of ezrin. A lysate from LLC-PK1 cells was incubated with either the beads alone (0), the immobilized peptide with the nonphosphorylated residue Tyr-353 (Y), or phosphorylated residue Tyr-353 (pY). Bound material was analyzed by Western blotting with polyclonal anti-p85 antibodies. (B) Binding of the carboxyl-terminal SH2 domain of p85 to the peptide containing the phosphorylated Tyr-353. The p85 amino-terminal SH2 (N) and the carboxyl-terminal SH2 (C) domains fused to GST were incubated with either the beads alone (O), the peptide with the nonphosphorylated residue Tyr-353 (Y), or the peptide with the phosphorylated residue Tyr-353 (pY). Bound material was analyzed by Western blotting with an anti-GST serum. (C) Analysis of Akt activity in three-dimensional cultures. Serum-starved LLC-PK1 clones were embedded in collagen gels. Lysates from P, E, and F cells treated with either LY294002 (+) or DMSO (−) were analyzed with either anti-phosphospecific Akt (Ser-473) (pAkt) or anti-Akt (Akt) polyclonal antibodies. Signals were analyzed by densitometry. The pAkt/Akt ratio was calculated and set to 100% for untreated P cells. Mean ± SEM of three independent experiments are presented in the histogram.
Akt is an essential component in signaling survival downstream of PI 3-kinase. Activation of Akt protects cells from apoptosis by phosphorylating and thereby inactivating Bad, a proapoptotic member of the Bcl-2 family (38, 39). To determine whether this PI 3-kinase/Akt pathway was defective in F cells, we analyzed the level of Akt activation by using a specific antibody that detects active, phosphorylated Akt (40). Akt activity was measured in the lysates of starved cells embedded into collagen type I. No Akt activity could be detected when cells were treated with LY294002, an inhibitor of PI 3-kinase. A 3-fold increase in Akt activity was observed in E cells over P cells, whereas F cells exhibited a low level of activity, 2- and 8-fold reduction over P and E cells, respectively (Fig. 3C). These results suggest that phosphorylated ezrin is involved in Akt activation through the PI 3-kinase pathway.
Expression of a cDNA Encoding a Constitutively Active PI 3-Kinase or of bcl-2 Rescues the Apoptotic Phenotype of F Cells.
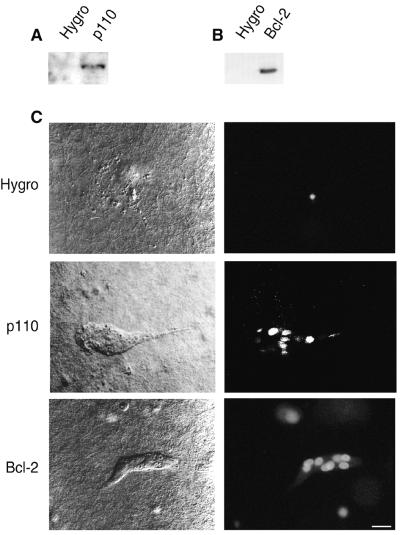
To confirm that a defect in the PI 3-kinase pathway was responsible for the apoptotic phenotype, we stably transfected F cells with a cDNA encoding a constitutively active form of PI 3-kinase, a chimera between the CD2 ectodomain and the catalytic p110 subunit (41) or with a cDNA encoding Bcl-2, an antiapoptotic protein (Fig. 4 A and B) (42). In the tubulogenesis assay, F cells transfected with the vector alone underwent apoptosis, whereas cells producing either the active PI 3-kinase or Bcl-2 survived (Fig. 4C). This suggested that ezrin activity is required upstream of PI 3-kinase for survival signaling. Reversion of the apoptotic phenotype led to the formation of small tubules of approximately 10 cells, compared with an average of 50 cells for tubules developed by P and E cells.
Figure 4.
Rescue of the F cell survival defect by constitutively active PI 3-kinase and Bcl-2. One F clone was stably transfected with either the control hygromycin-resistance plasmid (Hygro), a cDNA encoding a CD2-p110 chimera (p110), or a cDNA encoding human Bcl-2. (A) Immunodetection of the chimeric CD2-p110 protein with anti-CD2 mAb Ox34. (B) Immunodetection of exogenous Bcl-2 with a human-specific anti-Bcl-2 mAb. (C) Survival and tubulogenesis were assessed as described in Fig. 1. (Bar = 15 μm.)
DISCUSSION
Here we found that LLC-PK1 cells producing Y353F ezrin undergo apoptosis in a tubulogenesis assay. This observation suggests that, in this assay, ezrin conveys an antiapoptotic signal. The mechanism by which ezrin signals survival involves activation of the PI 3-kinase/Akt pathway.
We identified two distinct sites on ezrin that are required for interaction with p85, the amino-terminal domain corresponding to the first 309 aa, and the phosphorylated Tyr-353 site, which interacts with the carboxyl-terminal SH2 domain of p85. Consistent with the presence of two binding sites on ezrin, we observed a coimmunoprecipitation of the p85 subunit with wild-type ezrin as well as with Y353F ezrin. We propose that the ezrin amino-terminal domain may recruit the p85 subunit whereas the phosphorylation of Tyr-353 would activate the PI 3-kinase. The amino-terminal domain of ezrin is conserved among the ERM family members and the tumor suppressor gene product, Merlin/Schwannomin (43). However, Tyr-353 is not present in radixin or moesin. Therefore, this survival signaling function might be specific for ezrin.
Interaction between phosphotyrosine residues and p85 SH2 domains has been shown to be responsible for PI 3-kinase activation (44). In our study, two independent lines of evidence suggest that the carboxyl-terminal SH2 domain activates PI 3-kinase catalytic activity when engaged with ezrin phosphotyrosine 353. First, Akt activation was impaired in cells producing Y353F ezrin. Second, the survival defect of cells producing Y353F ezrin was rescued by the expression of a cDNA encoding an activated form of PI 3-kinase. The latter result establishes that ezrin functions upstream of PI 3-kinase. Moreover, our data expand the list of recognition motifs for p85 SH2 binding domains. Indeed, the motif pYEEK at position 353 in ezrin is different from the consensus sequence pY(M/V/I/E)XM defined in other PI 3-kinase-binding proteins (45).
Overproduced Y353F ezrin has a dominant-negative effect over the endogenous molecule on both cell survival and Akt activity. The two p85-binding sites might provide an explanation for this effect: the more abundant Y353F ezrin interacts with p85 via the amino-terminal domain, but is unable to activate PI 3-kinase by binding to the carboxyl-terminal SH2 domain of p85. Interestingly, the amino-terminal globular domain of ezrin binds to both p85 and phosphatidylinositol-(4,5)-bisphosphate (10). Therefore, it is possible that ezrin provides a mechanism to anchor PI 3-kinase in the proximity of its substrate. In addition, phosphoinositides might participate in the interaction of p85 with the amino-terminal domain of ezrin, as has been recently reported for the interaction of ezrin with CD44 or with ICAM-1 and ICAM-2 (14, 25).
The PI 3-kinase/Akt pathway is activated in response to growth factor stimulation and extracellular matrix adhesion (46–48). Apoptosis of F cells is observed only when cells are grown in a three-dimensional collagen type I matrix, but not on a collagen type I gel or in a three-dimensional matrigel matrix. Thus, ezrin, which is a substrate of tyrosine kinase receptors, might also be responsive to the matrix nature and its three-dimensional organization, or matrix nature and organization might determine cell susceptibility to apoptosis.
The requirement for an extracellular matrix in both cell survival and differentiation has been well documented for mammary gland epithelial cells. Basement membrane matrix triggers the differentiation of these cells into milk protein-secreting alveolar structures and prevents their apoptosis (49, 50). In contrast, tubulogenesis of kidney epithelial cells only occurs in a collagen type I matrix but not in matrigel (ref. 51, unpublished results). Similarly to tubulogenesis, apoptosis of F cells only occurs in a collagen type I matrix. This result suggests that ezrin may play an antiapoptotic role during epithelial cell differentiation in vivo.
Progression of an in situ carcinoma to a metastatic carcinoma requires invasion of the collagen type I containing connective tissue. The PI 3-kinase pathway is involved in motility and invasion of epithelial cells (52, 53). Because ezrin is also involved in cell motility (3) and cell survival in collagen type I matrix, an attractive hypothesis is that this novel ezrin/PI 3-kinase pathway may be relevant for metastasis progression.
In conclusion, ezrin, a structural linker between the plasma membrane and the actin cytoskeleton, is also a signal transducer. Through the phosphorylated Tyr-353, ezrin conveys an antiapoptotic signal for cells grown in a three-dimensional matrix.
Acknowledgments
We thank Drs. M. Thelen, D. Cantrell, S. Fischer, P. Mangeat, and D. Vaux for their generous gifts of antibodies and plasmids and Drs. B. Payrastre, R. Golsteyn, and T. Crepaldi for their helpful suggestions. The technical assistance of L. Del Maestro is gratefully acknowledged. This work was supported by grants from the Ligue Nationale Francaise Contre le Cancer, the Association pour la Recherche sur le Cancer (ARC 1825), and the Biomed program (BMH4-CT95–0090). A.G. was supported by a grant from the Ministère de l’Education Nationale de la Recherche et de la Technologie, and P.P. was supported by a grant from Association pour la Recherche sur le Cancer.
ABBREVIATIONS
- ERM
Ezrin–Radixin–Moesin
- PI 3-kinase
phosphatidylinositol 3-kinase
- GST
glutathione S-transferase
- HGF
hepatocyte growth factor
References
- 1.Bretscher A, Reczek D, Berryman M. J Cell Sci. 1997;110:3011–3018. doi: 10.1242/jcs.110.24.3011. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi K, Sato N, Kasahara H, Funayama N, Nagafuchi A, Yonemura S, Tsukita S. J Cell Biol. 1994;125:1371–1384. doi: 10.1083/jcb.125.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crepaldi T, Gautreau A, Comoglio P M, Louvard D, Arpin M. J Cell Biol. 1997;138:423–434. doi: 10.1083/jcb.138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Cohn J A, Mandel L J. Proc Natl Acad Sci USA. 1995;92:7495–7499. doi: 10.1073/pnas.92.16.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo T, Takeuchi K, Doi Y, Yonemura S, Nagata S, Tsukita S, Tsukita S. J Cell Biol. 1997;139:749–758. doi: 10.1083/jcb.139.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin M, Andréoli C, Sahuquet A, Montcourrier P, Algrain M, Mangeat P. J Cell Biol. 1995;128:1081–1093. doi: 10.1083/jcb.128.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helander T S, Carpén O, Turunen O, Kovanen P E, Vaheri A, Timonen T. Nature (London) 1996;382:265–268. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- 8.Lamb R F, Ozanne B W, Roy C, McGarry L, Stipp C, Mangeat P, Jay D G. Curr Biol. 1997;7:682–688. doi: 10.1016/s0960-9822(06)00295-8. [DOI] [PubMed] [Google Scholar]
- 9.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niggli V, Andréoli C, Roy C, Mangeat P. FEBS Lett. 1995;376:172–176. doi: 10.1016/0014-5793(95)01270-1. [DOI] [PubMed] [Google Scholar]
- 11.Serrador J M, Alonso-Lebrero J L, del Pozo M A, Furthmayr H, Schwartz-Albiez R, Calvo J, Lozano F, Sanchez-Madrid F. J Cell Biol. 1997;138:1409–1423. doi: 10.1083/jcb.138.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S, Tsukita S. J Cell Biol. 1998;140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legg J W, Isacke C M. Curr Biol. 1998;8:705–708. doi: 10.1016/s0960-9822(98)70277-5. [DOI] [PubMed] [Google Scholar]
- 14.Heiska L, Alfthan K, Grönholm M, Vilja P, Vaheri A, Carpén O. J Biol Chem. 1998;273:21893–21900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- 15.Reczek D, Berryman M, Bretscher A. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reczek D, Bretscher A. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- 17.Turunen O, Wahlström T, Vaheri A. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy C, Martin M, Mangeat P. J Biol Chem. 1997;272:20088–20095. doi: 10.1074/jbc.272.32.20088. [DOI] [PubMed] [Google Scholar]
- 19.Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gary R, Bretscher A. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gary R, Bretscher A. Proc Natl Acad Sci USA. 1993;90:10846–10850. doi: 10.1073/pnas.90.22.10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andréoli C, Martin M, Leborgne R, Reggio H, Mangeat P. J Cell Sci. 1994;107:2509–2521. doi: 10.1242/jcs.107.9.2509. [DOI] [PubMed] [Google Scholar]
- 23.Berryman M, Gary R, Bretscher A. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bretscher A, Gary R, Berryman M. Biochemistry. 1995;34:16830–16837. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- 25.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotani H, Takaishi K, Sasaki T, Takai Y. Oncogene. 1997;14:1705–1713. doi: 10.1038/sj.onc.1200998. [DOI] [PubMed] [Google Scholar]
- 27.Shaw R S, Henry M, Solomon F, Jacks T. Mol Biol Cell. 1998;9:403–419. doi: 10.1091/mbc.9.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay D J G, Esch F, Furthmayr H, Hall A. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshiro N, Fukata Y, Kaibuchi K. J Biol Chem. 1998;273:34663–34666. doi: 10.1074/jbc.273.52.34663. [DOI] [PubMed] [Google Scholar]
- 30.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Tsukita S, Tsukita S, Takai Y. J Biol Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- 32.Gould K, Cooper J, Bretscher A, Hunter T. J Cell Biol. 1986;102:660–669. doi: 10.1083/jcb.102.2.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bretscher A. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg J, Hunter T. J Biol Chem. 1992;267:19258–19265. [PubMed] [Google Scholar]
- 35.Berryman M, Franck Z, Bretscher A. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 36.Franke T F, Yang S-I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 37.Kreis T E. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.del Peso L, Gonzàlez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 39.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 40.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 41.Reif K, Nobes C D, Thomas G, Hall A, Cantrell D A. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 42.Vaux D L, Weissman I L, Kim S K. Science. 1992;258:1955–1957. doi: 10.1126/science.1470921. [DOI] [PubMed] [Google Scholar]
- 43.Arpin M, Algrain M, Louvard D. Curr Opin Cell Biol. 1994;6:136–141. doi: 10.1016/0955-0674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 44.Carpenter C L, Auger K R, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley L C. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 45.Kapeller R, Cantley L C. BioEssays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 46.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 47.Khwaja A, Rodriguez-Viciana P, Wennström S, Warne P H, Downward J. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulik G, Klippel A, Weber M J. Mol Cell Biol. 1998;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boudreau N, Sympson C J, Werb Z, Bissell M J. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roskelley C D, Desprez P Y, Bissell M J. Proc Natl Acad Sci USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos O F P, Nigam S K. Dev Biol. 1993;160:293–302. doi: 10.1006/dbio.1993.1308. [DOI] [PubMed] [Google Scholar]
- 52.Shaw L M, Rabinovitz I, Wang H H-F, Toker A, Mercurio A M. Cell. 1997;91:949–960. doi: 10.1016/s0092-8674(00)80486-9. [DOI] [PubMed] [Google Scholar]
- 53.Keely P J, Westwick J K, Whitehead I P, Der C J, Parise L V. Nature (London) 1997;390:632–635. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]