Abstract
Citrus tristeza virus (CTV) populations in citrus trees are unusually complex mixtures of viral genotypes and defective RNAs developed during the long-term vegetative propagation of the virus and by additional mixing by aphid transmission. The viral replication process allows the maintenance of minor amounts of disparate genotypes and defective RNAs in these populations. CTV is a member of the Closteroviridae possessing a positive-stranded RNA genome of ≈20 kilobases that expresses the replicase-associated genes as an ≈400-kDa polyprotein and the remaining 10 3′ genes through subgenomic mRNAs. A full-length cDNA clone of CTV was generated from which RNA transcripts capable of replication in protoplasts were derived. The large size of cDNA hampered its use as a genetic system. Deletion of 10 3′ genes resulted in an efficient RNA replicon that was easy to manipulate. To investigate the origin and maintenance of the genotypes in CTV populations, we tested the CTV replicase for its acceptance of divergent sequences by creating chimeric replicons with heterologous termini and examining their ability to replicate. Exchange of the similar 3′ termini resulted in efficient replication whereas substitution of the divergent (up to 58% difference in sequence) 5′ termini resulted in reduced but significant replication, generally in proportion to the extent of sequence divergence.
Citrus tristeza virus (CTV) isolates consist of unusually complex populations of distinct genotypes and defective RNAs (dRNAs). CTV populations have been structured over hundreds of years by the vegetative propagation of commercial citrus and are compounded by multiple aphid transmissions of virus from tree to tree. Characterized populations contain as many as 18 different genotypes, many of which are too divergent to be considered a quasispecies (1). The diversity of dRNAs within the populations is similar. There is limited information concerning whether different replication characteristics foster or inhibit population diversity. In a population with diverse genotypes and dRNAs, do these RNAs share replication machinery or are there mechanisms to segregate the processes? Do the specificities of interactions between RNAs and replicase complexes affect population structures?
CTV is a member of the complex Closteroviridae family that contains viruses with mono- and bipartite genomes with a range of vectors including aphids, whiteflies, and mealybugs (2–4). CTV is one of the more economically important viruses of plants because of the severity of damage it causes and the high value of individual citrus trees with normal productive life spans of up to 100 years (5). Decline isolates of CTV rapidly kill citrus trees, and stem-pitting isolates make citrus production noneconomical because of poor tree vigor and small fruit. During the last century, CTV has destroyed entire citrus industries in several countries: thus, the name “tristeza” (Portuguese and Spanish for “sadness”). The long flexuous virions (2,000 nm × 10–12 nm) of CTV are encapsidated by two coat proteins, one covering ≈95% of the virion and the other completing encapsidation of the other terminus (6). The single-stranded RNA genome of CTV is ≈20 kilobases (kb), divided into twelve ORFs (7, 8) (Fig. 1A). ORF 1a encodes a 349-kDa polyprotein containing two papain-like protease domains plus methyltransferase-like and helicase-like domains. Translation of the polyprotein is thought to occasionally continue through the polymerase-like domain (ORF 1b) by a +1 frameshift. Ten 3′ ORFs are expressed by 3′ coterminal subgenomic (sg) mRNAs that are promoted internally (9, 10). CTV characteristics are intermediate between those of alphaviruses and the Coronaviridae. CTV resembles members of the Coronaviridae because the replicase-associated polyprotein contains two putative proteases, the genomic RNA has large interdomain regions, the polymerase-like domain presumably is translated after a frameshift, and several sgRNAs are synthesized and have corresponding double-stranded RNAs that accumulate to high levels (11, 12). However, the amino acid sequence similarity of the polymerase-like domain and sgRNAs that do not contain a common 5′ leader are more similar to the alphavirus-like superfamily (8, 10).
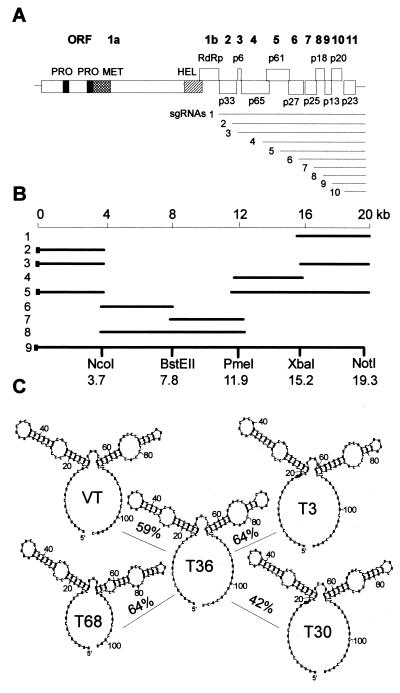
Figure 1.
(A) Schematic representation of the genome organization and expression of 3′-terminal ORFs of CTV, showing putative domains of papain-like proteases (PRO), methyl transferase (MET), helicase (HEL), RNA-dependent RNA polymerase (RdRp), and ORFs 2 through 11. The nested set of subgenomic mRNAs are shown below. (B) Diagrammatic representation of the steps involved in the construction of the full-length CTV T36 cDNA clone flanked by the SP6 RNA polymerase promoter at the 5′ end (solid box) and the NotI restriction site at the 3′ end. The intermediate clones used to make pCTV were, in step 1, pT36–3′Xba; 2, pT36–5′Nco; 3, pT36–5′Nco-3′Xba; 4, pT36-pme-Xba; 5, pT36–5′Nco-3′Pme; 6, pT36-Nco-Bst; 7, pT36-Bst-Pme; 8, pT36-Nco-Pme. The restriction endonuclease sites and their positions in the genome used to assemble pCTV are also indicated. (C) Predicted secondary structures of the 5′ NTR of CTV isolates T36, VT, T3, T68, and T30 using the mfold program (18). The homology of 5′ NTR of isolates VT, T3, T68, and T30 with 5′ NTR of isolate T36 are indicated between their respective secondary structures.
“Strains” of CTV that are considered members of the same virus species have unusual characteristics. First, the sequence variation among the different genotypes is large, with as little as 50–80% nucleotide identity in much of the genome. Also, although the sequence divergence between some isolates is uniform throughout the genome, the divergence of other isolates is asymmetrical and progressively increases toward the 5′ terminus to as little as 42% identity within the 5′ nontranslated region (NTR) (13–16). Second, CTV isolates have multiple dRNAs of various sizes and abundance (15, 17).
Do properties of CTV replication favor maintenance of unusually diverse populations? One functional measure of sequence compatibility is the interaction between the replicase components and specific cis-acting sequences that define which RNAs are replicated. In general, these elements reside in the 5′ and 3′ termini of positive-stranded RNA viruses. Recognition can be based on primary or secondary structure, or a combination of both. The only sequence segments retained by the various dRNAs of CTV are those at the termini, suggesting that cis-acting elements are similarly located in CTV. The 3′ NTR is the most conserved sequence of the CTV genotypes, usually with >95% identity. In contrast, the 5′ NTR sequences differ substantially. Although the 5′ NTRs have little sequence similarity, the mfold program (18) predicts similar secondary structures, consisting of two long stem loops (Fig. 1C) (14). There is no information concerning the specificity of interactions of closterovirus replicase complexes with heterologous cis-acting elements.
A requirement for examining replicase/cis-acting element interactions is a genetic system that allows manipulation of the viral genome. Here we report the development of a full-length cDNA clone of the CTV genome from which we can produce in vitro transcripts that infect plant protoplasts. However, the large size of this viral RNA and the complex ligation strategies required to clone its cDNA, the unusual length of the resulting in vitro transcripts, and the difficulties of inoculating protoplasts with such large RNAs were each near the limit of our capabilities, and the combination of all of these factors was even more difficult. Thus, for routine replication experiments in protoplasts, we created, by deletion of ≈40% of the genome, an autonomous CTV RNA replicon that provides a much simpler genetic system. Using this system, we examined the ability of the CTV replicase complex to recognize and replicate CTV RNAs with termini representing a wide range of sequence variation.
MATERIALS AND METHODS
Virus Isolates.
CTV isolates T36, T3, T68, and T30 were propagated in Madam vinous sweet orange [Citrus sinensis (L.) Osbeck] in a greenhouse in Florida. Isolate VT was propagated in Alemow citrus (Citrus macrophylla Wester) in Israel.
Construction of Full-Genomic cDNA Clones of CTV T36.
cDNA of ≈4.0-kb segments of the genome (Fig. 1B) were produced from denatured replicative form RNA (19) by reverse transcription and were amplified by PCR [reverse transcription (RT)–PCR] with Pfu DNA polymerase (Stratagene) with primers based on nucleotide sequence information (7, 8). The full-genomic cDNAs were ligated into pUC119 by a nine-step ligation and transformation strategy (Fig. 1B) that used unique and rare restriction sites within the genome. At least 10 cDNA clones for each portion of the genome were selected and used to construct 10 independent full-genomic cDNA clones. A pair of primers, one (C-57) complementary to nucleotides 19,277–19,296 plus a NotI restriction endonuclease site and the other corresponding to nucleotides 15,242–15,266 were used to amplify the 3′ end of the genome. The RT-PCR product was digested with restriction endonuclease XbaI (nucleotide 15,248) and was ligated into SmaI- and XbaI-digested pUC119 resulting in pT36–3′Xba (Fig. 1B, step 1). The 5′ end of the genome was amplified by using a primer (5′-ATTAGGCCTATTTAGGTGACACTATAGAATTTCACAAATTCAACC) containing a StuI site (underlined) followed by the SP6 RNA polymerase promoter sequence (in italics) and nucleotides 1–18 of CTV and a primer complementary to nucleotides 3,857–3,874. The RT-PCR product was digested with NcoI (nucleotide 3,691) and was ligated into pUC119 digested with NcoI and HpaI producing pT36–5′Nco (Fig. 1B, step 2). Internal regions of the genome were amplified by using sets of primers (i) complementary to nucleotides 15,383–15,397 and corresponding to nucleotides 11,649–11,670, (ii) complementary to nucleotides 7,782–7,809 and corresponding to nucleotides 3,565–3,583, and (iii) complementary to nucleotides 12,144–12,178 and corresponding to 7,753–7,778, and were ligated as shown in Fig. 1B, steps 3- 8. The full-genomic CTV cDNA clones were generated by ligating the step-8 insert into the step-5 plasmid at NcoI and PmeI sites (Fig. 1B, step 9).
Deletion mutants were constructed from the full-genomic cDNA clone pCTV9. The sequences between nucleotides 11,011–18,529 were deleted by digestion with ClaI, and religation resulted in pCTV-ΔCla. The remaining sequences of ORF 11 were deleted from pCTV-ΔCla by ligating the 3′ NTR amplified by RT-PCR with primers C-57 and C-341 (corresponding to nucleotides 19,018 to 19,042 plus a ClaI site) followed by ClaI and NotI digestion and ligation into ClaI/NotI-digested pCTV-ΔCla, resulting in pCTV-ΔCla-3′NTR.
Substitution of Termini of pCTV-ΔCla with Heterologous Termini from Other CTV Isolates.
Hybrids were constructed by exchanging the 5′ NTR of T36 in pCTV-ΔCla with 5′ NTRs of CTV isolates VT, T3, T68, and T30. The 5′ NTRs were amplified from replicative form RNA by RT-PCR by using specific primers based on the available sequence information (ref. 13; M.R.A.-M., unpublished data). The positive-sense primer contained HindIII and StuI restriction endonuclease sites followed by the SP6 RNA polymerase promoter sequence and isolate specific sequence. Because T36, VT, T68, T3, and T30 isolates start with 5′-AATTT… . , an additional nonviral guanosine was introduced as a transcriptional start site. Isolates T30, VT, and T36 all contained a BglII restriction endonuclease site at nucleotides 1,026 that was used to exchange a 5′ 1-kb region (including 5′ NTR). The BglII-digested RT-PCR products were ligated into pCTV-ΔCla between StuI and BglII sites.
The RT-PCR product corresponding to 3′ NTRs of VT and T30 were digested with ClaI and NotI and were ligated into ClaI- and NotI-digested pCTV-ΔCla-3′NTR to produce pCTV-ΔCla-VT-3′NTR and pCTV-ΔCla-T30–3′NTR or into pCTV-ΔCla containing respective 5′ NTRs to obtain pCTV-ΔCla-3′NTR with 5′ and 3′ NTRs of VT or T30.
RNA Transcription, Transfection of Mesophyll Protoplasts of Nicotiana benthamiana, and Analysis of RNAs.
SP6 RNA polymerase transcripts from CTV cDNAs were generated in vitro from NotI-linearized plasmid DNA as described by Peremyslov et al. (20) with minor modifications. The procedures for the isolation of mesophyll protoplasts from N. benthamiana and polyethylene glycol (PEG) mediated transfection were as described (21) except that 40% PEG 1540 (Polysciences) was used. Results presented represent three to four independent protoplast transfections for each construct. The isolation of total nucleic acids from the pelleted protoplasts and Northern hybridization analyses were essentially as described in Lewandowski and Dawson (22). The 3′-terminal 900 nucleotides of CTV T36 in pGEM-7fZ (Promega) were used to make positive- or negative-stranded RNA specific riboprobes with digoxigenin-labeled UTP using either SP6 or T7 RNA polymerase. Bands were quantified by scanning and densitometry with the os-scan program (Oberlin Scientific, Oberlin, OH).
RESULTS
Development of an in Vitro Genetic System.
The large size of the CTV genome (≈20.0 kb) made it impractical to synthesize full-length cDNA in a single step (Fig. 1A). A multistep cloning strategy using RT-PCR was used to build a full-length cDNA clone from the genomic-length replicative-form double-stranded RNA, which was easier to isolate as a homogenous preparation and in sufficient amounts than virion RNA. The presence of few unique restriction sites in the genome of CTV made ligation of cDNAs into a full-genomic cDNA complex, requiring a sequence of nine ligation and transformation steps (Fig. 1B). Full-genomic cDNA clones were sequentially assembled by ligating ≈4-kb fragments into a modified pUC119 (Fig. 1B). To limit the effects of possible errors within some components of the native T36 population and possible errors during RT-PCR and propagation of cDNA in Escherichia coli, independent clones were isolated at each step of the process and were used to assemble 10 independent full-genomic cDNAs (Fig. 1B, steps 1–9).
We chose to attempt to infect protoplasts with RNA transcripts produced in vitro from the cDNA clones. The resulting clones were linearized at the unique NotI site introduced downstream from the CTV cDNA and were transcribed in vitro by using SP6 RNA polymerase (23). Although the nonviral guanosine residue was added at the transcription start site to maximize the yield, only moderate amounts of the full-length transcripts were obtained and tested in protoplast transfection experiments.
We previously developed a N. benthamiana mesophyll protoplast system in which CTV replicated after inoculation with virions or virion RNA (21). Using this system, virion RNA was only ≈0.1% as effective as CTV virions in infecting protoplasts. CTV virion RNA also was substantially less effective in infecting protoplasts than were in vitro transcripts from an infectious cDNA of beet yellows virus (BYV) (20) and tobacco mosaic virus (24). There appeared to be an exponential decrease in infection of N. benthamiana protoplasts as sizes of the RNAs (tobacco mosaic virus, BYV, vs. CTV) increased, even though the molar quantities of RNAs were similar (data not shown).
In vitro transcripts from 10 independent full-genomic cDNAs were tested for infectivity in protoplasts by using CTV virion RNA and BYV in vitro transcripts as positive controls. Capped in vitro transcripts of only 1 of the 10 independent cDNA clones, pCTV9, initiated infection and replicated. Genomic and sgRNAs similar to those produced by transfection with CTV virions or virion RNA accumulated in infected protoplasts (Fig. 2). Protoplasts inoculated with transcripts synthesized in the absence of dinucleotide cap did not yield detectable positive- or negative-stranded genomic or subgenomic RNAs (data not shown). Therefore, all further experiments were carried out with capped transcripts.
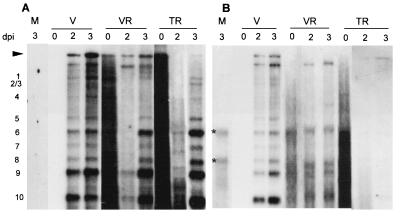
Figure 2.
Northern blot hybridization of the positive- (A) and negative-stranded (B) RNAs of CTV in N. benthamiana protoplasts 0, 2, and 3 dpi with CTV virions (V); virion RNA (VR); in vitro transcripts from full-length cDNA clone, pCTV9 (TR); or mock inoculation (M). The genomic (arrow head) and subgenomic mRNAs (1–10) corresponding to ORFs 2 through 11 are numbered in A. The asterisks in B indicate the position of background bands. Samples from virion-inoculated protoplasts were exposed for 10 min. All other lanes were exposed for 3 h.
When virion RNA or the transcripts from pCTV9 were used to transfect protoplasts, hybridization to residual in-put RNA (positive-stranded) at zero time was observed and obscured the detection of progeny genomic RNA at 1 day postinoculation (dpi). Replication of the input RNA was detected as an increase in the hybridization signal of the genomic positive-stranded RNA, but the production of virus-specific negative-stranded RNAs and the large amount of the precise set of sgRNAs were much more definitive measurements (Fig. 2). Progeny positive-stranded genomic and sgRNAs were visible at 2 dpi and increased substantially by 3 dpi in protoplasts inoculated with transcripts from pCTV9 (Fig. 2). Accumulation of these RNAs increased up to 5 dpi in experiments in which the protoplasts survived that long (data not presented). Although, the level of replication in protoplasts inoculated with CTV transcripts was never as high as in those inoculated with virions (exposed for a shorter time in Fig. 2), it was only slightly less than those inoculated with virion RNA.
Development of an Easier, More Manipulatable Genetic System.
The large size of the full-genomic cDNA clone of CTV was not amenable to efficient, routine genetic experiments. Mutation of the viral genome required subcloning the target area, mutagenesis, and then progressing back through a series of ligation steps to recreate the full-genomic cDNA clone. Additionally, in vitro transcription and transfection of protoplasts were near the limit of our capabilities. Related members of the Closteroviridae have been shown to replicate with much of their genomes removed. The lettuce infectious yellows virus genome is divided between two RNAs with the replicase-associated genes encoded on RNA 1 and the other genes, including structural proteins, on RNA 2. In protoplasts, RNA 1 alone replicates efficiently (25). Likewise, BYV replicates with most of the 3′ ORFs deleted (20). A similar deletion would simplify the construction of CTV mutants and increase the reliability of routine experiments.
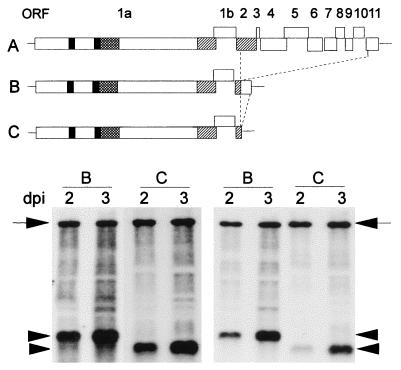
pCTV-ΔCla was a simple construct with sequences deleted from within ORF 2 to ORF 11 (Fig. 3B). In vitro transcripts from pCTV-ΔCla were highly infectious when used to inoculate protoplasts, and replication occurred faster and to higher levels than that of the full-genomic RNA (Fig. 3B). Higher yields of in vitro transcripts and a higher percentage of protoplasts infected by the smaller RNA probably contributed to this large increase in replication compared with that of the full-genomic RNA transcripts. Moreover, pCTV-ΔCla resulted in a much easier genetic system to examine replication of CTV. Mutations could be created directly within pCTV-ΔCla with only a one- or two-step ligation and transformation procedure, and inoculated protoplasts always had high levels of replication. Also, deletion of the remaining portion of ORF 11 (pCTV-ΔCla-3′NTR) producing in a virus with only the 3′ NTR from the 3′ region of the genome resulted in an RNA that replicated approximately the same as pCTV-ΔCla (Fig. 3C). Thus, with essentially all of the 3′ ORFs deleted, CTV-ΔCla-3′NTR replicated maximally, indicating that the 10 3′ genes did not provide functions required for replication of this virus.
Figure 3.
Schematic representation of CTV, CTV-ΔCla, and CTV-ΔCla-3′NTR and their replication in N. benthamiana protoplasts. Accumulation of positive- (Left) and negative-stranded (Right) RNAs at 2 and 3 dpi of protoplasts with in vitro transcripts from pCTV-ΔCla (B) or pCTV-ΔCla-3′NTR (C). The position of genomic RNA and sgRNA (promoted by ORF 2 sgRNA promoter) are indicated by arrows and arrow heads, respectively.
Function of Nonhomologous Terminal Sequences Substituted into the CTV Replicon.
There is considerable variation among the master sequences of different CTV isolates. To examine the ability of the T36 replicase to interact with different terminal sequences to initiate replication, the termini of CTV T36 were exchanged with corresponding sequences from a range of CTV isolates chosen to be representative based on sequence comparison to T36 (Fig. 1C). The VT, T3, and T68 isolates cause decline and stem pitting diseases whereas T30 is a symptomless isolate from Florida with the most disparate biology and sequence.
3′ terminal exchanges.
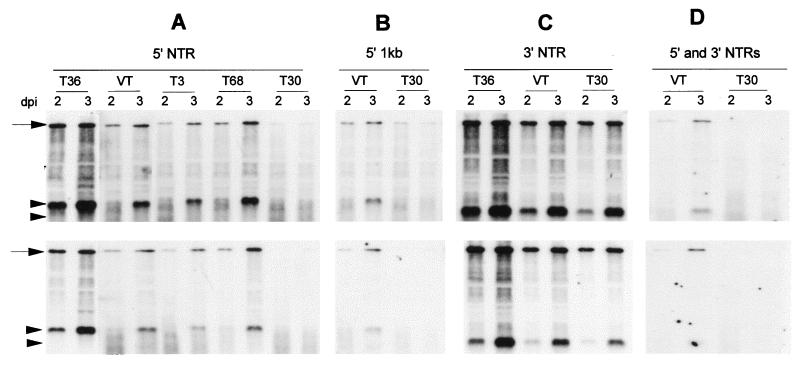
The most conserved region of all of the CTV sequences is the 3′ NTR. In this region, the sequence variation is generally <5%. However, single nucleotide differences have been shown to be sufficient to reduce or prevent replication among other viruses (ref. 26; R. Chandrika and W.O.D., unpublished work). The 3′ NTRs from the Israeli isolate VT and the Florida mild isolate T30, both of which are 97% identical to T36, were precisely exchanged into pCTV-ΔCla-3′NTR. In vitro transcripts from pCTV-ΔCla-VT-3′NTR and pCTV-ΔCla-T30–3′NTR were infectious and produced positive-stranded (top) and negative-stranded (bottom) RNAs at levels ≈70 and 50%, respectively (estimated by densitometry), of those of the wild-type T36 CTV-ΔCla-3′NTR (Fig. 4C).
Figure 4.
Replication of CTV hybrids containing 5′ and/or 3′ terminal exchanges. Shown is the accumulation of positive- (upper panels) and negative-stranded (lower panels) RNAs of CTV in protoplasts at 2 and 3 dpi with transcripts from pCTV-ΔCla (T36) and pCTV-ΔCla hybrids containing 5′ NTR sequences of VT, T3, T68, and T30 isolates (A), 5′ 1-kb sequences of VT and T30 (B), pCTV-ΔCla-3′NTR (T36) and pCTV-ΔCla-3′NTR hybrids containing heterologous 3′ NTRs (C), and both 5′ and 3′ NTRs of VT and T30 (D). The blots were exposed for 13 min except, T36 5′ NTR, which was exposed for 6 min. The position of genomic and sgRNAs in each panel are indicated by arrow and arrow heads, respectively.
5′ terminal exchanges.
López et al. (14) examined sequences of nine CTV isolates that contained 5′ NTR sequences that varied by as much as 58%. However, computer modeling folded all of these sequences into similar structures with two long stem loops (see Fig. 1C). This suggests that secondary structure might be the primary determinant involved in recognition and initiation of replication. To examine whether heterologous 5′ NTRs with substantially different nucleotide sequences would function with the T36 replicase, we built a series of cDNA constructs with precise 5′ NTR substitutions from a representative series of CTV sequences differing from T36 by 36–58% but that retain the ability to be folded into similar structures (Fig. 1C). Estimated replication levels of the hybrid RNAs containing VT, T3, and T68 5′ NTRs were ≈15, 7, and 23%, respectively, compared with the homologous T36 5′ NTR (CTV-ΔCla) (Fig. 4A). CTV-ΔCla containing T30 5′ NTR, which has only 42% sequence identity and has been classified into a third sequence group (14), replicated weakly compared with other isolates.
The 5′ sequences that function as cis-acting elements to initiate RNA replication likely involve more than the 5′ NTR. Compared with T36, VT and T30 have the closest and most divergent 5′ NTRs, respectively. We examined whether replication would differ if larger regions of 5′ sequences were exchanged by replacing the 5′ 1.0 kb of CTV-ΔCla with corresponding sequences from VT and T30. Even though the 5′ NTR of T30 was more divergent from T36 than was that of VT, over the entire 1 kb, both were similarly divergent, having ≈72% nucleotide identity and 55% amino acid identity. Although these constructs created hybrid polyproteins that might have failed to function in replication, the accumulations of genomic and subgenomic positive- and negative-stranded RNAs of these hybrids were almost the same as the hybrids containing only the heterologous 5′ NTRs (Fig. 4B).
Exchange of both 5′ and 3′ NTRs.
Because it is possible that 5′ and 3′ termini might interact to initiate RNA synthesis, we examined hybrid RNAs with both 5′ and 3′ NTR sequences of VT and T30 isolates exchanged into pCTV-ΔCla-3′NTR. The CTV-ΔCla-3′NTR hybrid with 5′ and 3′ NTRs from VT replicated ≈6% as well as the wild-type CTV-ΔCla-3′NTR (Fig. 4D) and only slightly less than the hybrid with only the VT 5′ NTR (Fig. 4A). Although the hybrid RNA with the T30 3′ NTR replicated well, the replication of the CTV-ΔCla-3′NTR hybrid with both 5′ and 3′ NTRs of T30 was detectable only after long exposures of the Northern hybridization blots. Matching heterologous termini at both ends did not function better with the T36 replicase, suggesting that the incompatible interaction between replicase and 5′ terminus was sufficient to prevent replication of the hybrid RNAs.
DISCUSSION
CTV has been such an innately difficult virus with which to work that development of an infectious cDNA clone of CTV was imperative to facilitate progress on the biological and physical characterization of this virus. The difficulties begin with an experimental host range restricted to woody plants in which the virus only occurs in phloem-associated tissues. Propagation of virus requires months, and some biological assays require years. The long flexuous virions are fragile and recalcitrant to purification. They can be purified only from bark tissues when they can be peeled from the stem and only from plants at certain stages of growth and under certain temperatures. CTV can be transmitted by aphids and also mechanically through bark slashes, but only at low efficiency. The ability to propagate and manipulate the viral genome in E. coli is a major advance for examination of this virus.
Although the infectious cDNA clone of CTV provides a system that will be useful in manipulating the virus, the size and limited number of unique restriction sites in this cDNA make creation and examination of mutants challenging. It was evident that a smaller, self-replicating subset of CTV, analogous to RNA 1 of lettuce infectious yellows virus (25), would be more amenable for examining replication in protoplasts. Similar to BYV (20), all of the 3′ ORFs of CTV could be deleted with continued replication. In contrast to BYV, in which the p21 gene product stimulated replication, none of the 3′ genes affected the levels of replication of CTV (data not presented). Moreover, deletion of the CTV 3′ ORFs greatly increased its levels of replication compared with that of the full-genomic RNA.
Citrus trees contain extraordinarily complex populations of CTV genotypes with divergent sequences along with an equally diverse collection of dRNAs. The CTV genotypes differ more than is normally expected for strains of the same virus. Based on analysis of ≈20 different sequences from representative isolates (14, 15), there presently appear to be two patterns of sequence divergence. The sequence variation within the large VT-like group occurs symmetrically throughout the genome whereas the T36-like group deviates asymmetrically, with high similarity throughout the 3′ half but progressively diverging toward the 5′ terminus. It has been suggested that T36-related sequences arose from a distant recombination event (13), perhaps between CTV and a different closterovirus, and gradually evolved away the abrupt recombination site. This also might have required an evolution of new specificities between replicase and cis-acting elements. Does the stringency of viral protein-RNA interactions regulate the breadth of recombinants as components of the population? There is little information concerning whether there might be mechanisms for discrimination that could affect interactions between different genotypes that could structure populations. One possible interaction would be the discrimination of the replicase complex for cis-acting elements of the RNAs within the population.
In general, there is considerable permissiveness for 3′ substitutions of sequences from strains of a virus or even different viruses within a group (see ref. 27), and exchanges of distantly related sequences can function minimally (28, 29). To examine the degree of specificity of the T36 replicase with divergent termini, which are expected to comprise at least part of the cis-acting elements for initiation of negative- and positive-stranded RNA synthesis, we created T36 hybrids with heterologous termini. We have found that 3′ sequences within the NTR of T36 constitute the complete 3′ cis-acting element (T.S., unpublished work). Replacement of the CTV T36 3′ terminus with that of VT or T30 resulted in an RNA that replicated efficiently, suggesting that there is little change in specificity within this area of relatively conserved sequences within different CTV genotypes. However, this is the most conserved region of CTV, and the sequence differences between these isolates were only 3% in this region.
The interactions between replicase and different 5′ terminal sequences have been examined in only a few virus groups. With bromoviruses, the interactions between the 5′ termini of RNA3 of brome mosaic virus and cowpea chlorotic mottle virus with the two different replicases resulted in some tolerance of heterologous sequences, but some reciprocal exchanges were not tolerated (30, 31), demonstrating a complex RNA–protein interaction. The 5′ cis-acting elements of bovine viral diarrhea virus RNA can be replaced with the 5′ termini of hepatitis C virus or encephalomyocarditis virus RNA with limited replication, suggesting marginal compatibility between replicase and heterologous RNA termini (32). The 5′ ends of the different CTV isolates examined differed in sequence to a degree similar to these viral exchanges. With CTV hybrids, there was a gradient of levels of replication generally proportional to sequence similarity. Although it appears that the different CTV 5′ NTRs can be folded into similar structures, the difference in levels of replication suggests that the specificity of the interaction between the replicase complex and 5′ terminal sequence might involve primary structure in addition to higher order structures.
The recognition of heterologous termini by the CTV T36 replicase suggests that the specificity of the RNA/protein interactions might contribute to the generation of complex populations. For example, these data make it probable that a T36 helper virus could replicate some heterologous dRNAs, but with different efficiencies. It is also possible that, if T36 were the major component of a population, its excess replicase could provide certain functions, for example, production of subgenomic mRNAs or amplification of positive-stranded RNAs, for other CTV genotypes in trans. The propagation of heterologous RNAs might allow survival of RNAs produced by recombination between disparate population components. However, the level of replicase discrimination for the 5′ terminal regions would probably reduce disparate components of the population if they had to rely on heterologous replication complexes. The availability of the infectious cDNA clone of CTV for the first time allows the production of pure cultures of the virus and the examination of populations constructed from defined parental sequences to further investigate the factors responsible for the diversity of CTV populations.
Acknowledgments
We thank Cecile Robertson for excellent technical assistance in preparation of protoplasts. This work was supported in part by an endowment from the J. R. and Addie S. Graves family and grants from the Florida Citrus Production Research Advisory Board, U.S. Department of Agriculture/Agricultural Research Service Cooperative Agreement 58-6617-4-018, the U.S.-Israel Binational Agricultural Research and Development Fund, and the National Citrus Research Council. This work is part of the University of Florida Agricultural Experiment Station Journal Series R-06880.
ABBREVIATIONS
- CTV
citrus tristeza virus
- dRNA
defective RNA
- kb
kilobase
- sg
subgenomic
- NTR
nontranslated region
- RT
reverse transcription
- BYV
beet yellows virus
- dpi
days postinoculation
References
- 1.Ayllón M A, Rubio L, Moya A, Guerri J, Moreno P. Virology. 1999;255:32–39. doi: 10.1006/viro.1998.9566. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Joseph M, Garnsey S M, Gonsalves D. Adv Virus Res. 1979;25:93–168. doi: 10.1016/s0065-3527(08)60569-2. [DOI] [PubMed] [Google Scholar]
- 3.Dolja V V, Karasev A V, Koonin E V. Annu Rev Phytopathol. 1994;32:261–285. [Google Scholar]
- 4.Agranovsky A A. Adv Virus Res. 1996;47:119–158. doi: 10.1016/S0065-3527(08)60735-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Joseph M, Lee R F. CMI/AAB Descriptions of Plant Viruses. Wellesbourne, Warwick, U.K.: Assoc. Appl. Biol.; 1989. , No. 353. [Google Scholar]
- 6.Febres V J, Ashoulin L, Mawassi M, Frank A, Bar-Joseph M, Manjunath K L, Lee R F, Niblett C L. Phytopathology. 1996;86:1331–1335. [Google Scholar]
- 7.Pappu H R, Karasev A V, Anderson E J, Pappu S S, Hilf M E, Febres V J, Eckloff R M G, McCaffery M, Boyko V, Gowda S, et al. Virology. 1994;199:35–46. doi: 10.1006/viro.1994.1095. [DOI] [PubMed] [Google Scholar]
- 8.Karasev A V, Boyko V P, Gowda S, Nikolaeva O V, Hilf M E, Koonin E V, Niblett C L, Cline K, Gumpf D J, Lee R F, et al. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 9.Hilf M E, Karasev A V, Pappu H R, Gumpf D J, Niblett C L, Garnsey S M. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- 10.Karasev A V, Hilf M E, Garnsey S M, Dawson W O. J Virol. 1997;71:6233–6236. doi: 10.1128/jvi.71.8.6233-6236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawicki S G, Sawicki D L. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai M M C, Cavanagh D. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph M. J Gen Virol. 1996;77:2359–2364. doi: 10.1099/0022-1317-77-9-2359. [DOI] [PubMed] [Google Scholar]
- 14.López C, Ayllón M A, Navas-Castillo J, Guerri J, Moreno P, Flores R. Phytopathology. 1998;88:685–691. doi: 10.1094/PHYTO.1998.88.7.685. [DOI] [PubMed] [Google Scholar]
- 15.Hilf M E, Karasev A V, Albiach-Marti M R, Dawson W O, Garnsey S M. Phytopathology. 1999;89:336–342. doi: 10.1094/PHYTO.1999.89.4.336. [DOI] [PubMed] [Google Scholar]
- 16.Vives M C, Rubio L, López C, Navas-Castillo J, Albiach-Marti M R, Dawson W O, Guerri J, Flores R, Moreno P. J Gen Virol. 1999;80:811–816. doi: 10.1099/0022-1317-80-3-811. [DOI] [PubMed] [Google Scholar]
- 17.Mawassi M, Karasev A V, Mietkiewska E, Gafny R, Lee R F, Dawson W O, Bar-Joseph M. Virology. 1995;208:383–387. doi: 10.1006/viro.1995.1165. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Science. 1989;224:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 19.Moreno P, Guerri J, Muñoz N. Phytopathology. 1990;80:477–482. [Google Scholar]
- 20.Peremyslov V V, Hagiwara Y, Dolja V V. J Virol. 1998;72:5870–5876. doi: 10.1128/jvi.72.7.5870-5876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navas-Castillo J, Albiach-Marti M R, Gowda S, Hilf M E, Garnsey S M, Dawson W O. Virology. 1997;228:92–97. doi: 10.1006/viro.1996.8369. [DOI] [PubMed] [Google Scholar]
- 22.Lewandowski D J, Dawson W O. Virology. 1998;251:427–437. doi: 10.1006/viro.1998.9420. [DOI] [PubMed] [Google Scholar]
- 23.Butler E T, Chamberlin M J. J Biol Chem. 1982;257:5772–5778. [PubMed] [Google Scholar]
- 24.Dawson W O, Beck D L, Knorr D A, Grantham G L. Pro Natl Acad Sci USA. 1986;83:5043–5047. doi: 10.1073/pnas.83.6.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaassen V A, Mayhew D, Fisher D, Falk B W. Virology. 1996;222:169–175. doi: 10.1006/viro.1996.0407. [DOI] [PubMed] [Google Scholar]
- 26.Duggal R, Rao A L N, Hall T C. Virology. 1992;187:261–270. doi: 10.1016/0042-6822(92)90314-f. [DOI] [PubMed] [Google Scholar]
- 27.Dreher, T. (1999) Annu. Rev. Phytopathol., in press. [DOI] [PubMed]
- 28.Ishikawa M, Kroner P, Ahlquist P, Meshi T. J Virol. 1991;65:3451–3459. doi: 10.1128/jvi.65.7.3451-3459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao A L N, Grantham G L. Virology. 1994;204:478–481. doi: 10.1006/viro.1994.1559. [DOI] [PubMed] [Google Scholar]
- 30.Allison R F, Janda M, Ahlquist P. J Virol. 1988;62:3581–3588. doi: 10.1128/jvi.62.10.3581-3588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacha R F, Ahlquist P. J Virol. 1991;65:3693–3703. doi: 10.1128/jvi.65.7.3693-3703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frolov I, McBride M S, Rice C M. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]