Abstract
The protein huntingtin (htt), aggregated in neuronal nuclear inclusions, is pathognomonic of Huntington’s disease (HD). Constructs, translated in vitro from the N terminus of htt, containing either polyQ23 from a normal individual, or polyQ41 or polyQ67 from an HD patient, were all soluble. Transglutaminase (TGase) crosslinked these proteins, and the aggregations did not have the staining properties of amyloid. More TGase-catalyzed aggregates formed when the polyglutamine domain of htt exceeded the pathologic threshold of polyQ36. Furthermore, shorter htt constructs, containing 135 aa or fewer, formed more aggregates than did larger htt constructs. TGase activity in the HD brain was increased compared with the control, with notable increases in cell nuclei. The increased TGase activity was brain specific. In lymphoblastoid cells from HD patients, TGase activity was decreased. TGase-mediated crosslinking of htt may be involved in the formation of the nonamyloidogenic nuclear inclusions found in the HD brain. The staining properties of nuclear inclusions in the HD brain revealed that they were not amyloid.
The protein huntingtin (htt) contains a stretch of glutamines near its N terminus. When the length of the polyQ domain exceeds 36Q in htt, the lethal neurological disease called Huntington’s disease (HD) occurs. Aggregated htt in the nuclei of neurons and in dystrophic neurites in the brain are the pathologic hallmarks of HD (1–3), although it remains unknown whether the aggregates are deleterious for neurons, or whether they represent an adaptive response to a desperate situation. Two hypotheses dominate our perception of how htt is aggregated: Perutz et al. (4) proposed that polyglutamine domains on neighboring proteins organize themselves into polar zippers. Such aggregations have the classic properties of amyloid (5, 6). Green proposed (7, 8), in the second hypothesis, that TGase polymerizes htt.
In Vitro Studies on Polar Zipper Formation with htt and PolyQ Peptides.
Synthetic polyglutamine polymers, containing polyQ domains far shorter than the pathologic threshold of 36Q in HD, form polar zippers and aggregate in an aqueous medium (9). Polymerized htt is not seen in the brains of normal individuals with htt < Q36. The polar zipper hypothesis cannot easily explain this. Scherzinger et al. (1) reported that there is a threshold for polar zipper formation, and that aggregates form in vitro only when the polyQ domain is above 36Q. They showed that glutathione S-transferase (GST) fusion proteins from exon 1 of htt, with Q > 36, aggregate, but only after proteolytic cleavage of the GST domain (1).
In Vitro Studies with htt and TGase.
Kahlem et al. (8) studied guinea pig TGase and TGase isolated from rat brain; they showed that htt isolated from the brains of juvenile HD patients could be crosslinked in vitro into aggregates. To date, no one has reported on the activity of TGase in the HD brain, on the biophysical properties of the aggregates catalyzed by TGase, or on the optical properties of inclusions in the HD brain.
MATERIALS AND METHODS
TGase Assay on HD Brains and Lymphoblastoid Cells.
Each assay (10, 11) contained 80 μg of brain extract, 4 mg/ml N,N-dimethylated casein, 50 mM Tris (pH 8.0), 5 mM CaCl2, 5 mM DTT, and 0.37 mM putrescine (1:5, [3H]putrescine:putrescine) in 80 μl. The reaction was incubated at 37°C for 30 min. After it was precipitated in 500 μl of 10% TCA and washed again in 100% EtOH, the reaction was resuspended in 220 μl of 0.1 M NaOH. The resuspended pellet was added to 10 ml of scintillation liquid. Specificity was demonstrated with 5 mM monodansylcadaverine (MDC). For lymphoid cells, 106 cells were suspended in 0.5 ml of buffer for 5 min, then centrifuged at 1,200 × g for 10 min.
Extracts from Human Brains.
Tissues were obtained from the Baltimore HD Project Brain Bank, Johns Hopkins School of Medicine. The HD material was from a 32-year-old patient with a score of 4 on the Vonsattel scale (1, 2, 8) and htt of Q60/Q19 (the number of Q residues encoded by each allele of the htt gene); a 43-year-old patient with a score of 4 on the Vonsattel scale and htt of Q56/Q19; a 38-year-old patient with a score of 3 on the Vonsattel scale and htt of Q63/Q26; a 43-year-old patient with htt of Q53/Q20; and a 75-year-old patient with htt of Q44/Q16. Control brains came from 30- to 80-year-old patients. Postmortem examinations were performed within 13 h. Approximately 500 mg of brain tissue was homogenized in 2 ml of 10 mM Hepes (pH 7.4) containing 150 mM NaCl, 0.2 mg/ml leupeptin, 0.2 mg/ml aprotinin, 0.2 mg/ml pepstatin, and 0.4 mM PMSF. The homogenate was centrifuged at 4°C for 10 min at 1,000 × g, and the supernatant was then centrifuged at 4°C for 10 min at 10,000 × g to separate the cytoplasmic proteins. The remaining nuclear pellet was washed twice, for 10 min each time, with the homogenization buffer at 4°C at 1,000 × g, and then suspended in 1 ml of 10 mM Tris-Cl, 140 mM NaCl, 3 mM MgCl2, 0.5 mM PMSF, 0.1% SDS, and 1% Nonidet P-40 (pH 7.4). The homogenate was spun at 4°C for 10 min at 8,000 × g. This procedure increased the level of β-actin (a protein found more frequently in the cytoplasm than in the nucleus [12]), enriching it 15.5-fold in the cytoplasm relative to the nucleus.
Electrophoresis and Western Blot Analysis.
One-hundred micrograms of protein was loaded onto 10% polyacrylamide/SDS gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes and detected using the enhanced chemiluminescence system (Amersham). Affinity-purified anti-TGase antibody was used at 1:1,250.
Affinity-Purified Antibodies Against TGase.
TGase C (300 μg; Sigma) was diluted in 500 μl of PBS and suspended in 500 μl of complete Freund’s adjuvant for the first two injections. For the third injection, the TGase in PBS was suspended in 500 μl of incomplete Freund’s adjuvant. Each rabbit was injected each time with a total of 300 μg of protein. The first two injections were given with an interval of 3 weeks, and the third injection was given 1 month after the second. Antisera were passed over an affinity column of AffiGel (Bio-Rad), coupled with TGase C.
htt DNA Constructs and In Vitro Translation.
cDNA constructs containing 330 aa of the N terminus of htt with 23 or 44 glutamine repeats were a gift of Christopher Ross. These were subcloned directionally as BamHI/NotI fragments into the vector pcDNA3.1(+) under the control of the T7 promoter (Invitrogen).
An htt cDNA construct containing the first (approximately) 135 aa of the N terminus with 67 glutamines and a large 5′-untranslated region was a gift of Richard Myers. A construct lacking the 5′-untranslated region was made by performing PCR, using the construct as a template and the primer pair 5′-GAATTCGCCATGGCGACCCTGGAAAAGCTGATGAAG-3′ and 5′-TCTAGACTATCGGTGCAGCGGCTCCTCAGCCACAGC-3′. The PCR product was cloned into pTargeT under the control of the T7 promoter (Promega) (13). The same PCR primer pair was also used on the previously mentioned Q23 and Q41 constructs.
For incubation with TGase, 5 μl of each of the these products was incubated for 45 min at 37°C in a 20-μl vol containing the following: 50 mM Tris (pH 8.0) 5 mM CaCl2, 5 mM DTT, and appropriate concentration of guinea pig liver TGase (Sigma). Inhibition of the TGase-mediated aggregation was demonstrated by co-incubation with a monoclonal antibody, CUB7402 (NeoMarkers, Union City, CA) at 80 μg/ml. For Western analysis, another monoclonal antibody against TGase, TG100 (NeoMarkers), was used at 1:2000.
Congo Red Staining of Human HD Tissue and Identification of Inclusions.
The neocortex of a juvenile HD patient from the Baltimore HD Project Brain Bank and an elderly male with Alzheimer’s disease from the University of New Mexico Brain Bank were studied. Sections were deparaffinized, stained with Congo red and hematoxylin counterstain, and photographed. Identical sections were then subjected to a polyclonal antibody to ubiquitin (DAKO, Carpinteria, CA). Sections were treated with hydrogen peroxide/methanol, microwaved for several minutes, blocked with 3% normal goat serum, incubated with primary antibody at room temperature overnight (16–20 h), and developed using avidin-biotin complex reagents (Vector Laboratories), 3,3′-diaminobenzidine chromagen, and a brief hematoxylin counterstain.
RESULTS
TGase Crosslinks a Fragment of Translated htt Containing the PolyQ Domain.
We examined whether soluble htt constructs could be crosslinked by TGase in vitro. We used a rabbit reticulocyte lysate system to translate transcripts containing portions of exon 1 of htt with polyQ23, polyQ41, or polyQ67. We first constructed a 310-aa fragment, beginning with the N-terminal methionine of htt, corresponding to a predicted 40-kDa protein. We chose this length because a 40-kDa band that is recognized by anti-htt antibodies in total protein homogenates and in nuclear extracts can be detected in an HD cortex but not in a control brain (2). Thus, httQ23 is a 310-aa fragment and httQ41 is a 330-aa fragment of the N terminus of htt (Fig. 1A). We also constructed a 90-aa fragment from the N terminus, httQ23; a 110-aa fragment, httQ41; and a 135-aa fragment, httQ67 (Fig. 2A).
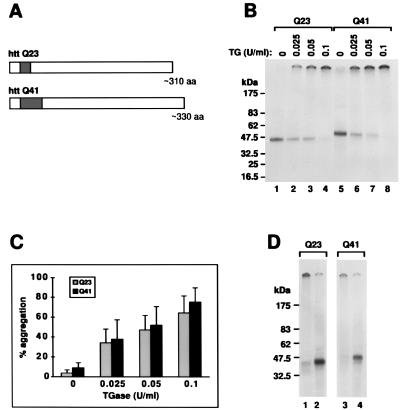
Figure 1.
httQ23 and httQ41are soluble and form aggregates in the presence of TGase. (A) Constructs used in in vitro translation. Each construct begins with the N-terminal methionine of the htt protein. The dark bar indicates the location and size of the polyglutamine repeats. (B) In vitro expression of httQ23 and httQ41. RNA synthesized from the T7 promoter of pCDNA3 containing the httQ23 or httQ41 constructs was translated in the presence of [35S]methionine in a rabbit reticulocyte lysate system, producing soluble products of the expected molecular weights (lanes 1 and 5, respectively). Incubation in the presence of increasing concentrations of TGase produced increasing amounts of an aggregate that remained at the top of the gel (lanes 2–4 for httQ23 and lanes 6–8 for httQ41). (C) Quantitation of htt aggregation in the presence of TGase. Densitometry was performed on three similar gels as in B. The percentage of products that remained at the top of the gel within each lane is shown as a function of TGase concentration. The percentage is a mean of three separate experiments ± 1 SEM. There is no significant difference in the aggregation percentages between the two constructs. (D) Inhibition of TGase-mediated aggregation with an antibody against TGase. Lanes 1 and 2 contain in vitro translated httQ23 incubated in the presence of 0.1 unit/ml TGase. Lanes 3 and 4 contain in vitro translated httQ41 incubated in the presence of 0.1 unit/ml TGase. In addition, for lanes 2 and 4 during incubation with TGase, a monoclonal antibody that inhibits the activity of TGase was present at a concentration of 80 μg/ml.
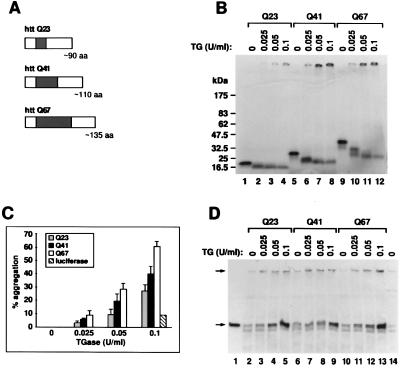
Figure 2.
TGase crosslinks httQ23, httQ41, and httQ67 and is found in the aggregates. (A) Constructs used in in vitro translation. Each construct begins with the N terminal methionine of the htt protein. The dark bar indicates the location and size of the polyglutamine repeats. (B) In vitro expression of httQ23, httQ41, and httQ67. RNA synthesized from the T7 promoter of pCDNA3 containing the httQ23, httQ41, or httQ67 constructs was translated in the presence of [35S]methionine in a rabbit reticulocyte lysate system, producing soluble products of the expected molecular weights (lanes 1, 5, and 9 respectively). Incubation in the presence of increasing concentrations of TGase produced increasing amounts of an aggregate that remained at the top of the 4% to 15% gradient gel (lanes 2–4 for httQ23, lanes 6–8 for httQ41, and lanes 10–12 for httQ67). (C) Quantitation of htt aggregation in the presence of TGase. Densitometry was performed on three similar gels as in B. The percentage of products that remained at the top of the gel within each lane is shown as a function of TGase concentration. The percentage is a mean of three separate experiments ± 1 SEM. The aggregation of an unrelated control protein, luciferase, at a single concentration of TGase is also shown. There is a significant increase in the amount of aggregation as a function of polyQ length (P < 0.002 httQ67 vs. httQ23 at 0.1 unit of TGase per ml; P < 0.01 httQ67 vs. httQ23 at 0.05 unit of TGase per ml; P < 0.02 httQ67 vs. httQ41 at 0.1 unit of TGase per ml). This result with the smaller htt constructs is in contrast to that with the larger htt constructs (Fig. 1C). (D) TGase comigrates with the htt aggregates. The httQ23, httQ41, and httQ67 constructs were translated and incubated in the presence of TGase as in B. The samples were loaded on a 4% to 15% gradient gel and blotted with a monoclonal antibody against TGase. Exposure times were such that none of the [35S]methionine-labeled htt were visible. The identities of the lanes are the same as in B except that an additional lane contains 2 milliunits of TGase alone (lane 1) and another lane contains an equivalent concentration of rabbit reticulocyte lysate alone (lane 14). Some of the TGase localized to the top of the gel (upper arrow) as did the htt aggregates in B. The lower arrow indicates a molecular mass of 80 kDa, the expected size of TGase alone (lane 1). A small amount of endogenous TGase can be found within this system (lane 14). No other significant immunoreactive bands are seen on this blot.
We intrinsically labeled httQ23, httQ41 or httQ67 with [35S]methionine. Fig. 1B shows that after the addition of TGase, httQ23 (310 aa) and httQ41 (330 aa) aggregated into a high molecular weight polymer within 45 min. There was no increase in aggregation with the longer httQ41 compared with httQ23 (results represent mean density ± 1 SEM, three experiments) (Fig. 1C).
Fig. 2B shows similarly that after the addition of TGase, httQ23 (90 aa), httQ41 (110 aa), and httQ67 (135 aa) aggregated into a high molecular weight polymer within 45 min. Fig. 2C shows increased aggregation with the constructs httQ41 (110 aa) and httQ67 (135 aa), containing polyQ domains exceeding the pathologic threshold of Q36. This is in comparison with httQ23 (90 aa), with (i) P < 0.002 httQ67 vs. 0.1 httQ23 unit of TGase per ml; (ii) P < 0.01 httQ67 vs. 0.05 httQ23 unit of TGase per ml; and (iii) P < 0.02 httQ67 vs. 0.1 httQ41 unit of TGase per ml; these three results represent the mean density ± 1 SEM of three experiments. Interestingly, there is a direct correlation between the amount of aggregation and the size of the polyQ domain. There was less aggregation of luciferase, the control, than of the various htt constructs. Luciferase contains 16 glutamines, but none are in tandem.
The ordered structure of amyloid allowed binding of the Congo red dye at regular intervals, producing a characteristic red-green birefringence under polarized light (1, 5, 6, 14). This red-green birefringence was noted in htt aggregates in vitro after enzymatic cleavage of the GST fusion protein (1). The TGase-mediated aggregates of htt in vitro, shown in Figs. 1 and 2, did not show green birefringence after staining with Congo red and thus cannot be considered amyloid, according to a widely accepted definition of amyloid (5, 6).
The aggregation of htt depends on TGase activity. By using a monoclonal antibody against TGase that is known to block its activity (CUB 7402; NeoMarkers), we were able to inhibit the aggregation of htt (Fig. 1D, lanes 2 and 4).
TGase was detected in the aggregates of httQ23, httQ41, or httQ67 (Fig. 2D) with the use of monoclonal anti-TGase antibodies. On Western blots with anti-TGase antibodies, products were seen that co-migrated with the [35S]httQ23, [35S]httQ41, or [35S]httQ67 aggregates (Fig. 2D, lanes 3–5, 7–9, and 11–13). A small amount of endogenous TGase was detected in the rabbit reticulocyte system (Fig. 2D, lanes 2, 6, 10, and 14).
It is noteworthy that httQ23, httQ41, and httQ67 were all soluble. In contrast, GST fusion constructs of htt with Q > 30 were insoluble after the GST protein was enzymatically cleaved (1). The in vitro translated material in our experiments was soluble and did not form aggregates when analyzed by SDS/PAGE after as long as 125 h at room temperature (data not shown).
TGase Activity Is Increased in Nuclei in HD Brains.
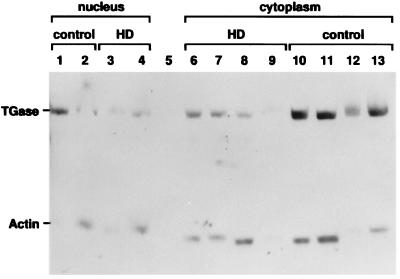
There is no information to date on whether TGase activity is present in the HD brain or in HD brain nuclei. First, we ascertained with Western blots that TGase had appeared in nuclei isolated from the brain of an HD patient (httQ63/Q26) and from a control brain, and that it could be seen in cytoplasm isolated from HD brains (httQ44/Q16; httQ63/Q26), and from a control brain (Fig. 3). Nuclear TGase has been described in neuroblastoma cells in vitro (15), but not heretofore in an HD brain.
Figure 3.
TGase is present in the nucleus and the cytoplasm of HD brains. TGase is seen in nuclei isolated from the brains of HD patients (as well as from a control brain), and from cytoplasm isolated from HD brains (as well as from a control brain). An 89-kDa band staining with the affinity-purified anti-TGase antibody, which migrates at the same position as recombinant guinea pig TGase (data not shown), and a 42-kDa band staining with a monoclonal anti-actin antibody are seen in both the nucleus and cytoplasm. Lanes 1 and 2 are control brain nuclear fractions, 50 and 100 μg of lysate; lanes 3 and 4 are HD brain nuclear fraction (httQ63/httQ26), 50 and 100 μg of lysate; lane 5 is BSA; lanes 6 and 7 are cytoplasmic extracts from an HD brain (httQ44/httQ16), 50 and 100 μg, respectively; lanes 8 and 9 are cytoplasmic extracts from another HD brain (httQ63/httQ26), 100 and 50 μg of lysate, respectively; lanes 10 and 11 are cytoplasmic extracts from control brain, 50 and 100 μg of lysate, respectively; lanes 12 and 13 are cytoplasmic extracts from another control brain, 50 and 100 μg of lysate, respectively.
In Table 1, we show that cytosolic extracts from an HD brain and from a control brain provided enzymatic activity for the incorporation of radiolabeled putrescine as an amine donor into dimethylated casein, which served as an amine acceptor (10, 11). Enzymatic activity was completely inhibited by MDC, which competed with radiolabeled putrescine as a substrate for TGase (Table 1). In the HD brain, neuronal loss is prevalent in the cortex, whereas areas such as the cerebellum are occasionally affected in adult HD brains (16) and routinely affected in juvenile HD brains. TGase activity was greater in the HD brain than in corresponding areas of the control brain (HD cortex 14,888 ± 2,863 cpm vs normal cortex 6,697 ± 1,410 cpm, mean ± 1 SEM, P < 0.009 for HD cortex vs normal cortex; HD cerebellum 11,221 ± 2,426 cpm vs control cerebellum 2,606 ± 719 cpm, mean ± 1 SEM, P < 0.001 for HD cerebellum vs control cerebellum).
Table 1.
TGase activity is increased in HD brain
| Source of brain sample | TGase activity, cpm
|
|||
|---|---|---|---|---|
| + Casein | + Casein + MDC(n = 2) | − Casein | − Casein + MDC(n = 2) | |
| HD cortex | 14,888 ± 2,863* (n = 3) | 94 ± 18 | 464 ± 104 | 81 ± 17 |
| HD cerebellum | 11,221 ± 2,426† (n = 3) | 186 ± 57 | 477 ± 61 | 58 ± 13 |
| Control cortex | 6,697 ± 1,410‡ (n = 5) | 63 ± 10 | 607 ± 167 | 151 ± 39 |
| Control cerebellum | 2,606 ± 719 (n = 3) | 83 ± 14 | 145 ± 50 | 117 ± 30 |
| HD cortical nuclear extract | 6,368 ± 764§ (n = 3) | ND | 153 ± 23 | ND |
| Control cortical nuclear extract | 2,357 ± 226 (n = 4) | ND | 189 ± 32 | ND |
TGase activities (mean ± SEM) were measured as described in the text; n, number of human patients analyzed. By unpaired Student’s t test: ∗, P < 0.009 comparing HD to control cortex; †, P < 0.001 comparing HD to control cerebellum; ‡, P < 0.04 comparing control cortex to control cerebellum; §, P < 0.0001 comparing HD to control cortical nuclear extract. ND, not done.
TGase activity was also greater in the extracts of nuclei from brains of HD patients than from control brains (HD brain cortical nuclear extract 6,368 ± 764 cpm vs. control brain cortical nuclear extract 2,357 ± 226 cpm, mean ± 1 SEM, P < 0.0001; Table 1). It is noteworthy that even in HD cortices that were damaged from the extensive degeneration typical of the disease, TGase activity was actually increased compared with controls. The increase in TGase activity in the HD cerebellum indicated that this brain region was also involved. At this level of analysis of the pathophysiology in adult HD, “Evidence for involvement of cerebellar Purkinje cells is convincing” (16). Neuronal inclusions are reported in the cerebella of patients with adult-onset HD, although less frequently than in the neocortex or striatum of such patients (17). In comparison, TGase activity is unchanged in the cerebellum in Alzheimer’s disease, compared with the control brain (18).
Tissue Specificity of TGase Activity.
Investigators have previously shown that TGase activity is decreased in lymphoid cells taken from HD patients (19). We wanted to confirm this observation and to compare TGase activity, which is elevated in the HD brain, to TGase activity in nonneurologic tissue. For this we used human Epstein–Barr virus-transformed lymphoblastoid diploid cell lines containing htt with various lengths of polyQ (20), and we measured the intrinsic TGase activity.
Representative results from one of four experiments show a negative correlation (P < 0.03) between endogenous TGase activity and the length of the polyQ stretch in the htt in the transformed cell lines. Thus, in a line from a normal individual (control; httQ18/httQ22), TGase activity was 938 ± 178 cpm vs 282 ± 66 cpm in a line from an HD patient (httQ48/httQ41); 446 ± 71 cpm in a line from another HD patient (httQ70/httQ19); and 190 ± 72 in yet another line from an HD patient (httQ46/httQ20). P < 0.03 for the control vs various HD lines, mean ± 1 SEM, repeated 6 times; cpm represent incorporation of [3H]putrescine into N,N-dimethylcasein. Thus, TGase activity was lower in the lymphoblastoid cell lines derived from patients with HD than in the cell lines from controls. These findings in lymphoid tissue are the opposite of those in HD brains, where TGase activity was elevated compared to activity in control brains.
Intranuclear Inclusions in Human HD Neurons Cannot Be Detected with Polarized Light When Stained with Congo Red.
To explore whether the intranuclear protein aggregates found in HD patients postmortem are capable of incorporating Congo red to produce green birefringence in polarized light, we developed a double staining method. Given that these inclusions can be demonstrated only by immunocytochemical (or electron microscopic) techniques, it is difficult to interpret a negative Congo red stain, either as a single stain (without knowing the sites of the inclusions), or as part of a double label on a previously reacted section that contains a large antigen–antibody–chromagen complex (17). Thus, we chose to stain a single section first with Congo red, photographing many neuronal nuclei with both brightfield and polarized light and then revealing the intranuclear aggregates by immunocytochemistry; this enabled a study of identical cells. The technique definitively demonstrates which nuclei that were visualized with polarized light (regardless of polarity) actually contained protein aggregates with the known, typical staining characteristics of HD inclusions. The applicability of this method was established through the simultaneous staining of similarly prepared neocortical tissue from a patient with Alzheimer’s disease, tissue that contained congophilic angiopathy (vascular amyloid) and senile plaques. This positive control for both amyloid and ubiquitin had abundant polarizable amyloid in both blood vessels and senile plaques, as well as ubiquitin immunoreactivity in neurites of senile plaques (Fig. 4).
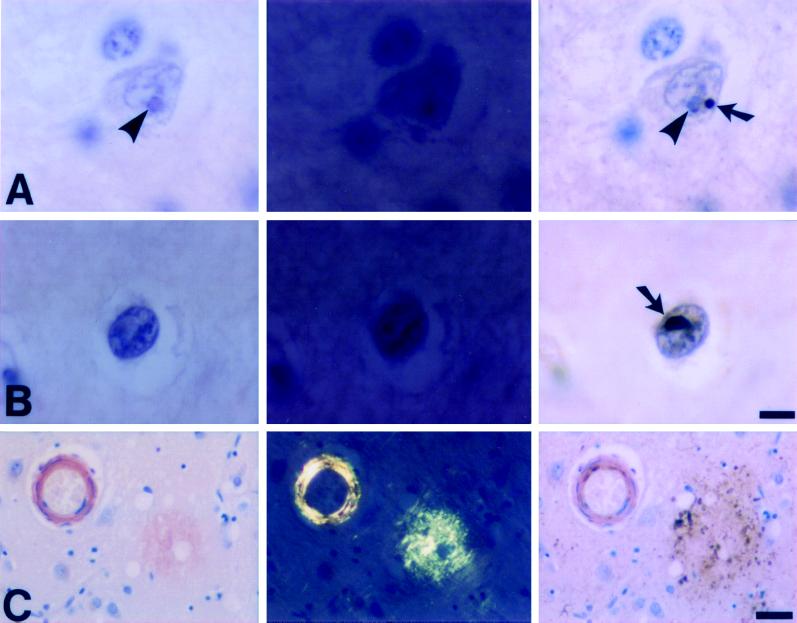
Figure 4.
Congo red staining of intranuclear inclusions in human HD neurons. Congo red stained intranuclear inclusions in cortical neurons (A, B) from a juvenile HD (genetically confirmed) patient do not show red staining when viewed by brightfield (Left) or display apple green birefringence when viewed with polarized light (Center). Sequential staining of the identical section with ubiquitin (Right) confirms the presence of a typical HD inclusion (arrows) which is distinct from the nucleolus (arrowheads) and is not visualized in the previous images. Amyloid in a blood vessel with congophilic angiopathy and a senile plaque from a patient with Alzheimer’s disease (C) serves as a positive control for both the Congo red (C, Left and Center) and ubiquitin (C, Right) stains. (Bars: A, B = 10 μm; C = 50 μm. Hematoxylin counterstain.)
With this method, no Congo red-stained material was identified in neuronal nuclei or in other neural components of the HD brain by either brightfield or polarized light (Fig. 4). These views included numerous nuclei that contained ubiquitin-immunoreactive protein aggregates in the same plane of focus (Fig. 4). Thus, in the configuration found in human HD neurons, the protein aggregates of HD do not incorporate Congo red to yield polarized light birefringence. It appears by immunocytochemistry that the protein aggregates of HD are well defined and large enough to be detected by a dye-based method such as Congo red. Therefore this negative result is not caused simply by the lack of protein that stains; rather it suggests, within the constraints of this postmortem experiment, that the proteins in these polyglutamine-rich aggregates do not have a classic amyloid-like configuration.
DISCUSSION
We find that TGase activity was elevated in the HD cortex and cerebellum, regions where htt aggregates into nuclear inclusions (2, 17). Moreover, TGase activity was increased in HD brain nuclei. Interestingly, TGase activity was reduced in lymphoid cells from HD patients, a region where aggregates do not occur in HD. Regional differences in TGase activity might help to explain the exquisite anatomic localization of HD in the brain.
A recent study (1) argued that amyloid formation could explain the nuclear inclusions in HD tissue. There are essential differences between the in vitro aggregates reported in ref. 1 and the aggregates actually seen in neuronal nuclear inclusions in an HD brain (2), in a dentatorubral-pallidoluysian atrophy (DRPLA) brain (21), and in an spinocerebellar ataxia type 3 brain (22). The aggregates formed in vitro after cleavage of GST as reported in ref. 1 have the properties of amyloid, are able to stain with Congo red, and show green birefringence under polarized light (1). Congo red binding is characteristic of amyloid aggregates (1, 5, 6, 14). “It has been generally accepted that naturally occurring mammalian protein polymers that exhibit a rod or fibril appearance by EM and show green birefringence after Congo Red staining should be classified as amyloid” (6). Similar definitions appear elsewhere (1, 5, 14).
However, in the HD brain no amyloid inclusions have been reported: “previous searches for amyloid deposits in brains of HD patients have been negative” (23). We show in this paper that inclusions in the HD brain do not stain with Congo red and therefore should not be considered amyloid. Thus, neither TGase-catalyzed polymers of htt (Figs. 1 and 2) nor the polymers that appear in HD tissue have the optical characteristics of amyloid. The covalent linkages of htt polymerized with TGase probably do not give enough order for the periodic binding of Congo red that is necessary for red-green birefringence (5, 6). It is possible that after purification, some degree of Congo red staining might be visible in aggregates if regions of the aggregates consist of proteins bound by polar zippers, but this is certainly not evident in tissue sections from the HD brain.
We find other differences between our results and those of Scherzinger et al. (1). We note that full-length htt, including htt with polyglutamine expansions in the pathologic range, does not aggregate in vitro without TGase. Short in vitro-translated fragments of 90 to 330 aa from the N terminus of htt (present study) and longer in vitro-translated portions of the N-terminal portion (50–60 kDa) of htt containing Q91 do not aggregate in vitro (24).
Can concentration differences reconcile opposing data and conclusions? One potential reason that aggregation was not seen with in vitro translated httQ41 or httQ67 (present study), or with the larger 50- to 60-kDa fragment of httQ91 reported by Goldberg et al. (24) could be that the concentration of the translated protein was not high enough to start the aggregation process. It is known that a concentration of approximately 30 to 100 μM (25) is necessary for the formation of fibrillar aggregates. In yeast, for amyloid aggregates of prion protein above a concentration of 6 μM, the rate-limiting step in conversion from oligomers to aggregates is unaffected by protein concentration (14).
We note the importance of the protein context of htt in aggregate formation: The degree of enzyme-catalyzed aggregation is a function of the size of the portions of htt outside the polyQ domain. There was an increased amount of aggregation with the constructs httQ41 (110 aa) and httQ67 (135 aa), which contained polyQ domains that exceed the pathologic threshold of Q36, compared with httQ23 (90 aa) (P < 0.02, httQ41 or httQ67 vs httQ23; results represent mean density ± 1 SEM of three experiments). This threshold is not observed for larger fragments of htt that exceed 300 aa. In Martindale et al. (26), the degree of apoptosis and death in fibroblasts and the number of aggregates in monkey kidney cells and in cortical neuronal cultures was related to the size of the htt fragment, independent of the length (greater than 36Q) of the polyQ domain. Perinuclear aggregates are also more frequent in COS cells transfected with truncated DRPLA protein that contain a pathologic polyQ domain than they are with full-length DRPLA protein that contain a pathologic polyQ domain (21). In Saudou et al. (27), shorter fragments of htt with a pathologic polyQ domain formed aggregates more readily than longer fragments with a pathologic domain.
These results may lead to the development of novel therapies for HD and other diseases involving polyglutamine expansions. Recently, Igarashi et al. (21) showed suppression of aggregate formation and apoptosis in COS cells by competitors of TGase-mediated crosslinking, including cystamine and MDC. This study and that of Kahlem et al. (8) open the very real possibility of treating diseases that are mediated by proteins with polyglutamine domains by inhibiting TGase activity in the brain (28). Recent studies (27) argue that apoptosis in striatal cell cultures transfected with mutant htt occurs after the peak of aggregate formation. It is claimed that blocking aggregation may even worsen, not help, HD patients (27). One should note, however, that the crosslinking of htt into polymers (not necessarily the formation of gross aggregates) may be sufficient to initiate the pathologic cascade of events.
It is not possible at present to resolve the issue of whether aggregation formation in HD is by polar zipper formation, by a TGase-catalyzed reaction, by a combination of both mechanisms, or by other mechanisms. One of the hallmarks of polar zipper formation, the appearance of amyloid caused by the order inherent in hydrogen-bonded β-sheets, is simply not present in HD tissue sections or in other polyglutamine diseases. Final proof of the nature of inclusions will come from ongoing studies on the composition of the chemical bonds in the aggregations.
Acknowledgments
We thank Charles Yanofsky for his critical review. This work was supported by the National Institutes of Health (Grant NS16367 to J.G., Grant K08AI01492 to H.G., Grant K08NS02027 to M.W.B., and Javits Award R0118235 to L.S.) and by a grant from the Hereditary Disease Foundation to M.W.B. The Weizmann Institute of Science provided support for M.V.K.
ABBREVIATIONS
- DRPLA
dentatorubral-pallidoluysian atrophy
- HD
Huntington’s disease
- htt
huntingtin
- GST
glutathione S-transferase
- MDC
monodansylcadaverine
- TGase
transglutaminase
References
- 1.Scherzinger E, Lurz R, Turmaine M, Mangiarine L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 2.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 3.Davies S W, Turmaine M, Cozens B, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 4.Perutz M, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glenner G G. N Engl J Med. 1980;302:1283–1292. doi: 10.1056/NEJM198006053022305. and 1333–1343. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 7.Green H. Cell. 1993;74:955–956. doi: 10.1016/0092-8674(93)90718-6. [DOI] [PubMed] [Google Scholar]
- 8.Kahlem P, Green H, Djian P. Mol Cell. 1998;1:595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 9.Stott K, Blackburn J M, Butler P J, Perutz M. Proc Natl Acad Sci USA. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C G, Lorand L. Methods Enzymol. 1976;45:177–191. doi: 10.1016/s0076-6879(76)45018-8. [DOI] [PubMed] [Google Scholar]
- 11.Cooper A J, Sheur K-F R, Burke J R, Onodera O, Strittmater W J, Roses A D, Blass J P. J Neurochem. 1997;69:431–434. doi: 10.1046/j.1471-4159.1997.69010431.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhao K, Wang W, Rando O, Xue Y, Swiderek K, Duo A, Crabtree G. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 13.Ebens A J, Garren H, Cheyette B N R, Zipursky S L. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- 14.DePace A, Sartoso A, Hillner P, Weissman J. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 15.Lesort M, Attanavanich K, Zhang J, Johnson G V W. J Biol Chem. 1998;273:11991–11994. doi: 10.1074/jbc.273.20.11991. [DOI] [PubMed] [Google Scholar]
- 16.Robitaille Y, Lopes-Cendes I, Becher M, Rouleau G, Clark A W. Brain Pathol. 1997;7:901–926. doi: 10.1111/j.1750-3639.1997.tb00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 18.Johnson G V W, Cox T M, Lockhart J P, Zinnerman M D, Miller M L, Powers R E. Brain Res. 1997;751:323–329. doi: 10.1016/s0006-8993(96)01431-x. [DOI] [PubMed] [Google Scholar]
- 19.Cariello L, de Cristofaro T, Zanetti L, Cuomo T, Di Maio L, Campanella G, Rinaldi S, Zanetti P, Di Lauro R, Varrone S. Hum Genet. 1996;98:633–635. doi: 10.1007/s004390050273. [DOI] [PubMed] [Google Scholar]
- 20.Anderson M A, Gusella J F. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 21.Igarashi S, Koide R, Shomohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 22.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J L, Fischbeck K H, Pittman R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 23.Lunkes A, Mandel J L. Nat Med. 1997;3:1201–1202. doi: 10.1038/nm1197-1201. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg Y P, Nicholson D W, Rasper D M, Kalchman M A, Koide H B, Graham R K, Bromm M, Kazemi-Esfarjani P, Thornberry N A, Vaillancourt J P, et al. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 25.Harper A, Lansbury J L. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 26.Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim S, et al. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- 27.Saudou P, Finkbeiner S, Devys D, Greenberg M. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 28.Lorand L. Proc Natl Acad Sci USA. 1996;93:14310–14313. doi: 10.1073/pnas.93.25.14310. [DOI] [PMC free article] [PubMed] [Google Scholar]