Abstract
The p53 gene was sequenced in 100 primary human lung cancers by using direct dideoxynucleotide cycle sequencing and compared with sequence analysis by using the p53 GeneChip assay. Differences in sequence analysis between the two techniques were further evaluated to determine the accuracy and limitations of each method. p53 mutations were either detected by using both techniques or, if only detected by one technique, were confirmed by using mutation-specific oligonucleotide hybridization. Dideoxynucleotide sequencing of the conserved regions of the p53 gene (exons 5–9) detected 76% of the mutations within this region of the gene. The GeneChip p53 assay detected 81% of all (exons 2–11) mutations, including 80% of the mutations within the conserved regions of the gene. The GeneChip assay detected 46 of 52 missense mutations (88%), but 0 of 5 frameshift mutations. The specificity of direct sequencing and of the p53 GeneChip assay at detecting p53 mutations were 100% and 98%, respectively. The GeneChip p53 assay is a rapid and reasonably accurate approach for detecting p53 mutations; however, neither direct sequencing nor the p53 GeneChip are infallible at p53 mutation detection.
The p53 tumor-suppressor gene is the most frequently mutated gene in human cancer. Between 30% and 70% of cancers of almost every organ and histologic subtype have a point mutation in one of the two p53 gene copies and loss of the other allele (1, 2). Mutation of the p53 gene detected by using gene sequencing and/or loss of function as determined by the accumulation of nonfunctional p53 protein in the cell nucleus have been reported to be associated with poor prognosis in numerous human cancers (2, 3). Moreover, recent pilot studies have suggested a role for p53 in determining the responsiveness of a tumor to chemotherapy or radiation therapy (4, 5). Despite many studies examining the role of p53 gene mutations or protein expression as a prognostic factor in human cancer, their use has not been recommended for routine use in clinical practice for several reasons. p53 immunohistochemical staining is not currently performed in any standardized fashion (2). A variety of antibodies has been used to detect p53 protein accumulation without standardization among antibodies, and the methods used for interpretation of positivity differ among various studies. In addition, p53 protein does not accumulate with all types of p53 mutations (6, 7). Detection of p53 mutations by using direct sequencing is labor-intensive, involves the use of radioactive isotopes or sophisticated software analysis with fluorescent detection, and is thus beyond the capability of almost all clinical laboratories. Moreover, identification of mutations in primary tumors is further complicated by the dilution of neoplastic cells among many normal, nonmutated cells. These factors have contributed to the paucity of large prospective studies of sufficient statistical power to provide conclusive evidence of the role of p53 inactivation in predicting patient outcome.
Rapid mutation analysis of the p53 gene sequence has recently been developed utilizing an oligonucleotide probe array (8, 9). Oligonucleotide probe arrays have been used previously to detect germ-line mutations in BRCA1 and the cystic fibrosis transmembrane conductance regulator gene (10, 11). However, this approach has not previously been used to detect somatic mutations in a large number of human cancers. This technique involves a single PCR amplification followed by fragmentation and fluorescent labeling of the PCR product and hybridization of the labeled product with an oligonucleotide probe array. The array contains oligonucleotide probes with the wild-type p53 sequence in addition to the sequences of the most commonly occurring p53 mutations. The relative binding of fragmented, labeled template DNA to each probe in the array is then determined with a laser scanner and evaluated with software that relies on an algorithm to score for p53 mutations. This technique is potentially rapid, adaptable to a clinical laboratory setting, and permits the analysis of a large volume of clinical samples.
In the current study, the results of p53 sequence analysis of 100 surgically resected lung cancers by conventional dideoxynucleotide sequencing methods were compared with the mutation analysis of the same tumors by the GeneChip p53 assay. Differences in sequence analysis between the two approaches were further evaluated to determine the accuracy and limitations of each method.
METHODS
Sample Collection.
Primary tumor, blood, and normal lung were collected from 100 patients undergoing surgical resection of lung cancer at The Johns Hopkins Hospital and The Johns Hopkins Bayview Medical Center. Lymphocytes were collected from blood and used a source of normal DNA. Tumor samples were promptly frozen at −80°C after initial gross pathological examination.
Portions of the primary tumor were cut into 7-μm sections, stained with hematoxylin and eosin, and examined by light microscopy. Additional 12-μm sections were cut and placed in a mixture of 1% SDS and proteinase K at 48°C overnight. Tumors with low neoplastic cellularity (<70%) were further microdissected to remove contaminating normal cells. DNA was extracted with phenol/chloroform and precipitated with ethanol.
Manual p53 Sequencing.
A 1.8-kb fragment of the p53 gene (exons 5–9) was amplified from primary tumor DNA in all 100 patients by using PCR as described (12, 13). The PCR products were purified and sequenced directly by using cycle sequencing (AmpliCycle sequencing kit, Perkin–Elmer) and appropriate sequencing primers (12, 13). In addition, exon 6 was amplified with the antisense primer, 5′-GAGACCCAGTTGCAAACCA-3′ and exon 7 with the antisense primer 5′-GAGGCAAGCAGAGGCTGG-3′ in selected cases. The products of the sequencing reactions were then separated by electrophoresis on a 6% denaturing polyacrylamide gel (Genomyx, Foster City, CA) by using the Genomyx LR DNA sequencing system, and exposed to film. Dideoxynucleotide lanes were grouped together (e.g., all As, all Cs, etc.) and read by two independent observers (S.A. and J.C.) All mutations were confirmed on a second sequencing gel after reamplification of the 1.8-kb fragment from tumor DNA.
Automated p53 Sequencing.
The same oligonucleotides used for manual sequencing (above) were synthesized on a Perkin–Elmer Applied Biosystems Division (PE/ABD) 394 DNA Synthesizer by using the standard 0.2 μM scale. All synthesis reagents were obtained from PE/ABD. The samples were sequenced by using the fluorescent dideoxy terminator method of cycle sequencing on a Perkin–Elmer Applied Biosystems Division (PE/ABD) 377 automated DNA sequencer, following ABD protocols at the DNA Analysis Facility of Johns Hopkins University (14, 15). All wild-type and mutant sequences were initially identified by using the sequencher Software from Gene Codes (Ann Arbor, MI). Electropherograms were also read manually to identify mutations below the detection threshold of the software. All mutations identified were previously detected and thus confirmed by manual sequencing.
GeneChip p53 Assay.
Tumor DNA from all 100 patients was also sequenced by using the GeneChip p53 assay (Affymetrix, Santa Clara, CA) per the manufacturer’s protocol. Exons 2–11 of the p53 gene from each tumor and the normal reference DNA were amplified as 10 separate amplicons in a single PCR reaction. Each PCR reaction contained 250 ng of genomic DNA, 5 μl of the p53 primer set (Affymetrix), 10 units of AmpliTaq Gold (Perkin–Elmer), PCR buffer II (Perkin–Elmer), 2.5 mM MgCl2, and 0.2 mM each dNTP in a final volume of 100 μl. The reaction tubes were then heated to 95°C for 10 min followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 45 sec followed by a final extension of 10 min at 72°C. Amplified tumor and reference DNA (45 μl) was then fragmented with 0.25 units of fragmentation reagent (Affymetrix) at 25°C for 18 min in 2.5 units of calf intestine alkaline phosphatase, 0.4 mM EDTA, and 0.5 mM Tris⋅acetate (pH 8.2) followed by heat inactivation at 95°C for 10 min.
The fragmented amplicons were then 3′-end labeled with fluoresceinated dideoxy(AMP). Fragmented DNA (50 μl) was incubated at 37°C for 45 min in a 100-μl reaction containing 25 units of terminal transferase (Boehringer Mannheim), TdTase buffer, and 10 μM fluorescein-N6-ddATP followed by heat inactivation at 95°C for 5 min. The fluorescein-labeled sample was then hybridized in a 0.5-ml reaction containing 6× SSPE [standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA] 0.05% Triton X-100, 1 mg of acetylated BSA, and 2 nM control oligonucleotide F1 (Affymetrix) to the p53 probe array for 30 min at 45°C. The probe array was washed four times with wash buffer A (3× SSPE, 0.005% Triton X-100) and then scanned by laser (HP GeneArray Scanner, Hewlett–Packard). The emitted light intensity was proportional to bound tumor DNA at each location on the probe array.
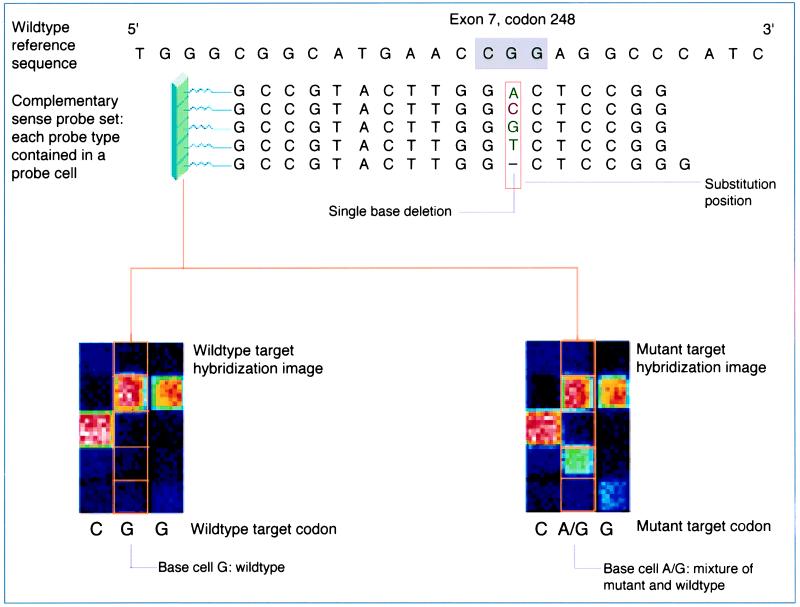
A mixture-detection algorithm was used to facilitate detection of p53 mutations in heterogeneous samples. The algorithm utilized a sample with wild-type p53 sequence as a reference, which had been hybridized and scanned under identical conditions to samples with unknown p53 sequence. Mutations were detected based on the differences in hybridization intensities between the reference and unknown sample. The ability to detect differences in intensity patterns was enhanced by including redundancy at each base position. Each site on the GeneChip corresponding to a base in the p53 sequence was covered by at least two probe sets (one sense and one antisense)(Fig. 2). In addition, 300 known missense mutations in the p53 gene are also covered by an additional 14 probe sets. Possible base calls for each site included “N” (unable to make a call); “−” (single base deletion); “A;” “C;” “G;” or “T” (wild-type bases); or a mixture call (e.g., “A/G,” indicating the presence of a mutant base).
Figure 2.
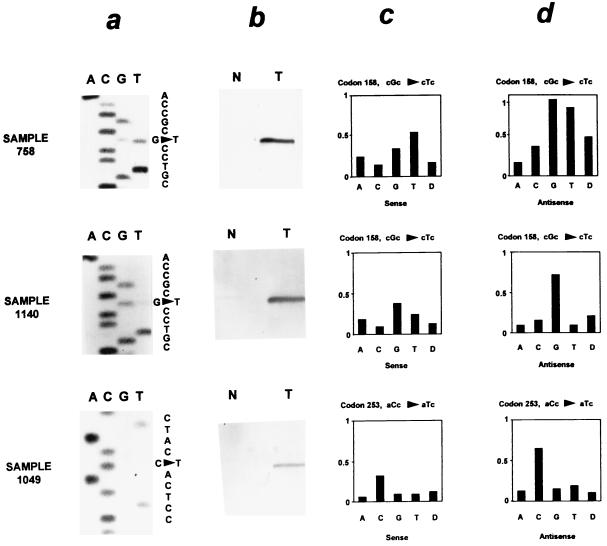
Comparison of direct dideoxynucleotide sequencing (a), mutation-specific oligonucleotide hybridization (b), and the p53 GeneChip assay (c and d) in sequence analysis of the p53 gene. Relative fluorescent intensity is shown for the sense (c) and antisense (d) oligonucleotide probe sets of the GeneChip but not for the 14 additional probe sets present at each of these base pairs. Sample 758 contains a missense mutation (cgc → ctc at codon 158) detected by all three techniques. Sample 1,140 contains the same mutation as sample 758 present as a faint band on the sequencing gel. However, it was not detected by the p53 GeneChip assay. A mutation at codon 253 in sample 1,049 was detected by the p53 GeneChip assay and confirmed by mutation-specific oligonucleotide hybridization despite its absence by direct sequencing.
The algorithm also assigned a score for each site containing a mutation or deletion. These scores were derived from the sum of mixture variables calculated from the contributing probe sets and were affected by the number of probe sets contributing to the call for a specified position. The higher the score for a given position, the greater the likelihood that site contained a mutant base. For sites containing only sense and antisense probe sets, possible scores were 1–7 for point mutations and 1–4 for single base pair deletions. For sites covered by additional probe sets, possible scores were 1–32 for point mutations and 1–10 for single base pair deletions. For this analysis, only scores exceeding 13 (score that correlated with the presence of a mutation in preliminary experiments) were called a mutation. Samples with mutations detected by the GeneChip p53 assay that were not present on the direct sequencing gel were further analyzed by repeating the direct sequencing of the involved exon.
Mutation-Specific Oligonucleotide Hybridization.
Tumors with a p53 mutation identified by using either manual sequencing or the GeneChip p53 assay and not by using the other technique were further evaluated with mutation-specific oligonucleotide hybridization. The same 1.8-kb fragment of p53 was amplified from the primary tumor and normal lymphocyte or lung tissue DNA with primers containing EcoRI sites as described (12). The PCR products were then either electrophoresed on a 1% agarose gel and transferred to nylon membranes or cloned into a λ bacteriophage vector (Stratagene) and amplified further in Escherichia coli cells (12). Between 1,000 and 2,000 clones were then transferred to nylon membranes (NEN). Both normal and tumor DNA were then hybridized with 32P-end labeled oligonucleotide probes specific for the p53 mutation identified in each patient’s primary tumor by one of the sequencing methods as described (12). A >90% difference in hybridization between normal and tumor DNA confirmed the presence of the p53 mutation.
Statistical Analysis.
GeneChip scores are given as mean ± SEM. Comparisons between GeneChip scores were performed by using Mann–Whitney U test. Comparisons between patient groups were performed by using the χ2 test.
RESULTS
p53 Mutations in Lung Cancer.
Direct manual p53 sequencing initially detected 42 mutations in 41 of the 100 lung cancers within exons 5–9 of p53 and in the adjacent intronic sequences. Automated dideoxynucleotide sequence analysis with fluorescence labeling was much less sensitive (see below) and was therefore not used for direct comparison to the GeneChip assay. The GeneChip p53 assay (Fig. 1) initially detected 45 mutations in the same 100 lung cancers within exons 5–9 and the adjacent flanking introns. The two techniques gave conflicting results about the presence of a mutation in 26 of the 100 samples (Fig. 2, samples 1,140 and 1,049). Identical results were obtained with both assays in 74 of the 100 samples (43 wild-type, 31 with a mutation)(Fig. 2, sample 758).
Figure 1.
Schematic representation of chip array system. Probes on the array are arranged in sets of five. Each probe in the set is complementary to the reference sequence except for a mismatch position, called the “substitution” position. At the substitution position, each of the four possible nucleotides (A, C, G, T) and a single base pair deletion are represented in the probe set. Assay conditions optimize hybridization of the fluorescently labeled DNA target to the probe that best matches its sequence. This hybrid yields a higher fluorescence intensity relative to the other four target–probe hybrids in the set. There are probe sets complementary to every base in the p53 gene (figure courtesy of Affymetrix).
Analysis of Mutations Detected Only by Direct Sequencing.
Eleven p53 mutations detected by direct sequencing in 10 of the tumors were not detected by the GeneChip p53 assay (Table 1). Five of these were frameshift mutations (three 1-bp deletions, one 11-bp deletion, and one complex rearrangement). Two of these five mutations (samples 792 and 1,044) were recognized by the GeneChip software but were assigned a score below our threshold for calling a mutation (13). One mutation (sample 1,044) was detected only on the second analysis by the GeneChip. The other six mutations were missense mutations, and only one of these six mutations (sample 826) was detected by the software, being assigned a score just below the threshold (13) for calling a mutation (see Methods). All 6 missed missense mutations were at sites with 14 oligonucleotide probe sets. Eight of the 11 mutations were confirmed by strong hybridization of a mutation-specific oligonucleotide probe to tumor DNA and weak or no hybridization to normal lymphocyte DNA from the same patient (Fig. 1, sample 1,140). Two tumors (samples 829 and 1,217) had a frameshift mutation clearly present on two different sequencing gels and therefore were not hybridized with a mutant-specific oligonucleotide probe. Paired normal DNA was not available for oligonucleotide hybridization for one tumor (sample 1,263) containing a missense mutation.
Table 1.
p53 gene mutations detected by direct dideoxynucleotide sequencing and not detected by the GeneChip p53 assay
| Sample | p53 gene mutation∗ | p53 GeneChip score† | Confirmed‡ |
|---|---|---|---|
| 741 | 1-bp deletion at 154 | 0 | Yes |
| 792 | 1-bp deletion at 154 | 6§ | Yes |
| 826 | TGC → GGC at 176 | 12§ | Yes |
| 829 | 11-bp deletion at 132 | 0 | ND |
| 856 | GGC → GTC at 154 | 0 | Yes |
| 970 | GGA → TGA at 199 | 0 | Yes |
| 1,044 | 1-bp deletion at 299 | 0/5§ | Yes |
| 1,140 | CGC to GTC at 158 | 0/0 | Yes |
| 1,217 | complex rearrangement, exon 6 | 0/0 | ND |
| 1,217 | ATC → ACC at 255 | 0 | Yes |
| 1,263 | t → a, intron 8 | 0 | ND |
p53 mutation gene mutation at given codon determined by direct dideoxynucleotide sequencing.
Samples 1,044, 1,140, and 1,217 were analyzed twice from two different PCR amplifications.
Mutation confirmed by hybridization of mutant specific oligonucleotide probe to PCR products from tumor DNA and not to normal DNA. ND, not done.
Scores of 12 or less are not considered definite mutations (see Methods).
Analysis of Mutations Detected Only by the GeneChip p53 Assay.
Sixteen mutations detected by the GeneChip p53 assay were not identified on initial review by two observers of the manual p53 sequencing gels (Table 2). On review of the direct sequencing gels after completing the GeneChip analysis, a third observer with knowledge that a mutation was present was able to identify 6 of the 16 mutations. Two mutations (samples 847 and 1,174) were not visible on the initial direct sequencing gel but were clearly present after repeat sequencing of the involved exon. One of these mutations (sample 847) was only apparent with the antisense primer for exon 6. Four of the 16 mutations (samples 864, 1,049, 1,052, and 1,113) identified by the GeneChip p53 assay were not seen on manual sequencing gels despite by using both sense and antisense primers [exon 6 (n = 3) and exon 7 (n = 1)]. The GeneChip analysis was repeated and again identified a mutation at the same site in three of the four tumors. Mutant-specific oligonucleotide primers were obtained for each of these mutations and confirmed the presence of the mutation as called by the p53 GeneChip in each of these tumors. The remaining tumor (sample 1,049) identified as p53 mutant by the GeneChip p53 assay had a score (12) just below the threshold on repeat analysis but did hybridize with a mutant-specific oligonucleotide confirming the presence of a mutation (Fig. 2). The GeneChip p53 assay scores for these 4 samples (17 ± 2) were significantly (P < 0.02, Mann–Whitney U test) lower than the scores from those tumors identified by both direct sequencing and the GeneChip (21 ± 1). The mean GeneChip software score assigned to all twelve samples with mutations in the conserved region missed by direct sequencing (18 ± 2) was lower than the mean score of the mutations detected by both manual sequencing and GeneChip analysis, but this did not reach statistical significance. Two of the 16 mutations (samples 1,011 and 1,102) fell outside the conserved regions analyzed by direct sequencing (exon 10), and one mutation (sample 1,023) was in intron 8 and not included on the direct sequencing gel. Finally, one mutation (sample 1,318) identified by the GeneChip was not evident on the manual sequencing gels or on repeat analysis of the same sample using the GeneChip.
Table 2.
p53 gene mutations detected by the GeneChip p53 assay and not detected by dideoxynucleotide sequencing
| Sample | p53 gene mutation* | p53 GeneChip score† | Confirmed‡ |
|---|---|---|---|
| Mutation missed on first sequencing (gel-reader error) | |||
| 778 | g → t, intron 8 | 14 | ND |
| 901 | GAG → TAG at 294 | 23 | Yes |
| 938 | CAG → TAG at 165 | 14 | ND |
| 971 | a → g, intron 6 | 32 | Yes |
| 1,105 | CGA → TGA at 213 | 15 | Yes |
| 1,133 | TGT → TTT at 277 | 17 | ND |
| Mutation not visible on first sequencing gel but present on repeat gel | |||
| 847 | ATC → TTC at 195 | 24/20 | Yes |
| 1174 | GCC → CCC at 159 | 17/17 | Yes |
| Mutation not visible on first or on repeat sequencing gels | |||
| 864 | CCT → CTT at 190 | 26/24 | Yes |
| 1,049 | ACC → ATC at 253 | 15/12 | Yes |
| 1,052 | TAT → TGT at 220 | 13/17 | Yes |
| 1,113 | GAC → GTC at 208 | 14/13 | Yes |
| Mutation outside of conserved regions evaluated (not by dideoxynucleotide sequencing) | |||
| 1,011 | GAG → TAG at 339 | 15 | Yes |
| 1,102 | GGG → TGG at 334 | 32 | Yes |
| Mutation present in intron 8 and not included on sequencing gel | |||
| 1,023 | a → t, intron 8 | 31 | ND |
| Mutation not present on sequencing gel or on repeat GeneChip analysis | |||
| 1,318 | GGC → GGT at 154 | 13/0 | ND |
p53 mutation gene mutation at given codon determined by the GeneChip p53 assay.
Samples 847, 864, 1,049, 1,052, 1,113, 1,174, and 1,318 were analyzed twice from two different PCR amplifications.
Mutation confirmed by hybridization of mutant specific oligonucleotide probe to tumor DNA and not to normal DNA. ND, not done.
In several cases where a mutation was identified by the GeneChip and missed by direct sequencing, we have amplified and cloned the PCR products. Sequence analysis of individual clones revealed that these cases generally contained less than 15% mutant alleles, accounting for the increased sensitivity of the GeneChip assay (data not shown).
Comparison of Direct Sequencing with the GeneChip p53 Assay.
Analysis of 100 primary lung cancers by using both direct sequencing of exons 5–9 and the GeneChip p53 assay detected a combined total of 57 mutations in 56 tumors. Direct sequencing detected 42 of these 57 (74%) p53 mutations. Only 2 of the 57 (4%) mutations detected were outside the conserved regions of the p53 gene usually covered in direct sequencing approaches. Manual sequencing detected 42 of 55 (76%) mutations within the conserved region of the p53 gene.
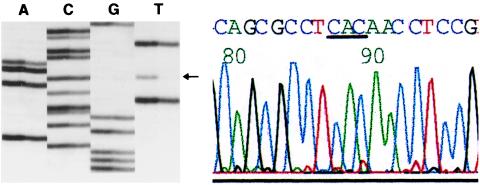
In a further comparison, we proceeded with automated sequence analysis of exons 5–9 in 48 of these p53 mutant tumors. Automated analysis detected only 33 of 48 (68%) mutations and did not identify any new mutations (Fig. 3). The poor sensitivity of automated sequence analysis is consistent with our previous observations and those of others (see Discussion).
Figure 3.
Comparison of manual and automated sequence analysis. A CAC → CAT mutation is immediately evident on manual sequencing (Left) but is “missed” by automated analysis even after manual reading because of the very low height of the mutant T peak (red).
The GeneChip p53 assay detected 46 of the 57 (81%) p53 mutations, including 44 of the 55 (80%) mutations within the conserved region of the p53 gene. The types of mutations observed in this group of 100 lung cancers were similar to those previously described [deletions/insertions (9%), G/C to A/T (12%), G/C to A/T at CpG (7%), G/C to T/A (32%), G/C to C/G (7%), and changes at A/T (33%)] in lung cancer. There was no significant difference χ2 in the ability of direct sequencing and the GeneChip p53 mutation detection assay to detect missense mutations (74% versus 88%) or a specific type of missense mutation within the conserved region of the p53 gene. However, frameshift mutations and deletions were significantly more likely to be detected with manual sequencing than with the GeneChip mutation detection assay (100% versus 0%, P < 0.05, χ2). All of the mutations (with the exception of 1319) detected by using direct sequencing or the p53 GeneChip were subsequently confirmed with the other modality or oligonucleotide hybridization. The specificity of direct sequencing and the p53 GeneChip assay at detecting p53 mutations were 100% and 98%, respectively.
DISCUSSION
The p53 tumor suppressor gene is among the most frequently mutated genes in human cancer (1, 2). Mutations in the p53 gene may play an important role in the diagnosis, staging, and management of the cancer patient. A rapid, accurate means of identifying p53 mutations in clinical samples would expedite the use of this information in clinical practice as well as facilitate studies further defining its role in the management of patients with cancer. The p53 GeneChip was more sensitive than direct sequencing at detecting p53 mutations, but neither technique was infallible at detecting mutations.
Currently, the standard method for detecting p53 mutations is PCR amplification of the p53 gene followed by direct DNA sequencing (2). Sequencing of the p53 gene in most studies has been limited to the conserved region of the gene (exons 5–9), where 87% of reported mutations occur (1). A prescreening step of the PCR products by sensitive gel assays (single-strand conformation analysis, denaturing gradient gel electrophoresis) can reduce the labor-intensive work of identifying mutations by direct genetic analysis but does not identify specific mutations. Direct DNA sequencing still has several limitations including the need to maximize neoplastic (mutant) DNA free from contamination with wild-type DNA from infiltrating normal cells, the inherent error rate of Taq polymerase, the potential for false-positive and false-negative results because of PCR product contamination, and the inability to detect mutations or deletions outside of the region sequenced. Mutations are currently identified with direct sequencing by either labeling the sequencing primer with a radioisotope or fluorescent dye and then separating the products on a polyacrylamide gel. We used radioactive-based direct cycle sequencing to evaluate the results of the GeneChip, because as shown here and in the experience of others (B. Vogelstein and S. Thibodeau, personal communications), radioactive based direct sequencing is more sensitive than fluorescent based automatic sequencing in detecting mutations within mixed populations of mutant and wild-type cells (primary tumors). Moreover, the low intensity of mutant peaks in these samples often dictates manual reading of fluorescent gels greatly increasing the required labor in “automated” systems Some of the pitfalls can be overcome by using dye-primer sequencing, but this is an expensive approach not readily available to most clinical laboratories.
In the present study, our laboratory, with considerable experience in sequencing the p53 gene, detected mutations in 41% of patients with early stage lung cancer, a result similar to other series of patients with resectable lung cancer (6, 16–21). Nevertheless, direct sequencing missed many of the mutations detected in this series by the p53 mutation detection GeneChip. Similar to other studies, only 4% of the mutations detected by analysis of the entire coding region of the p53 gene by the GeneChip were outside of the conserved region (exons 5–9), and therefore were missed by direct sequencing (1). Half of the mutations missed by direct sequencing were subtle changes on the autoradiograph and were not seen or mistaken for stop or extra bands, were in poor quality areas of the autoradiograph, or were at compressed intron–exon boundaries. The remaining half of the missed mutations were simply not evident on good-quality sequencing gels. Several of these mutations were detected by using an opposite-sense sequencing primer after knowing the site of the mutation from the GeneChip p53 assay. However, two-thirds of the mutations not detected on the first sequencing gel were also not detectable by running additional sequencing gels despite knowing the site of the mutation and by using both sense and antisense primers. The mutation rate in this group of early lung cancers is the highest reported supporting the contention that dideoxynucleotide sequencing has a substantial false-negative rate. In the present study, 14 of the 58 (24%) tumors determined to be wild type by direct sequencing had a mutation detected by the GeneChip. Similar false-negative rates would clearly confound the results of clinical studies examining the role of the p53 gene in prognosis or responses to treatment.
In addition to the technical limitations inherent in the technique, direct sequencing has not proven readily adaptable to the clinical setting. The technique is time-consuming, requiring DNA preparation, two PCR amplification steps, and polyacrylamide gel electrophoresis of the sequencing reaction products for 5 separate exons followed by reanalysis of any suspicious or technically inadequate areas of the sequencing gel. Automated dideoxynucleotide sequencing obviates the need for radioisotopes but is still limited by the dilution of mutant alleles and software analysis of primary tumor samples.
The GeneChip p53 assay detects mutations by comparing the emitted light intensity from fragmented, fluoresceinated sample DNA hybridized with an oligonucleotide probe array with that from a control sample with wild-type p53. The entire coding region of the p53 gene is covered by at least two oligonucleotide probe sets (one sense and one antisense). In addition, the GeneChip contains 14 probe sets corresponding to ≈300 missense mutations that appear more than once in the p53 gene and have been previously entered in the European Molecular Biology Laboratory Data Library (22). The emitted light intensity corresponding to bound fragmented, labeled sample DNA from each oligonucleotide in the array is detected by laser scanner and further evaluated by the GeneChip software to determine the p53 sequence. The software is designed to detect missense mutations complementary to the mutant oligonucleotides in the array as well as single base pair deletions. The GeneChip was not designed to detect deletions of >1 bp because these represent a small percentage of p53 mutations reported to date. Inclusion on the chip of probe sets to detect larger deletions as in other chip designs may have decreased the sensitivity at detecting the more common p53 mutations (10). Mutations detected at sites covered with 14 probe sets have scores ranging from 0 to 32, whereas mutations at sites covered by only 2 probe sets rarely have scores exceeding 7.
In the current series of 100 patients, the GeneChip detected 81% of the p53 mutations and was more sensitive in detecting mutations in the conserved region of the p53 gene than direct sequencing (80% versus 76%). The GeneChip was unable to identify any of the frameshift mutations in this group of cancers. Two of the five frameshift mutations were detected by the software, but were both assigned a score below our threshold for defining them as mutations. Assigning a different, lower threshold for frameshift mutations would have led to the identification of two of the four single base pair deletions. The lower sensitivity at detecting frameshift mutations appears to be the greatest weakness of the GeneChip p53 assay and would be more significant when analyzing a tumor type with more frequent frameshift mutations such as head and neck cancer (19%), sarcomas (17%), or skin cancer (31%) (1). Lowering the threshold for frameshift mutations should improve the ability to detect frameshift mutations with the GeneChip assay. The six missed missense mutations were spaced throughout the conserved region of the gene and included mutations at evolutionarily conserved “hot spots” (codons 154, 176, and 255) (1). One of the six missed missense mutations (sample 1140) also occurred in a different sample in the series and was identified as a mutation in that sample. Mutations were also detected at the same codon as five of the six missed missense mutations elsewhere in the group of 100 lung cancers. Although the sensitivity of the p53 GeneChip is slightly lower than the reported sensitivity of similar arrays at detecting germ-line mutations (10), the demands placed on the p53 chip to detect somatic mutations against a background of wild-type DNA from tumor-infiltrating cells are considerably greater.
The GeneChip p53 assay provides several distinct advantages over direct DNA sequencing. The assay is accurate, detecting more mutations in this group of 100 lung cancers than direct sequencing. Furthermore, analysis of the p53 gene with the GeneChip assay saves considerable time over direct sequencing. The assay involves a single PCR reaction, followed by agarose gel confirmation of successful amplification, several hours to fragment and fluorescein-label the product, and 1–2 hours to run the hybridization, laser scanning, and sequence analysis. All 100 samples were amplified and analyzed with the GeneChip p53 assay by a single investigator in 6 weeks. Conventional sequencing of these 100 samples consumed almost 1 year of an investigator’s time.
The p53 GeneChip provides rapid, accurate sequence analysis of the p53 tumor suppressor gene in clinical samples. Neither the p53 GeneChip or direct sequencing were able to detect all of the p53 mutations in a group of 100 lung cancers. This result alone is important when considering how often mutations in tumors with loss of heterozygosity or in families with linkage to a particular locus are likely to be missed. However, the p53 GeneChip provides a rapid screen, detecting >80% of the mutations with a very low false-positive rate (2%). For samples with mutations assigned a GeneChip score just above the threshold for mutation identification (13–15), repeating the GeneChip p53 assay may distinguish those samples not containing a mutation. In addition, mutations assigned a GeneChip score <13 at sites with two probe sets may be real and also warrant further analysis. All samples with a score >15 had a mutation also confirmed by direct sequencing and/or oligonucleotide hybridization. In settings where the accurate determination of p53 status in every tumor is mandatory, direct sequencing should be performed on the tumors diagnosed as wild-type by the GeneChip to identify tumors with frameshift or deletion mutations.
Acknowledgments
This work was supported by a Lung Spore Grant CA 58184-02 (D.S.); grants from Edwin and Barbara Pearlstine, Efrat and David Muller Lefkowitz, the American Physician Association for Medicine in Israel, and the Israel Anticancer Fund (S.H.).
References
- 1.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2.Sidransky D, Hollstein M. Annu Rev Med. 1996;47:285–301. doi: 10.1146/annurev.med.47.1.285. [DOI] [PubMed] [Google Scholar]
- 3.Harris C C, Hollstein M. N Engl J Med. 1993;329:1318–1327. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 4.Rusch V, Klimstra D, Venkatraman E, Oliver J, Martini N, Gralla R, Kris M, Dmitrovsky E. Cancer Res. 1995;55:5038–5042. [PubMed] [Google Scholar]
- 5.Lowe S W, Bodis S, McClatchey A, Remington L, Ruley H E, Fisher D E, Housman D E, Jacks T. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Oyama T, Nishida K, Ogami A, Osaki T, Nakanishi R, Sugio K, Yasumoto K, Sugimachi K. Ann Oncol. 1995;6, Suppl. 3:S9–S13. doi: 10.1093/annonc/6.suppl_3.s9. [DOI] [PubMed] [Google Scholar]
- 7.Carbone, D. P., Mitsudomi, T., Chiba, I., Piantadosi, S., Rusch, V., Nowak, J. A, McIntre, D., Slamon, D., Gazdar, A. & Minna, J. (1994) Chest106, Suppl., 377S–381S. [PubMed]
- 8.Fodor S P A, Read J L, Pirrung M C, Stryer L, Lu A T, Solas D. Science. 1993;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 9.Pease A C, Solas D M, Sullivan E J, Cronin M T, Holmes C P, Fodor S P A. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacia J G, Brody L C, Chee M S, Fodor S P, Collins F S. Nat Genet. 1996;14:441–447. doi: 10.1038/ng1296-441. [DOI] [PubMed] [Google Scholar]
- 11.Cronin M T, Fucini R V, Kim S M, Masino R S, Wespi R M, Miyada C G. Hum Mutat. 1996;7:244–255. doi: 10.1002/(SICI)1098-1004(1996)7:3<244::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 12.Sidransky D, von Eschenbach A, Tsai Y C, Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton S R, Frost P, et al. Science. 1991;252:706–709. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 13.Boyle J O, Hakim J, Koch W, van der Riet P, Hruban R H, Roa A, Correo R, Eby Y J, Ruppert J M, Sidransky D. Cancer Res. 1993;53:4477–4480. [PubMed] [Google Scholar]
- 14.Smith L M, Sander J Z, Kaiser R J, Hughes P, Dodd C, Connel C R, Heiner C, Kent S B, Hood L E. Nature (London) 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 15.McCombie W R, Heiner C, Kelly J M, Fitzgerald M G, Gocayne J D. DNA Seq. 1992;2:289–296. doi: 10.3109/10425179209030961. [DOI] [PubMed] [Google Scholar]
- 16.Casson A G, McCuaig S, Craig I, Ayed A, Inculet R, Kerkvliet N, O’Malley F. J Surg Oncol. 1994;56:13–20. doi: 10.1002/jso.2930560105. [DOI] [PubMed] [Google Scholar]
- 17.Horio Y, Takahashi T, Kuroishi T, Hibi K, Suyama M, Niimi T, Shimokata K, Yamakawa K, Nakamura Y, Veda R, et al. Cancer Res. 1993;53:1–4. [PubMed] [Google Scholar]
- 18.Kishimoto M, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Cancer Res. 1992;52:4799–4804. [PubMed] [Google Scholar]
- 19.Kitagawa Y, Wong F, Lo P, Elliott M, Verburgt L M, Hogg J C. Am J Respir Cell Mol Biol. 1996;15:45–54. doi: 10.1165/ajrcmb.15.1.8679221. [DOI] [PubMed] [Google Scholar]
- 20.Miller C W, Simon K, Aslo A, Kok K, Yokota J, Buys C H C M, Terada M, Koeffler H-P. Cancer Res. 1992;52:1695–1698. [PubMed] [Google Scholar]
- 21.Mitsudomi T, Oyama T, Kusano T, Osaki T, Nakanishi R, Shirakusa T. J Natl Cancer Inst. 1993;85:2018–2023. doi: 10.1093/jnci/85.24.2018. [DOI] [PubMed] [Google Scholar]
- 22.Hollstein M L, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3351–3355. [PMC free article] [PubMed] [Google Scholar]