Abstract
With respect to conveying useful comparative information, current biological classifications are seriously flawed because they fail to (i) standardize criteria for taxonomic ranking and (ii) equilibrate assignments of taxonomic rank across disparate kinds of organisms. In principle, these problems could be rectified by adopting a universal taxonomic yardstick based on absolute dates of the nodes in evolutionary trees. By using procedures of temporal banding described herein, a simple philosophy of biological classification is proposed that would retain a manageable number of categorical ranks yet apply them in standardized fashion to time-dated phylogenies. The phylogenetic knowledge required for a time-standardized nomenclature arguably may emerge in the foreseeable future from vast increases in multilocus DNA sequence information (coupled with continued attention to phylogeny estimation from traditional systematic data). By someday encapsulating time-dated phylogenies in a familiar yet modified hierarchical ranking scheme, a temporal-banding approach would improve the comparative information content of biological classifications.
Keywords: systematics, taxonomy, molecular clocks, comparative biology
If systematics is to be a science it must bow to the self-evident requirement that objects to which the same label is given must be comparable in some way.
Hennig (1)
No scientific enterprise, least of all one that considers the promotion of nomenclatural universality as one of its primary objectives, can accept the inconsistencies and ambiguities current in biological taxonomy.
de Queiroz and Gauthier (2)
The Linnaean system of classification (3) has served biologists for more than two centuries. Originally designed to catalogue diverse works of the Creator, the hierarchical categories in this ordering scheme later became interpretable as natural outcomes of the nested branching structures in evolutionary trees. Yet most classifications in current use continue to group species according to some unspecified mix of similarity by resemblance (phenetic grades) and similarity by descent (phyletic clades). Apart from this epistemological flaw, the kinds of empirical data used to recognize grades or clades vary greatly among organismal groups, with no explicit attempt to normalize assayed characters, to equilibrate taxonomic assignments, or even to adopt any universally standardized criteria for taxonomic ranking (4, 5).
Hennig (ref. 1, pp. 154–161) proposed more than 30 years ago that the categorical rank of any taxon should denote its geological age, but this suggestion has been neglected (see refs. 6 and 7), perhaps because of a widespread perception that the nodes in evolutionary trees cannot be dated with reasonable assurance. However, recent studies suggest that multilocus DNA sequence data (complemented by fossil evidence) may help to resolve once intractable issues on the approximate absolute nodal depths in phylogenetic reconstructions (8–11). For example, Kumar and Hedges (12) analyzed sequences from 658 nuclear genes representing 207 vertebrate species to estimate divergence times for mammalian orders and other vertebrate lineages.
In that study, gene-specific molecular clocks (calibrated by using well dated anchor points from fossil evidence) were used to assign provisional times to other internal nodes in the vertebrate tree. Although divergence times from single-gene clocks have large statistical errors, reliable dates presumably emerged from estimates accumulated across hundreds of loci (but see also refs. 13–15). Given the current explosive growth in DNA sequence information (16–19), such multilocus phylogenetic treatments may one day become commonplace, and it is not overly fanciful to imagine that much of the tree of life (D. R. Maddison and W. P. Maddison, http://phylogeny.arizona.edu/tree/phylogeny.html.) will be reconstructed in the ensuing decades. Without necessarily endorsing the particular conclusions of recent molecular phylogenetic appraisals, we address here a broader question that such assessments logically raise: How should time-dated phylogenies, once available, be translated into biological classifications?
For the sake of current discussion, we adopt as a starting point a fundamental assumption [which itself has been debated elsewhere (see refs. 2 and 20)] that some sort of ranked classification scheme is desirable as an informational shorthand for encapsulating the more complete information in the phylogenetic trees they summarize. Any classification system (in biology or elsewhere) is arbitrary to some extent because its purposes are to condense, organize, convey, and permit the retrieval of usable information (1, 6, 21), and many different artifices can be envisioned toward these ends. However, many biologists now adopt the view that propinquity of descent is a particularly useful criterion for classifying organisms because phylogenetic relationships convey a great deal of information about known or yet-to-be-discovered characteristics of the members of a taxon (2, 22). This is the view herein adopted and logically extended.
Shortcomings of Conventional Taxonomic Practice
A primary limitation of conventional taxonomy is that extant taxa placed at the same Linnaean rank are not necessarily equivalent in age, diversity, disparity, or any other consistent property of their biology or evolutionary histories. Current taxonomic anachronisms communicate almost no information as to whether, for example, a rank such as genus, tribe, family, or order in mammals is equivalent to its counterpart rank in fishes, insects, or any other assemblage. Thus, the ranks in current use do little to aid, and indeed often may hinder, comparative evolutionary studies.
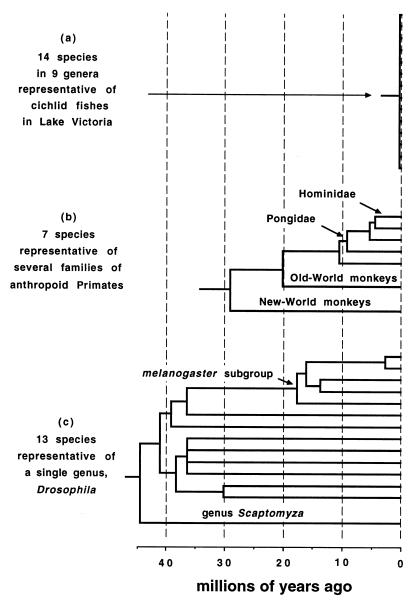
For example, current classifications can be grossly misleading if interpreted to imply absolute or relative dates of evolutionary separation. Some species of fruit flies in the genus Drosophila last shared common ancestors >40 million years ago whereas some primates currently placed in different families separated within the last few million years, and some cichlid fishes placed in different genera diverged within the last few thousand years (Fig. 1). Even if intragroup taxonomies generally scale with time because of the hierarchical nature of branched lineages within them, current taxonomic ranks clearly do not scale in a time-standardized fashion across organismal groups.
Figure 1.
Examples of gross disparities of taxonomic assignments in current classifications. The phylogenies depicted, based on an integration of molecular and paleontological evidence, come from information in refs. 43 (a), 26 (b), and 44 (c).
Nor is it demonstrable that current taxonomies scale consistently with any other features of biological interest, such as magnitudes of divergence in morphology, physiology, behavior, or ecology. For the conventional characters used by systematists, this state of affairs exists because morphological or other phenotypic features useful in, for example, fish systematics (e.g., fin placement, pharyngeal apparatus, etc.) often lack clear homologies to phylogenetically informative features in mammals (teeth arrangement, penile structure) or insects (configuration of body segments, wing venation). The great diversity of life precludes standardized organismal-level comparisons.
For molecular characters such as those involved in biochemical pathways nearly universal to life, this situation differs dramatically. With sufficient effort, it is possible to compare homologous and often orthologous gene sequences in nearly any species and to accumulate phylogenetic information across hundreds of comparable loci (12). Given the quasi-clocklike behavior of many biological macromolecules (12, 23–26), temporal as well as cladistic aspects of phylogenetic trees can be estimated. However, our focus here is not on the reliability of molecular dating but, rather, on how standardized classifications might be erected should well dated trees become widely available in the future.
Proposal for a Standardized Classification Scheme
We propose that the approximate dates of nodes in evolutionary trees should be the universal criterion according to which taxonomic classifications above the level of biological species are erected. Decisions about the particular window of time to be associated with each taxonomic rank are arbitrary, but the conventions adopted should reflect some agreed-to consensus (7) among practicing systematists reaching this initial consensus may be the most difficult part of the entire endeavor).
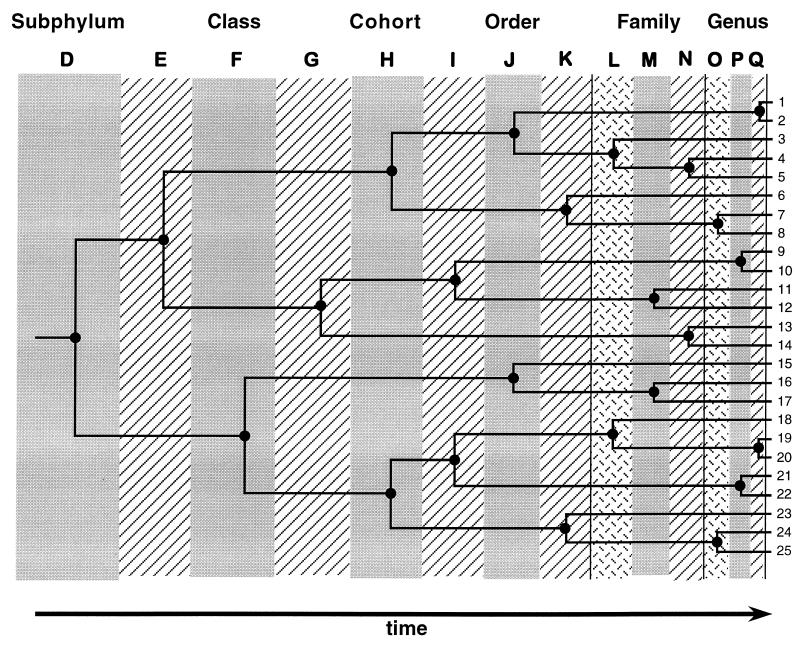
Once these taxonomic conventions are adopted, the procedural rules for implementing temporal banding are exceedingly simple (Fig. 2): (i) Overlay the universal temporal bands (or columns, as in Fig. 2) on any “right-justified” pictorial representation of a time-dated phylogeny. (ii) For each phylogenetic node falling within a designated window of time, unite the extant species belonging to that clade into a taxon to be recognized at the particular hierarchical rank specified by that temporal column. (iii) Looking back from the present, any pair of extant species whose ancestral lineages traverse particular temporal bands without joining to form a clade are to be placed in different named taxa at each of the ranks defined by those temporal bands.
Figure 2.
Hypothetical phylogeny explaining the concept of temporal banding (see text).
To illustrate the application of these rules, consider in Fig. 2 the clade comprised of species 9–14. This clade would be designated as a subclass because that taxonomic rank corresponds to the temporal band in which the node for that clade falls. Within that subclass, extant species 9–10 would be placed in the same genus, species 13–14 in one subfamily, species 11–12 in one taxonomic family, and species 9–12 in a superorder. Overall, the temporal-banding scheme as applied to the phylogeny in Fig. 2 would yield named clades corresponding to (for example) 21 taxonomic genera, 15 families, 9 orders, 5 cohorts, 3 classes, and 1 subphylum.
Several points should be made about such exercises in temporal banding. First, depending on the original convention adopted, the names of the taxonomic ranks used may be those of the conventional Linnaean framework, or they may be any standardized alternative such as lettered designations (e.g., D through Q in Fig. 2). A classification for the 25 extant species in Fig. 2 is shown in alphanumeric format in Table 1.
Table 1.
Complete temporal-banded classification, shown in alphanumeric code, for the 25 extant species in Fig. 2
| Extant species | Classification | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D1 | E1 | F1 | G1 | H1 | I1 | J1 | K1 | L1 | M1 | N1 | O1 | P1 | Q1 |
| 2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 3 | — | — | — | — | — | — | — | K2 | L2 | M2 | N2 | O2 | P2 | Q2 |
| 4 | — | — | — | — | — | — | — | — | — | M3 | N3 | O3 | P3 | Q3 |
| 5 | — | — | — | — | — | — | — | — | — | — | — | O4 | P4 | Q4 |
| 6 | — | — | — | — | — | I2 | J2 | K3 | L3 | M4 | N4 | O5 | P5 | Q5 |
| 7 | — | — | — | — | — | — | — | — | L4 | M5 | N5 | O6 | P6 | Q6 |
| 8 | — | — | — | — | — | — | — | — | — | — | — | — | P7 | Q7 |
| 9 | — | — | F2 | G2 | H2 | I3 | J3 | K4 | L5 | M6 | N6 | O7 | P8 | Q8 |
| 10 | — | — | — | — | — | — | — | — | — | — | — | — | — | Q9 |
| 11 | — | — | — | — | — | — | J4 | K5 | L6 | M7 | N7 | O8 | P9 | Q10 |
| 12 | — | — | — | — | — | — | — | — | — | — | N8 | O9 | P10 | Q11 |
| 13 | — | — | — | — | H3 | I4 | J5 | K6 | L7 | M8 | N9 | O10 | P11 | Q12 |
| 14 | — | — | — | — | — | — | — | — | — | — | — | O11 | P12 | Q13 |
| 15 | — | E2 | F3 | G3 | H4 | I5 | J6 | K7 | L8 | M9 | N10 | O12 | P13 | Q14 |
| 16 | — | — | — | — | — | — | — | K8 | L9 | M10 | N11 | O13 | P14 | Q15 |
| 17 | — | — | — | — | — | — | — | — | — | — | N12 | O14 | P15 | Q16 |
| 18 | — | — | — | G4 | H5 | I6 | J7 | K9 | L10 | M11 | N13 | O15 | P16 | Q17 |
| 19 | — | — | — | — | — | — | — | — | — | M12 | N14 | O16 | P17 | Q18 |
| 20 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 21 | — | — | — | — | — | — | J8 | K10 | L11 | M13 | N15 | O17 | P18 | Q19 |
| 22 | — | — | — | — | — | — | — | — | — | — | — | — | — | Q20 |
| 23 | — | — | — | — | — | I7 | J9 | K11 | L12 | M14 | N16 | O18 | P19 | Q21 |
| 24 | — | — | — | — | — | — | — | — | L13 | M15 | N17 | O19 | P20 | Q22 |
| 25 | — | — | — | — | — | — | — | — | — | — | — | — | P21 | Q23 |
Dashes indicate identity at a given taxonomic rank to the species in the preceding row. Letters refer to the temporal bands in Fig. 2, and each number signifies a different taxon name at the indicated rank. For any two extant species, the rightmost alphanumeric designation that they share denotes the taxonomic rank of the clade to which they belong.
Second, the windows of time agreed to (in the original ratified convention) need not be of equal width for different taxonomic ranks. Indeed, to maintain a manageable number of hierarchical ranks, as well as to acknowledge the poorer temporal resolution (wider absolute confidence limits on date estimates) normally expected from empirical data on more ancient nodes in a phylogenetic tree, the temporal bands probably should be increasingly wide for more inclusive taxonomic categories (i.e., those further to the left in Fig. 2).
Third, the number of temporal bands to be recognized is arbitrary in the initial convention. However, that number should reflect some agreed-on compromise among various considerations, such as the degree of temporal resolution likely to be achieved with molecular or other data and the desirability of naming all clades versus the need for a simple and usable taxonomic summary. If the temporal bands were indefinitely small and the resolving power in the assays infinitely great, every clade could be named, but such a taxonomy surely would be no less cumbersome as a “summary” than would a direct pictorial representation of the phylogeny itself. In the taxonomic summaries envisioned in this proposal, all named taxa would be clades, but not all clades would be named taxa (because multiple bifurcations within a temporal band are subsumed into a single taxon at that rank).
Fourth, a universal convention (that we tend to favor) could match each temporal band (and, hence, taxonomic category) to a geological episode. For example, each taxonomic genus (or equivalent lettered rank) might signify membership in a clade whose lineages shared a most recent common ancestor in the Pliocene, and each taxonomic order could indicate a clade whose coalescent node fell in the Jurassic (Table 2). A scaling of taxonomic ranks to conventional geological windows is appealing because the latter are well known, are fortuitously about equal in number to conventional Linnaean ranks (Table 2), and often are associated with important evolutionary events such as mass extinctions (e.g., the Permian and Cretaceous) and adaptive radiations (perhaps in the Cambrian).
Table 2.
One proposal for matching temporal bands of classification rank to the geological time scale
| Taxonomic rank* | Geological episode | Temporal band† |
|---|---|---|
| Domain(A) | Archaean | 2.5–3.6 Bya |
| Kingdom(B) | Proterozoic | 0.55–2.5 Bya |
| Phylum(C) | Cambrian | 500–550 Mya |
| Subphylum(D) | Ordovician | 440–500 Mya |
| Superclass(E) | Silurian | 410–440 Mya |
| Class(F) | Devonian | 350–410 Mya |
| Subclass(G) | Carboniferous | 290–350 Mya |
| Cohort(H) | Permian | 250–290 Mya |
| Superorder(I) | Triassic | 205–250 Mya |
| Order(J) | Jurassic | 145–205 Mya |
| Suborder(K) | Cretaceous | 65–145 Mya |
| Superfamily(L) | Paleocene | 56–65 Mya |
| Family(M) | Eocene | 33–56 Mya |
| Subfamily(N) | Oligocene | 24–33 Mya |
| Tribe(O) | Miocene | 5–24 Mya |
| Genus(P) | Pliocene | 2–5 Mya |
| Subgenus(Q) | Pleistocene | 0–2 Mya |
| Species‡— | — | — |
Mya, million years ago; Bya, billion years ago.
All categories except Domain are from ref. 6.
Within which the relevant phylogenetic node falls (from ref. 22).
The current proposal does not extend to species-level taxonomic assignments, where biological criteria including reproductive isolation should be applied (see ref. 42).
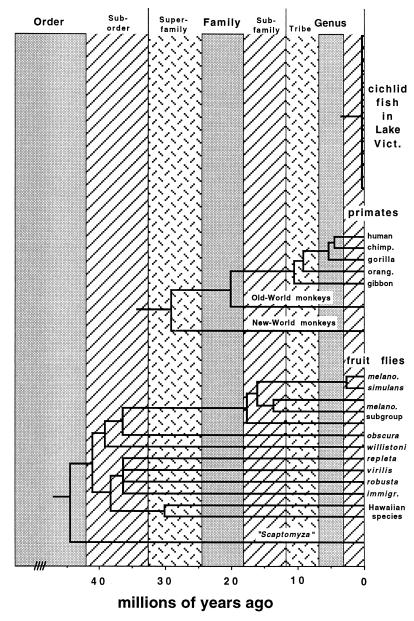
Fig. 3 illustrates a time-standardized classification scheme based on temporal banding for the three organismal groups in Fig. 1 that currently have wildly nonstandardized taxonomies. Under the universal temporal windows as drawn, all assayed species of cichlid fishes in Lake Victoria would be placed into a single subgenus whereas the fruit flies shown (current genus Drosophila) would be split into 12 genera and 9 families all united into a clade at the level of suborder. Humans, chimpanzees, and gorillas would be placed in one genus, all great apes (including orangutan, gibbon, and human) would be in one tribe, Old World monkeys would join the great-ape clade at the family level, and New World monkeys would join all of these at the rank of superfamily.
Figure 3.
The temporal-banding concept as applied to produce a time-standardized classification for the three groups of organisms in Fig. 1.
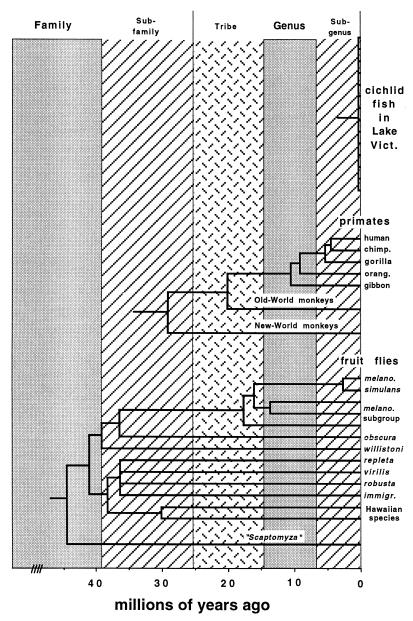
Fig. 4 shows a time-standardized classification under an alternative convention in which the temporal bands were stretched to greater width. A comparison of Figs. 3 and 4 illustrates two major points: The particular choice of temporal bands to be sanctioned is definitional at the outset, but, once the convention is ratified and applied universally, the classification scheme adopted will enable time-standardized comparisons of taxonomic rank anywhere.
Figure 4.
The temporal-banding concept as applied to produce an alternative time-standardized classification for the three groups of organisms in Fig. 1.
Merits and Demerits of a Time-Standardized Classification
A universal time-based taxonomy would both prompt and facilitate comparative evolutionary studies (as per refs. 27–31). By providing standardized information on the approximate dates when particular taxa separated phylogenetically, the classification itself immediately would suggest many otherwise obscure research opportunities in fields such as macroevolution, biogeography, and conservation biology.
For example, one immediate boon would be to promote comparisons of evolutionary rates in any traits of interest, both within and among organismal groups and in asexual as well as sexual taxa. Much effort in evolutionary biology goes into rate assessments, be they measured in the currency of a “Darwin” [a change in morphology by a factor of e per million years (32)] or the parameter λ [rate of nucleotide substitution in DNA (18)]. With the approximate time of divergence of particular taxa as a known denominator in rate equations, comparisons of evolutionary tempos in any molecular or organismal traits would be assisted by the time-standardized classification itself. Also, the age-old issue of whether to “split” or “lump” supraspecific taxa would vanish because the ratified standards in the temporal-banding convention would be the final arbiter.
A time-based classification also would benefit researchers involved in “genetic prospecting.” A goal of many private and government research ventures is to explore the biological world for new genes or gene products of use, for example, as diagnostic or therapeutic agents. With a universal taxonomy based on divergence times, the phylogenetic landscape available for exploration would be far more evident. For similar reasons, a time-standardized classification would inform conservation efforts aimed at preserving phylogenetic diversity (33–35).
The major concern in any transition to a new classification scheme is that taxonomic alterations will compromise (at least temporarily) the retrieval and communication of biological information. Although a time-scaled classification might be far preferable to the current system in principle, there are undeniable costs of taxonomic confusion in the short term, and these might be perceived to outweigh any longer-term benefits. For this reason, and because relatively few time-dated phylogenies are as yet available anyway, the temporal-banding scheme should probably be implemented only after much of the tree of life has been resolved.
At that future date, various procedures can be envisioned that would honor traditional classifications to the extent possible and thereby minimize taxonomic upheaval. First, as mentioned, familiar Linnaean ranks might be retained (albeit in renovated form to accommodate the temporal-banding convention). Second, the particular temporal bands adopted could be chosen according to the explicit criterion that they minimally disrupt current classifications. Another possibility is a dual scheme of classification (either in a transitional period or permanently). Thus, the current Linnaean classification could be retained while a second, time-based classification using alternative labels (e.g., alphanumeric characters) is adopted as well. Additional matters of convention will have to be considered collectively by systematists, such as the always-thorny issues (under any classification scheme) of how to classify fossils (2, 36), how to deal with taxonomic instability as new information comes to light, and how technically to prescribe nomenclature (37, 38).
A primary objection that may be voiced to the current proposal is that the goal of dating nodes in evolutionary trees is unachievable for most taxa. If true, that itself would be an extremely important message for the fields of molecular biology, evolutionary biology, and systematics. However, at least some leading systematists believe that it is only a matter of time before the complete phylogenetic histories of most groups will be illuminated with considerable clarity (39–41). Although this remains to be seen, the vision of someday erecting a time-standardized taxonomy could only add incentive to desirable attempts to identify the temporal positions as well as cladistic arrangements of the nodes in evolutionary trees.
Acknowledgments
We thank Andrew DeWoody, David Hibbitt, Richard Olmstead, and DeEtte Walker for useful comments on the manuscript. Work in the Avise laboratory is supported by a Pew Foundation Award and by funds from the University of Georgia.
References
- 1.Hennig W. Phylogenetic Systematics. Urbana, IL: Univ. of Illinois Press; 1966. [Google Scholar]
- 2.de Queiroz K, Gauthier J. Annu Rev Ecol Syst. 1992;23:499–480. [Google Scholar]
- 3.Linnaeus C. Systema Naturae. Stockholm: Laurentius Galvius; 1758. [Google Scholar]
- 4.Anderson C. Classification of Organisms: Living and Fossil. Lancaster, OH: Golden Crowns Press; 1992. [DOI] [PubMed] [Google Scholar]
- 5.Wheelis M L, Kandler O, Woese C R. Proc Natl Acad Sci USA. 1992;89:2930–2934. doi: 10.1073/pnas.89.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayr E, Ashlock P A. Principles of Systematic Zoology. New York: McGraw–Hill; 1991. [Google Scholar]
- 7.Goodman M, Porter C A, Czelusniak J, Page S L, Schneider H, Shoshani J, Gunnell G, Groves C P. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle R F, Feng D-F, Tsang S, Cho G, Little E. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 9.Burmester T, Massey H C, Jr, Zakharkin S O, Benes H. J Mol Evol. 1998;47:93–108. doi: 10.1007/pl00006366. [DOI] [PubMed] [Google Scholar]
- 10.Gu X. J Mol Evol. 1998;47:369–371. doi: 10.1007/pl00013150. [DOI] [PubMed] [Google Scholar]
- 11.Huynen M A, Bork P. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 13.Bromham L, Phillips M J, Penny D. Trends Ecol Evol. 1999;14:113–118. doi: 10.1016/s0169-5347(98)01507-9. [DOI] [PubMed] [Google Scholar]
- 14.Foote M, Hunter J P, Janis C M, Sepkowski J J., Jr Science. 1999;283:1310–1314. doi: 10.1126/science.283.5406.1310. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons A. Science. 1998;280:675–676. doi: 10.1126/science.280.5364.675. [DOI] [PubMed] [Google Scholar]
- 16.Avise J C. Molecular Markers, Natural History and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 17.Hillis D M, Moritz C, Mable B K. Molecular Systematics. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 18.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 19.Page R D M, Holmes E C. Molecular Evolution: A Phylogenetic Approach. Oxford: Blackwell; 1998. [Google Scholar]
- 20.de Queiroz K, Gauthier J. Trends Ecol Evol. 1994;9:27–31. doi: 10.1016/0169-5347(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 21.Simpson G G. Principles of Animal Taxonomy. New York: Columbia Univ. Press; 1961. [DOI] [PubMed] [Google Scholar]
- 22.Futuyma D J. Evolutionary Biology. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 23.Zuckerkandl E, Pauling L. In: Evolution of Genes and Proteins. Bryson V, Vogel H J, editors. New York: Academic; 1965. pp. 97–166. [Google Scholar]
- 24.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 25.Sibley C G, Ahlquist J E. Phylogeny and Classification of Birds. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 26.Easteal S, Collet C, Betty D. The Mammalian Molecular Clock. Austin, TX: R. G. Landes; 1995. [Google Scholar]
- 27.Wilson A C, Maxson L R, Sarich V M. Proc Natl Acad Sci USA. 1974;71:2843–2847. doi: 10.1073/pnas.71.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson A C, Sarich V M, Maxson L R. Proc Natl Acad Sci USA. 1974;71:3028–3030. doi: 10.1073/pnas.71.8.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King M-C, Wilson A C. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 30.Cherry L M, Case S M, Wilson A C. Science. 1978;200:209–211. doi: 10.1126/science.635583. [DOI] [PubMed] [Google Scholar]
- 31.Johns G C, Avise J C. Mol Biol Evol. 1998;15:1481–1490. doi: 10.1093/oxfordjournals.molbev.a025875. [DOI] [PubMed] [Google Scholar]
- 32.Haldane J B S. Evolution. 1949;3:51–56. doi: 10.1111/j.1558-5646.1949.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 33.Vane-Wright R I, Humphries C J, Williams P H. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 34.Faith F P. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 35.Avise J C, Hamrick J L, editors. Conservation Genetics: Case Histories from Nature. New York: Chapman & Hall; 1996. [Google Scholar]
- 36.Patterson C, Rosen D E. Bull Am Mus Nat Hist. 1977;158:81–172. [Google Scholar]
- 37.Cantino P D, Olmstead R G, Wagstaff S J. Syst Biol. 1997;46:313–331. [Google Scholar]
- 38.Lee M S Y. Syst Biol. 1998;47:719–726. doi: 10.1080/106351598260707. [DOI] [PubMed] [Google Scholar]
- 39.Mayr E. Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyffeler R. Trends Ecol Evol. 1999;14:168–170. doi: 10.1016/s0169-5347(99)01630-4. [DOI] [PubMed] [Google Scholar]
- 41.Waddell P J, Okada N, Hasegawa M. Syst Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- 42.Avise J C, Wollenberg K. Proc Natl Acad Sci USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer A, Kocher T D, Basasibwaki P, Wilson A C. Nature (London) 1990;347:550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 44.Powell J R. Progress and Prospects in Evolutionary Biology: The Drosophila Model. New York: Oxford Univ. Press; 1997. [Google Scholar]