Abstract
In mammalian females, most genes on one X chromosome are transcriptionally silenced as a result of X chromosome inactivation. Whereas it is well established that some X-linked genes “escape” X inactivation and are expressed from both active (Xa) and inactive (Xi) X chromosomes, most models for the chromosomal control of X-linked gene expression assume that the X inactivation status of a given gene is constant among different females within a population. In this report, we test the expression of human X-linked genes in primary cell lines from females with complete nonrandom X inactivation, by using transcribed polymorphisms to distinguish Xa and Xi expression. Six X-linked genes used to document this assay system showed monoallelic expression in all informative cell lines, consistent with X inactivation. However, a novel pattern of expression was observed for another gene, REP1; monoallelic expression, indicating inactivation, was detected in some lines, whereas biallelic expression, indicating escape from inactivation, was detected in others. Furthermore, levels of Xi expression varied among cell lines that expressed REP1. The cellular basis of Xi expression was examined by expression assays in single cells. These data indicate that REP1 is expressed from the Xi in all cells, but that the level of expression relative to Xa levels is reduced. These findings suggest that Xi gene expression is under a previously unsuspected level of genetic or epigenetic control, likely involving local or regional changes in chromatin organization that determine whether a gene escapes or is subject to X inactivation.
Early in female development in eutherian mammals, X chromosome inactivation transcriptionally silences most genes on one X chromosome as a mechanism of dosage compensation. However, a number of X-linked genes “escape” X inactivation and are expressed from both the active (Xa) and inactive (Xi) X chromosomes (1, 2). Although it is well known that there are differences between species in Xi gene expression [for example, some genes that escape inactivation in humans are inactivated in mouse or vice versa (3–6)], it is generally assumed that the X inactivation status of a given gene is constant among different females within a species. However, this assumption is based on studies of a limited number of genes; with the exception of HPRT (7, 8) and G6PD (9), no other X-linked genes have been analyzed in a large enough number of individuals to address the possibility of heterogeneous expression on the Xi.
Historically, a number of approaches have been used to address whether a particular X-linked gene is subject to or escapes from inactivation (reviewed in refs. 1 and 10). Protein polymorphisms have been used to distinguish Xa and Xi expression in clonal cell lines from heterozygous individuals (11). In heterozygous carriers of X-linked disorders, the mosaic expression of a protein in a subset of cells has provided evidence that the gene involved is expressed only on the Xa chromosome (12–14). In contrast, the nonmosaic expression of steroid sulfatase, STS, in clonal cell lines demonstrated that this gene escapes inactivation (15).
Approaches that require the identification of protein isozymes or heterozygous carriers of individual X-linked disorders are not amenable to testing the inactivation status of a large number of X-linked genes. An alternative approach has been to examine the dose-dependence of protein or transcript levels between individuals with different numbers of X chromosomes. Strict dose-dependence suggests that a gene is not dosage compensated and therefore escapes X inactivation (16, 17). This approach can identify genes that fully escape inactivation, but is more difficult to interpret for others, such as the human STS gene (18) or the murine Smcx gene (19, 20), that are only partially expressed from the Xi chromosome relative to the allele on the Xa.
In response to the need to examine the X inactivation status of the complete battery of X-linked genes, a number of studies have relied on a model system using rodent/human somatic cell hybrids that retain either human Xa or Xi chromosomes (21–25). Although such studies have demonstrated that many X-linked genes are stably inactivated in hybrids (23), the possibility remains that the inactivation status of other genes may not be adequately reflected in this model system because certain epigenetic features of the inactivation process are not fully maintained in hybrids (26, 27).
To develop a complementary approach to the study of X inactivation directly in human diploid cells and to avoid the potentially confounding aspects of the somatic cell hybrid system, we describe here an assay system that tests the expression of X-linked genes by using transcribed polymorphisms to distinguish Xa and Xi expression in an extensive panel of primary human cell lines from females with nonrandom X inactivation caused by the presence of a structurally abnormal X. To demonstrate the utility of this approach, seven X-linked genes were tested by using this system. Surprisingly, expression of one of these genes, REP1, was heterogeneous; monoallelic expression, consistent with X inactivation, was detected in some lines, whereas biallelic expression, indicating escape from X inactivation, was detected in others. We conclude that the inactivation status of at least some X-linked genes varies among different females and may reflect chromatin differences among different X chromosomes that determine whether a given gene escapes from or is subject to X inactivation.
MATERIALS AND METHODS
Cell Lines, DNA, and RNA Preparations.
The fibroblast cell lines used in this study are described in Table 1; they were obtained from the NIGMS Human Genetic Mutant Cell Repository (http://locus.umdnj.edu/nigms/) or from the references listed. Cell lines were maintained and RNA and DNA were prepared as described (23). Methylation assays were performed as described (28, 29).
Table 1.
Panel of human diploid fibroblasts demonstrating complete nonrandom X inactivation
| Case no. | Karyotype* | Evidence of nonrandom inactivation
|
NIGMS or ref. no. | ||
|---|---|---|---|---|---|
| Cytogenetic | Methylation | Monoallelic expression | |||
| 5 | 46,X,t(X;14)(q13;q32) | Late replication | AR | FMR1, KIAA0128 | GM0073 |
| 48 | 46,X,t(X;11)(q11.1;p13) | Late replication | AR, FMR1 | KIAA0128 | GM2859A |
| 49 | 46,X,t(X;11)(q22;q13) | Late replication | AR, FMR1 | G6PD, FMR1, DXS6673E | GM3322 |
| 50 | 46,X,t(X;7)(q21;p22) | Late replcation | AR | XIST | GM1696 |
| 51 | 46,X,t(X;19)(q22;q13.3) | Late replication | FMR1 | G6PD, DXS6673E | GM0089 |
| 53 | 46,X,t(X;14)(q22;q24.3) | AR | DXS6673E | Ref. 67 | |
| 63 | 46,X,t(X;20)(p10;q10) | Late replication | AR, FMR1 | XIST, DXS6673E, KIAA0128 | GM7792 |
| 67 | 46,X,t(X;9)(q13.1;p24) | G6PD, XIST, KIAA0128 | GM0705 | ||
| 68 | 46,X,t(X;22)(q12;p11) | Late replication | AR | G6PD, DXS6673E | GM4628 |
| 86 | 46,X,t(X;11)(p21;q13) | Late replication | AR, FMR1 | XIST, ZXDA, DXS6673E | GM1695A |
| 95 | 46,X,t(X;Y)(q11;q11) | Late replication | AR, FMR1 | XIST, DXS6673E | GM2103 |
| 128 | 46,X,t(X;10)(p11.2;q24.3) | AR | GM7693 | ||
| 144 | 46,X,t(X;3)(q26;p12) | Late replication | AR | GM1533B | |
| 145 | 46,X,t(X;16)(q26;q24) | Late replication | XIST, DXS6673E | GM3884 | |
| 146 | 46,X,t(X;13)(p22jq12) | Late replication | AR | G6PD, XIST, KIAA0128 | GM2971 |
| 147 | 46,X,t(X;5)(p21.2;q35.3) | Late replication | AR | G6PD, XIST | GM5835 |
| 148 | 46,X,t(X;1)(q26;q21) | Late replication | DXS6673E | GM00097A | |
| 149 | 46,X,t(X;12)(q22;q24) | Late replication | AR, FMR1 | FMR1, ZXDA, DXS6673E | GM2621A |
| 150 | 46,X,t(X;11)(q26;q23) | Late replication | AR | DXS6673E | GM3552A |
| 155 | 46,X,t(X;21)(q11;p11) | AR, FMR1 | XIST | GM1411 | |
| 156 | 46,X,t(X;9)(q13;q34) | G6PD, DXS6673E | GM1429 | ||
| 158 | 46,X,t(X;3)(p22.1;q23) | XIST, ZXDA, DXS6673E, KIAA0128 | GM11459 | ||
| 159 | 47,XY,t(X;7)(q24;q32) | AR | GM0324 | ||
| 160 | 46,X,t(X;21)(q22.3;q11) | AR | GM8135 | ||
| 52 | 46,X,der(X)t(X;14)(q22;q24.3) | Late replication | AR | Ref. 67 | |
| 94 | 46,X,der(9)t(X;9)(q34;q12) | Late replication | AR | DXS6673E, KIAA0128 | GM1414 |
| 77 | 46,X,del(X)(q27;q27) | G6PD, DXS6673E | Ref. 68 | ||
| 87 | 46,X,del(X)(q13q22) | Late replication | AR | DXS6673E | GM3923 |
| 139 | 45,X,dic(X;22)(p11;p12) | Late replication | XIST, KIAA0128 | GM5396 | |
| 140 | 45,X,dic(X;22)(p22.1;p11.2) | Late replication | KIAA0128 | GM7149 | |
| 46 | 46,X,i(X)(q26) | Late replication | AR | DXS6673E, KIAA0128 | GM03935 |
| 47 | 46,X,i(X)(p11) | FMR1, DXS6673E, KIAA0128 | GM0735 | ||
| 90 | 46,X,i(X)(p11.21) | AR | XIST, DXS6673E, KIAA0128 | GM0088 | |
| 92 | 46,X,i(X)(q10) | XIST, DXS6673E, KIAA0128 | GM2595 | ||
| 117 | 46,X,i(X)(q22) | AR | ZXDA, DXS6673E | GM6960 | |
| 118 | 46,X,i(X)(p11.4) | Late replication | AR | XIST, ZXDA, DXS6673E | GM8944 |
| 129 | 46,X,i(X)(q28) | Late replication | XIST | GM7213 | |
| 525 | 46,X,i(X)(p11.21) | Late replication | AR | G6PD, XIST, ZXDA | Ref. 69 |
| 126 | 45,X/46,X,i(X)(p11) (60%/40%) | KIAA0128 | GM339 | ||
| 152 | 45,X/46,X,i(X)(p22) (30%/70%) | Late replication | G6PD, XIST, ZXDA, DXS6673E | GM0314 | |
Nonrandom inactivation in the cell lines listed were determined indirectly by late replication (as reported in the references listed) or by methylation assays at the androgen receptor (AR) or fragile X (FMR1) loci, and/or directly with expression assays at the XIST, G6PD, FMR1, ZXDA, DXS6673E, or KIAA0128 loci.
For each cell line, the relevant X chromosome portion of the karyotype is listed. Complete karyotypes are listed by the NIGMS cell repository at http://locus.umdnj.edu/nigms/.
PCR-Based Expression Assays.
Primers identified with an asterisk were 32P or fluorescently labeled. An A to G transition at position 381 from the published REP1 sequence (30) correlates with an ApaLI or HhaI restriction enzyme site. Primers to identify the polymorphism were REP1–10: CAGGATTTGCATGAAGATGTCG and *REP1–11: TCGCTGCTTGGAGTTTGTTC, and allele ratios in HhaI-digested products were confirmed by reciprocal ratios in samples cut with ApaLI. For XIST, G6PD, and FMR1, the polymorphisms did not correlate with endogenous restriction site differences. Underlined nucleotides in primers indicate that a mismatch was incorporated at a nonpolymorphic nucleotide, with the 3′ end of each of these oligonucleotides positioned just upstream of the polymorphic site. PCR products amplified with this primer create a restriction site when one of the alleles is present. An XIST polymorphism (31) was detected by using the primers *XIST-150F: AGCTATATCTGCTGAATGATCATTGATTAC and XIST-150R: TCATTCCTATCTGTATAGAACTGTAGGATT, and alleles were distinguished by digestion with HinfI. To analyze G6PD (32, 33), samples were amplified with G6PD-1311R: GTGAAGCTCCCTGACGCGTA and *G6PD-E: TTCTCCAGCTCAATCTGGTG, and labeled products were digested with RsaI to distinguish alleles. To genotype DNA samples for a single nucleotide polymorphism in the FMR1 gene (34), samples were amplified with FMR1-TaqI: GATGTGCCAGAAGACTTTCG and *FMR1-DNA: CATCAGACACGTGTATAGCCA, and expression was tested by amplification with FMR-TaqI and *FMR1-cDNA: GACTCCGAAAGTGCATGTCA. Alleles were differentiated by TaqI digestion. Size polymorphisms were present in three other genes. A polymorphic CA repeat in KIAA0128 (35) was identified by comparing sequences deposited in GenBank, and alleles were distinguished by amplification with *F: ACAAGCACATATTAACAGCCACA and R: CAAGAAGTTTGCTTTCCCTAGC. Similarly, the CA repeat in ZXDA was assayed by using primers ZXDA-cpx210d: TCAATTAAGGTGGGAGGCAG and *ZXDA-cpx210c: TGTGAGGTAATTATGGCAAAGTC (36). For DXS6673E, primers *1f: AGACAAGGACAGAAAGGGGG and 2: GGAGTTTTCCTCCCTCACCA were used to resolve alleles of a highly polymorphic transcribed repeat (37).
To establish quantitative conditions under which the relative abundance of each allele could be determined, products were amplified for an initial 25–30 cycles with standard conditions, except for DXS6673E, which required 30–35 cycles of amplification with touchdown PCR (38) with an annealing range of 68°C to 52°C. Subsequently, DXS6673E products were resolved and analyzed on an ABI 373. For XIST, G6PD, FMR1, and REP1, a primer extension reaction that used an aliquot from each sample was then performed with a 32P end-labeled primer. These products were subsequently digested with the appropriate restriction enzyme to distinguish alleles. By using this strategy, only products from the final extension are analyzed and quantitative conditions are not compromised by heteroduplex formation. Because the detection of each allele does not require separate reactions, this approach eliminates potential error caused by sample loading. Relative band intensities were quantitated on a PhosphorImager using imagequant software (Molecular Dynamics). Mixing controls established a level of sensitivity for each assay of < 1% to 5% relative to the intensity of the other allele.
Single-Cell Reverse Transcription-PCR (RT-PCR) Analysis.
Trypsinized fibroblasts were resuspended at very low density and visualized on a Nikon SMZ-U dissecting microscope equipped with a 1.5× objective. Individual cells were isolated in a volume of ∼1 μl by using finely drawn Pasteur pipettes, then placed into PCR tubes on ice. Cell lysis, RT reactions (using the primer REP1–15: 5′-CCTGTCACTTCAGCACCATT-3′) and nested PCRs were performed as described (39). cDNA was initially amplified for 40 cycles with primers REP1–15 and REP1–16: 5′-TTGCTCTTAGCAGGAAGGAC-3′. Subsequently 3-μl aliquots were further amplified for 30 cycles with internal primers REP1–14: 5′-GTTATGCCAGTCAGGATTTGC-3′ and REP1–11: 5′-TCGCTGCTTGGAGTTTGTTC-3′. Both primer pairs exclusively amplify cDNA. Positive samples were assayed as described above.
To control for contamination, additional tubes containing lysis buffer but no cell were simultaneously processed, and, appropriately, no amplification products were seen in any of these control tubes. Experiments were scored only when >70% of cells were successfully amplified.
For mixing controls, single cells from homozygous individuals were mixed 1:1 in all wells to mimic full Xi expression in every cell (model A), 1:3 in each well to represent reduced Xi expression in all cells (model B), or 1:1 in one-third of wells (with the other wells containing cells of only a single allele) to mimic full Xi expression, but in only a proportion of cells (model C). Cells were lysed and a volume of lysate was removed (half of the lysate for models A and for the wells that contained cells mixed 1:1 in model C, and three-fourths of the lysate for model B), leaving the equivalent of a single cell for subsequent analysis.
RT-PCR Analysis in Somatic Cell Hybrids.
The somatic cell hybrids used for these studies have been previously described or were generated by using established protocols (23). PCR primers for PGK1, MIC2, and XIST have been published (23). REP1 and DXS6673E primers were designed from published sequence, REP1–5: ACAGTGCCAGCAGAGGAA, REP1–4: TCAGCCTGTTTGACTGCA (40), DXS6673E-4: TGCTCGTCATCCTGCCTCAC, DXS6673E-5: ACACTGGTCACAACAGTTGG (37). All primer pairs were annealed at 55°C and amplified for 30 cycles.
RESULTS
Assaying X Inactivation in Nonrandomly Inactivated Primary Fibroblast Lines.
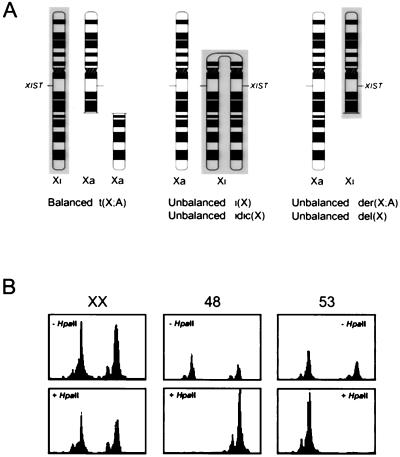
The fibroblast cell lines used for this study were derived from females carrying a variety of structurally abnormal X chromosomes (Fig. 1A). In cell lines with abnormal Xs, complete nonrandom X inactivation ensures proper dosage of X-linked gene products (41, 42). With these fibroblasts, transcribed polymorphisms could be analyzed to test whether gene expression was monoallelic (expressed only from the Xa chromosome, indicating that the gene is subject to X inactivation) or biallelic (expressed from both X chromosomes, indicating that the gene escapes inactivation). A panel of 40 primary cell lines was assembled (Table 1). Nonrandom X inactivation was established either by previous cytogenetic studies of late replication or by standard PCR-based methylation assays (Fig. 1B) (29, 43). With the PCR assays, we determined that all (of 28 informative lines) were nonrandomly inactivated at a sensitivity of >95:5 (Table 1).
Figure 1.
Nonrandom inactivation in females with structurally abnormal Xs. (A) Fibroblast cell lines were studied from individuals carrying balanced [t(X;A)] or unbalanced [der(X;A)] X; autosome translocations, iso- or isodicentric chromosomes [i(X), idic(X)], or deleted X chromosomes [del(X)]. X chromosomes (normal or with structural rearrangements) that contain the XIST gene can be inactivated (shaded). In carriers of balanced X;A translocations, inactivation of the normal X ensures proper dosage of X-linked gene products (41). In other cases, structurally abnormal Xs are inactivated in females, establishing proper dosage from the active normal X (42). (B) Methylation assay at the androgen receptor gene (AR) to establish nonrandom inactivation. An HpaII site is adjacent to the polymorphic trinucleotide repeat. Digestion of genomic DNA with HpaII prior to amplification digests the unmethylated allele on the Xa chromosome. Subsequent PCR amplifies only the methylated allele(s) on Xi chromosomes. The relative representation of each allele in the HpaII-digested samples (+HpaII), as compared with the amplified undigested samples (−HpaII), demonstrates the randomness (as seen in the sample from a normal XX female) or nonrandomness of X inactivation (cases 48 and 53).
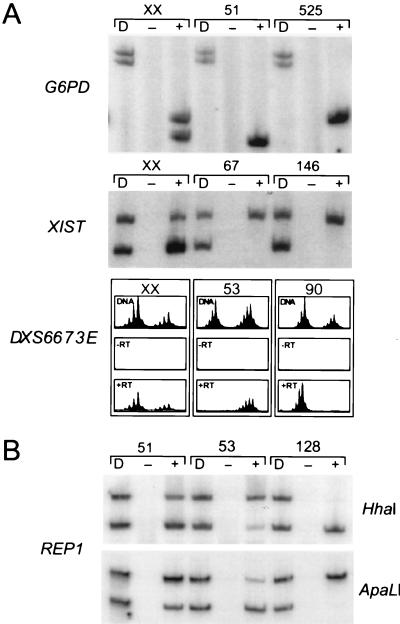
To evaluate X-linked gene expression in these cell lines, we initially assayed loci whose X inactivation status is well established: G6PD, FMR1, and XIST (11, 15, 23, 44, 45). Ten fibroblast cell lines were heterozygous for a polymorphism at position 1311 of the G6PD transcript (32, 33), and RT-PCR analysis indicated that only one allele was expressed for all 10 nonrandomly inactivated cell lines (Fig. 2A), consistent with G6PD being subject to X inactivation. Similarly, monoallelic expression of an FMR1 polymorphism was seen for each of the four heterozygous cell lines (34). Additionally, an XIST polymorphism (31) was informative in 17 cell lines, and all showed monoallelic expression (Fig. 2A), consistent with previous data demonstrating that XIST is expressed only from Xi chromosomes (31, 45).
Figure 2.
Gene expression of X-linked genes. (A) XIST, G6PD, and DXS6673E show monoallelic expression in all nonrandomly inactivated fibroblast lines tested. D indicates amplification of DNA, and a + or − refers to RNA that has been amplified with or without prior reverse transcription. For XIST and G6PD, samples are digested with HinfI or RsaI (respectively) to differentiate alleles. The DXS6673E PCR products were separated on an ABI 373, and the electropherogram traces are shown. Different normal control female samples (XX) showing various degrees of random and skewed inactivation are shown, in contrast to the nonrandomly inactivated cell lines from the females with structural abnormalities (case numbers indicated). (B) Quantitative amplification of REP1 alleles in fibroblast cell lines from cases 51, 53, and 128. PCR products at the top are digested with HhaI and on the bottom with ApaLI. D, − and +, as in A.
Expression was next tested for genes whose inactivation states were unknown or had been established previously solely on the basis of Xi expression in somatic cell hybrids. Assays were developed to examine expression of polymorphisms in DXS6673E (37), ZXDA (36), and KIAA0128 (sequence from ref. 35). All 23 heterozygous cell lines showed monoallelic DXS6673E expression, as did seven informative cell lines for ZXDA and 14 heterozygous cell lines for KIAA0128. Together with the above data, these studies indicate that the X chromosome inactivation patterns of many genes are consistent among different individuals. Combined with the results of the cytogenetic and methylation assays, these data for XIST, G6PD, FMR1, ZXDA, DXS6673E, and KIAA0128 comprise strong evidence that all 40 informative lines show essentially complete nonrandom inactivation (Table 1).
REP1 Shows Heterogeneous Expression in Nonrandomly Inactivated Fibroblast Lines.
Expression was also analyzed for the gene REP1, mutations in which are responsible for the X-linked eye disorder choroideremia (40). REP1 encodes a Rab escort protein (46, 47) and is expressed in many cell types, including skin fibroblasts. Nine lines in the panel were heterozygous for a transcribed REP1 polymorphism that results in reciprocal cleavage by the restriction enzymes HhaI and ApaLI (30). As shown in Fig. 2B, REP1 expression in fibroblasts was heterogeneous among different cell lines, with some showing clear evidence of biallelic expression and others showing only monoallelic expression. We developed a quantitative assay (see Materials and Methods) to measure the relative expression of the Xa and Xi alleles in heterozygous lines. Among the nine informative cell lines, three showed essentially monoallelic expression (Xi expression ≤1% of Xa levels; e.g., line 128 in Fig. 2B), whereas REP1 was biallelically expressed in the other lines, with Xi expression levels ranging from 5% to 42% of Xa levels (Table 2; Fig. 2B). Importantly, for all lines that showed biallelic REP1 expression (Table 2), expression assays for XIST, G6PD, DXS6673E, ZXDA, and/or KIAA0128 confirmed complete nonrandom inactivation (Table 1). That REP1 Xi expression does not simply reflect aberrant expression from structurally abnormal chromosomes is indicated by the finding that six of the nine lines examined carry balanced X;autosome translocations (Fig. 1A); thus, in these lines, the intact normal X is the Xi. Of the six REP1-informative lines in this category, four showed clear evidence of REP1 Xi expression whereas the other two showed only Xa expression (Table 2).
Table 2.
Levels of REP1 expression from Xi chromosomes
| Cell line | Xi/Xa | Inactive X |
|---|---|---|
| 51 | 0.42 ± 0.06 | Normal X |
| 67 | 0.22 ± 0.02 | Normal X |
| 53 | 0.16 ± 0.03 | Normal X |
| 152 | 0.11 ± 0.02 | i(X)(p22) |
| 140 | 0.06 ± 0.01 | dic(X;22) |
| 146 | 0.05 ± 0.01 | Normal X |
| 48 | ≤0.01 | Normal X |
| 52 | ≤0.01 | der(X) |
| 128 | ≤0.01 | Normal X |
Levels of Xi expression are indicated relative to Xa expression, as determined in nonrandomly inactivated fibroblast cell lines (see Materials and Methods). Each cell line was assayed at least twice, and averages ± one SD are indicated.
Single-Cell Analysis of REP1 Expression.
The levels of REP1 Xi expression that we observed could represent reduced expression from each cell within a cell line or cellular heterogeneity if the gene escapes inactivation in only a subset of cells. Therefore, because primary (i.e., untransformed) fibroblast cultures are established by the outgrowth of multiple cells, heterogeneous escape from inactivation could reflect inter-individual differences, either in levels of Xi expression or in the percentage of cells escaping inactivation. Because REP1 Xi transcript levels are not high enough to use RNA fluorescence in situ hybridization of nascent transcripts to distinguish these possibilities, we used single-cell RT-PCR to examine REP1 Xi expression.
Initially, a series of controls were performed to determine the sensitivity of the assay. Single cells from homozygous individuals were mixed in proportions to mimic full Xi expression in all cells (model A), reduced Xi expression in all cells (model B), or Xi expression in only a proportion of cells (model C). Cells were lysed and a volume of lysate was removed, leaving the RNA equivalent of a single cell for subsequent processing (see Materials and Methods). Expression of both alleles in the samples mixed 1:1 (model A) was detected in 51% of samples with detection of only one or the other allele in the remaining 49% (Table 3). Alleles expressed at a reduced level (representing a partially expressed Xi allele, as in model B) were less frequently detected; if detected, they were usually in samples that also expressed the Xa allele (Table 3). In contrast, in a mosaic sample consisting of some cells that express only the Xa allele and others that express both Xa and Xi alleles (model C), both the frequency of Xi allele detection and the proportion of samples expressing both alleles were reduced (Table 3). Thus, the distinct amplification profiles from these controls could be used to infer the cellular basis for REP1 heterogeneity in the fibroblast cell lines.
Table 3.
Analysis of REP1 expression in single cells
| Sample | No. cells | Allele detection, %
|
||
|---|---|---|---|---|
| Xa only | Xi only | Xa and Xi | ||
| Model A | 37 | 22 | 27 | 51 |
| Model B | 41 | 47 | 9 | 44 |
| Model C | 39 | 72 | 15 | 13 |
| Line 51 | 127 | 30 | 9 | 61 |
Single-cell RT–PCR detection of REP1. For each sample, the percentage of successfully amplified cells (or, for the control experiments, the number of wells containing the RNA equivalent of a single cell) that showed expression of one or the other allele (or both) is indicated. For the controls, homozygous cell lines were mixed to mimic Xi expression that was: equivalent to Xa expression in all cells (model A); reduced to 25% of Xa expression levels in all cells (model B); or equivalent to Xa expression, but only in one-third of cells (model C). Independent experiments gave similar results.
RT-PCR was then performed on single cells from the t(X;19) fibroblast cell line that showed the highest level of REP1 Xi expression (line 51). Biallelic REP1 expression was detected in >60% of successfully amplified cells (Table 3). This result is most consistent with models A or B, in which all cells express REP1 from the Xi; it is incompatible with model C. That the ratio of cells expressing only the Xa allele to those expressing only the Xi allele was different from 1:1 contradicts the expectations of model A and suggests that the level of Xi expression is reduced in this line. This estimate is consistent with the ≈40% estimate derived from the intensity of the two alleles, as seen in Fig. 2B. Thus, we conclude that, at least in line 51, REP1 is expressed from the Xi in all cells, but at levels that are reduced relative to the Xa allele.
REP1 Shows Heterogeneous Expression from Inactive Xs in Somatic Cell Hybrids.
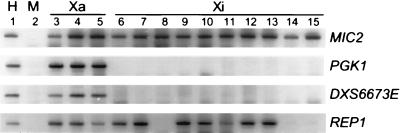
Not all X-linked genes show uniform expression patterns in Xi-containing somatic cell hybrids (23). Whether the observed heterogeneity reflects a similar degree of heterogeneity in human cells or indicates instability of the inactive state for some genes in somatic cell hybrids (26) is currently unknown. To examine this question for REP1, we analyzed expression of the REP1 gene by RT-PCR in 10 independent mouse-human somatic cell hybrids that retain a human Xi. As in human cells, REP1 showed heterogeneous expression among the hybrids tested; the gene was well expressed from seven Xi hybrids, but showed little or no expression from three other hybrids (Fig. 3).
Figure 3.
Expression from Xa and Xi chromosomes in mouse/human somatic cell hybrids for DXS6673E and REP1. Negative images of ethidium bromide-stained PCR products are shown. As controls, PGK1 is subject to X inactivation and verifies the inactivation status of the hybrids, and MIC2, a gene that escapes inactivation, demonstrates the presence of amplifiable human X chromosome cDNA from each hybrid sample (23). Amplification products in lanes 1 and 2 are from human (H) and mouse (M) controls. Lanes 3–5, hybrids carrying Xa chromosomes; lanes 6–15, hybrids carrying Xi chromosomes.
The Xi for two of the somatic cell hybrids tested were derived from two of the primary fibroblast lines described in Table 1. Importantly, results for the somatic cell hybrids were similar to those seen in the fibroblasts; the Xi allele of REP1 in cell line 48 showed undetectable expression and was also not expressed in a hybrid derived from line 48, whereas REP1 was well expressed from Xi in cell line 51 and from the same Xi isolated in a hybrid (Fig. 3, lanes 8 and 12, and Table 2).
DISCUSSION
Previously, three different expression patterns have been recognized for X-linked genes: those subject to X inactivation, those escaping inactivation, and a single gene, XIST, that is expressed exclusively from the Xi in somatic cells. Here, we describe a fourth class of gene on the X chromosome, represented by REP1, that shows heterogeneous Xi expression, being subject to inactivation on some X chromosomes and escaping inactivation to different extents on others. The existence of genes in this category has implications for understanding both the chromosomal control of gene expression on the Xi and varied clinical manifestations in heterozygous carriers of X-linked disorders.
What is the basis for the heterogeneity of REP1 expression observed in both human cells and somatic cell hybrids? Inactivation of human genes is believed to be extraordinarily stable (reviewed in ref. 48). However, this conclusion is based on studies of a relatively small number of genes containing a strong CpG island (7, 49). As the number of genes that escape inactivation or show heterogeneous escape from inactivation increases, the possibility that such patterns reflect reactivation of previously inactivated genes (e.g., ref. 50) may need to be investigated more comprehensively. Our single-cell RT-PCR data for cell line 51 support a model in which all cells express REP1 from the Xi, yet the level of expression, relative to Xa, is reduced (Table 3). This experiment suggests that reactivation in a subset of cells is not a likely mechanism in this cell line, at least not for REP1.
REP1 heterogeneity instead likely points to inter-individual differences in the organization of heterochromatin on inactivated X chromosomes. Concerted mapping and expression studies have identified clusters of genes that escape inactivation, suggesting that at least portions of the X are comprised of coordinately controlled domains (51). Whether REP1 maps to such a domain is unknown, because the region in Xq21.3 is relatively gene-poor and the inactivation status of the few genes identified that map within a few megabases of REP1 is unknown. If adjacent genes do escape X inactivation, REP1 heterogeneous expression patterns could be explained by variability in the location of domain boundaries or in the effectiveness of heterochromatin spreading (52, 53). Alternatively, factors specific to REP1 (e.g., enhancers) may influence transcription variably within a heterochromatic environment (54).
It may be feasible to distinguish among some of these possibilities by determining whether patterns and levels of REP1 Xi expression are heritable or stochastic. Although family studies using the assay presented here would not be straightforward (because this would require nonrandom X inactivation in multiple family members), mapping of this trait may lead to the identification of trans-acting factors involved in Xi heterochromatin formation or of cis-acting sequences necessary for Xi gene control. Notably, there are important parallels between the REP1 data as reported here and inter-individual variability as observed for some imprinted genes (55–59). As many imprinted genes also cluster in specific chromosomal domains (reviewed in ref. 60), studies of epigenetic variability in both imprinting and X inactivation may lead to greater understanding of global mechanisms of gene silencing and heterochromatin formation. Whether the heterogeneous pattern of Xi expression described here for REP1 could reflect an imprinted state is currently unclear, because the parental origins of the X chromosomes analyzed here are, in most instances, unknown. Nonetheless, this question could be addressed in future studies.
The current panel of nonrandomly inactivated human lines was assembled in part to complement the somatic cell hybrid model system for studying X inactivation (23). Although in most instances Xi expression patterns in hybrids are also seen in primary fibroblasts, it is possible that expression states of some genes in fibroblast cell lines will not be well maintained in somatic cell hybrids. Proper maintenance of X chromosome inactivation may depend on a redundant system of late replication, DNA methylation, histone hypoacetylation, and XIST RNA association with the Xi chromosome (reviewed in ref. 61). Although dozens of the genes that we assayed are stably inactivated in all hybrids tested (23), the systems for maintenance of inactivation of some genes in inter-specific hybrids may not function as well as in primary human cell lines (26, 27). If so, such epigenetic instability would be of considerable interest mechanistically.
It is important to consider potential clinical consequences of X inactivation heterogeneity. Skewed inactivation patterns are often implicated to explain manifesting or attenuated phenotypes in females that are heterozygous for X-linked defects (reviewed in refs. 42 and 62). Whether variable Xi expression, such as that documented here, could similarly impact clinical presentation in heterozygotes will depend on the gene involved and on the levels of Xi expression. Heterozygous carriers of REP1 are described as having variable degrees of patchiness of the retinal pigment epithelium (63, 64), a phenotype that has been widely interpreted as reflecting random X inactivation (46, 63). Given the data reported here, it will be necessary to reevaluate these conclusions and to determine whether REP1 Xi levels influence the clinical presentation in this disorder.
Last, given the range of patterns of X-linked gene expression, a comprehensive survey of inactivation patterns for all widely expressed X-linked genes, using the panel of primary cell lines described here, would clearly be of interest. A complete X inactivation profile will provide relevant information for clinical cytogenetics and genetic counseling and give insight into the genomic and epigenetic organization of the X chromosome. Although, to date, the number of highly informative, transcribed X-linked polymorphisms limits this approach, genome sequencing efforts are anticipated to generate large numbers of single nucleotide polymorphisms (65, 66) that should greatly facilitate these studies.
Acknowledgments
We are grateful to members of the Willard lab for helpful discussions. We thank G. Compitello and A. Cottle for technical assistance, A. Cadle, J. Amos-Landgraf, and C. Hodges for participation in the early stages of this project and I. MacDonald for information before publication. This work was supported by National Institutes of Health Grant GM45441 to H.F.W.
ABBREVIATIONS
- RT–PCR
reverse transcription–PCR
- Xa
active X chromosome
- Xi
inactive X chromosome
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Willard H F. In: The Metabolic Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw-Hill; 1995. pp. 719–735. [Google Scholar]
- 2.Disteche C. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- 3.Ashworth A, Rastan S, Lovell-Badge R, Kay G. Nature (London) 1991;351:406–408. doi: 10.1038/351406a0. [DOI] [PubMed] [Google Scholar]
- 4.Zinn A R, Bressler S L, Beer-Romero P, Adler D A, Chapman V M, Page D C, Disteche C M. Genomics. 1991;11:1097–1101. doi: 10.1016/0888-7543(91)90037-f. [DOI] [PubMed] [Google Scholar]
- 5.Adler D A, Bressler S L, Chapman V M, Page D C, Disteche C M. Proc Natl Acad Sci USA. 1991;88:4592–4595. doi: 10.1073/pnas.88.11.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dal Zotto L, Quaderi N A, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen C H, Chapman V, Kalcheva I, et al. Hum Mol Genet. 1998;70:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 7.Migeon B R. Am J Hum Genet. 1971;23:199–210. [PMC free article] [PubMed] [Google Scholar]
- 8.Migeon B R, Axelman J, Beggs A H. Nature (London) 1988;335:93–96. doi: 10.1038/335093a0. [DOI] [PubMed] [Google Scholar]
- 9.Migeon B R. Isozymes Curr Top Biol Med Res. 1983;9:189–200. [PubMed] [Google Scholar]
- 10.Brown C J, Willard H F. Adv Dev Biol. 1993;2:37–72. [Google Scholar]
- 11.Davidson R G, Nitowsky H M, Childs B. Proc Natl Acad Sci USA. 1963;50:481–485. doi: 10.1073/pnas.50.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown R M, Dahl H H M, Brown G K. Genomics. 1989;4:174–181. doi: 10.1016/0888-7543(89)90297-8. [DOI] [PubMed] [Google Scholar]
- 13.Arahata K, Ishihara T, Kamakura K, Tsukahara T, Ishiura S, Baba C, Matsumoto T, Nonaka I, Sugita H. N Engl J Med. 1989;320:138–142. doi: 10.1056/NEJM198901193200302. [DOI] [PubMed] [Google Scholar]
- 14.Hurko O, Hoffman E P, McKee L, Johns D R, Kunkel L M. Am J Hum Genet. 1989;44:820–826. [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro L J, Mohandas T, Weiss R, Romeo G. Science. 1979;204:1224–1226. doi: 10.1126/science.156396. [DOI] [PubMed] [Google Scholar]
- 16.Schneider-Gadicke A, Beer-Romero P, Brown L G, Nussbaum R, Page D C. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 17.Fisher E M C, Beer-Romero P, Brown L G, Ridley A, McNeil J A, Lawrence J B, Willard H F, Bieber F R, Page D C. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 18.Migeon B R, Shapiro L J, Norum R A, Mohandas T, Axelman J, Dabora R L. Nature (London) 1982;299:838–840. doi: 10.1038/299838a0. [DOI] [PubMed] [Google Scholar]
- 19.Carrel L, Hunt P A, Willard H F. Hum Mol Genet. 1996;5:1361–1366. doi: 10.1093/hmg/5.9.1361. [DOI] [PubMed] [Google Scholar]
- 20.Sheardown S, Norris D, Fisher A, Brockdorff N. Hum Mol Genet. 1996;5:1355–1360. doi: 10.1093/hmg/5.9.1355. [DOI] [PubMed] [Google Scholar]
- 21.Willard H F, Brown C J, Carrel L, Hendrich B, Miller A P. Cold Spring Harb Symp Quant Biol. 1993;58:315–322. doi: 10.1101/sqb.1993.058.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Tribioli C, Mancini M, Plassart E, Bione S, Rivella S, Sala C, Torri G, Toniolo D. Hum Mol Genet. 1994;3:1061–1067. doi: 10.1093/hmg/3.7.1061. [DOI] [PubMed] [Google Scholar]
- 23.Brown C J, Carrel L, Willard H F. Am J Hum Genet. 1997;60:1333–1343. doi: 10.1086/515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esposito T, Gianfrancesco F, Ciccodicola A, D’Esposito M, Nagaraja R, Mazzarella R, D’Urso M, Forabosco A. Genomics. 1997;43:183–190. doi: 10.1006/geno.1997.4797. [DOI] [PubMed] [Google Scholar]
- 25.D’Esposito M, Matarazzo M R, Ciccodicola A, Strazzullo M, Mazzarella R, Quaderi N A, Fujiwara H, Ko M S, Rowe LB, Ricco A, et al. Hum Mol Genet. 1997;6:1917–1923. doi: 10.1093/hmg/6.11.1917. [DOI] [PubMed] [Google Scholar]
- 26.Kahan B, DeMars R. Proc Natl Acad Sci USA. 1975;72:1510–1514. doi: 10.1073/pnas.72.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemson C M, Chow J C, Brown C J, Lawrence J B. J Cell Biol. 1998;142:13–23. doi: 10.1083/jcb.142.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plenge R M, Hendrich B D, Schwartz C, Arena J F, Naumova A, Sapienza C, Winter R A, Willard H F. Nat Genet. 1997;17:353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 29.Carrel L, Willard H F. Am J Med Genet. 1996;64:27–30. doi: 10.1002/(SICI)1096-8628(19960712)64:1<27::AID-AJMG3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Nesslinger N, Mitchell G, Strasberg P, MacDonald I. Ophthalmic Genet. 1996;17:47–52. doi: 10.3109/13816819609057870. [DOI] [PubMed] [Google Scholar]
- 31.Rupert J L, Brown C J, Willard H F. Eur J Hum Genet. 1995;3:333–343. doi: 10.1159/000472322. [DOI] [PubMed] [Google Scholar]
- 32.De Vita G, Alcalay M, Sampietro M, Cappellini M D, Fiorelli G, Toniolo D. Am J Hum Genet. 1989;44:233–240. [PMC free article] [PubMed] [Google Scholar]
- 33.Beutler E, Kuhl W. Am J Hum Genet. 1990;47:1008–1012. [PMC free article] [PubMed] [Google Scholar]
- 34.Kunst C B, Warren S T. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 35.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 36.Mahtani M M, Willard H F. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 37.van der Maarel S M, Scholten I H, Huber I, Philippe C, Suijkerbuijk R F, Gilgenkrantz S, Kere J, Cremers F P, Ropers H H. Hum Mol Genet. 1996;5:887–897. doi: 10.1093/hmg/5.7.887. [DOI] [PubMed] [Google Scholar]
- 38.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaynor E M, Mirsky M L, Lewin H A. Biotechniques. 1996;21:286–291. doi: 10.2144/96212rr02. [DOI] [PubMed] [Google Scholar]
- 40.Cremers F P M, van de Pol D J R, van Kerkhoff L P M, Wieringa B, Ropers H-H. Nature (London) 1990;347:674–677. doi: 10.1038/347674a0. [DOI] [PubMed] [Google Scholar]
- 41.Therman E, Patau K. Humangenetik. 1974;25:1–16. doi: 10.1007/BF00281002. [DOI] [PubMed] [Google Scholar]
- 42.Belmont J W. Am J Hum Genet. 1996;58:1101–1108. [PMC free article] [PubMed] [Google Scholar]
- 43.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 44.Kirchgessner C, Warren S, Willard H. J Med Genet. 1995;32:925–929. doi: 10.1136/jmg.32.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 46.Seabra M C, Brown M S, Goldstein J L. Science. 1993;259:377–381. doi: 10.1126/science.8380507. [DOI] [PubMed] [Google Scholar]
- 47.Andres D A, Seabra M C, Brown M S, Armstrong S A, Smeland R E, Cremers F P M, Goldstein J L. Cell. 1993;73:1091–1099. doi: 10.1016/0092-8674(93)90639-8. [DOI] [PubMed] [Google Scholar]
- 48.Gartler S M, Goldman M A. Dev Genet. 1994;15:504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- 49.Migeon B R. Nature (London) 1972;239:87–89. doi: 10.1038/239087a0. [DOI] [PubMed] [Google Scholar]
- 50.Lingenfelter P A, Adler D A, Posllinski D, Thomas S, Elliott R W, Chapman V M, Disteche C M. Nat Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 51.Miller A P, Willard H F. Proc Natl Acad Sci USA. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geyer P K. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 53.Dillon N, Grosveld F. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- 54.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y, Goodyer C G, Deal C, Polychronakus C. Biochem Biophys Res Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 56.Jinno Y, Yun K, Nishiwaki K, Kubota T, Ogawa O, Reeve A E, Niikawa N. Nat Genet. 1994;6:305–309. doi: 10.1038/ng0394-305. [DOI] [PubMed] [Google Scholar]
- 57.Giannoukakis N, Deal C, Paquette J, Kukuvitis A, Polychronakos C. Biochem Biophys Res Commun. 1996;220:1014–1019. doi: 10.1006/bbrc.1996.0524. [DOI] [PubMed] [Google Scholar]
- 58.Dao D, Frank D, Qian N, O’Keefe D, Vosatka R J, Walsh C P, Tycko B. Hum Mol Genet. 1998;7:597–608. doi: 10.1093/hmg/7.4.597. [DOI] [PubMed] [Google Scholar]
- 59.Jiang S, Hemann M A, Lee M P, Feinberg A P. Genomics. 1998;53:395–399. doi: 10.1006/geno.1998.5511. [DOI] [PubMed] [Google Scholar]
- 60.Bartolomei M S, Tilghman S M. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 61.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 62.Puck J M, Willard H F. N Engl J Med. 1998;338:325–328. doi: 10.1056/NEJM199801293380611. [DOI] [PubMed] [Google Scholar]
- 63.Jay B. Trans Ophthalmol Soc U K. 1985;104:836–844. [PubMed] [Google Scholar]
- 64.Karna J. Acta Ophthalmol Suppl. 1986;176:15–60. [PubMed] [Google Scholar]
- 65.Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 66.Chakravarti A. Nat Genet. 1998;19:216–217. doi: 10.1038/885. [DOI] [PubMed] [Google Scholar]
- 67.Markovic V D, Cox D W, Wilkinson J. Am J Med Genet. 1985;20:87–96. doi: 10.1002/ajmg.1320200111. [DOI] [PubMed] [Google Scholar]
- 68.Clarke J T R, Greer W L, Strasberg P M, Pearce R D, Skomorowski M A, Ray P N. Am J Hum Genet. 1991;49:289–297. [PMC free article] [PubMed] [Google Scholar]
- 69.Sharp C B, Bedford H M, Willard H F. Hum Genet. 1990;85:330–336. doi: 10.1007/BF00206757. [DOI] [PubMed] [Google Scholar]