Abstract
Vα14 NKT cells express an invariant antigen receptor encoded by Vα14 and Jα281 gene segments as well as natural killer (NK) markers, including NK1.1. Here, we describe a precursor population of NKT cells (pre-NKT) that expresses NK1.1, T cell antigen receptor β, pTα, and RAG1/2 but not Vα14 and surface CD3ɛ. Such pre-NKT cells were differentiated successfully in vitro into mature CD3ɛ+ Vα14+ NKT cells by IL-15 and granulocyte/macrophage colony-stimulating factor (GM-CSF) in conjunction with stroma cells. Interestingly, only GM-CSF without stroma cells induced the Vα14-Jα281 gene rearrangement in the pre-NKT cells. This also was confirmed by the findings that the number of mature Vα14 NKT cells and the frequency of Vα14-Jα281 rearrangements were decreased significantly in the mice lacking a GM-CSF receptor component, common β-chain. These results suggest a crucial role of GM-CSF in the development of Vα14 NKT cells in vivo.
Vα14 natural killer T (NKT) cells constitute a unique lineage of lymphocytes distinct from conventional αβ T cells, natural killer (NK) cells, and B cells and are characterized by the coexpression of NK cell marker NK1.1 and the invariant antigen receptor encoded by Vα14 and Jα281 gene segments preferentially associated with the Vβ8.2 chains (1, 2). Two lines of evidence clearly demonstrate Vα14 NKT cells as a novel lineage of lymphocytes: (i) Vα14 NKT cells develop at the embryo body but not yolk sac as early as day 9.5 of gestation, before thymus formation (3), and (ii) a targeted disruption of the invariant Vα14 NKT receptor gene causes a selective loss of the Vα14 NKT cells, leaving other lymphocytes intact (4), whereas a transgenic expression of the invariant Vα14 NKT receptor and/or Vβ8.2 genes with T cell antigen receptor α (TCRα)-deficient and recombination activating gene (RAG)-deficient background results in an exclusive development of Vα14 NKT cells (5, 6). These studies suggest an essential role for the invariant Vα14 NKT receptor in the development and lineage commitment of Vα14 NKT cells but not other populations.
Besides their unique developmental properties, Vα14 NKT cells are shown to recognize glycolipid antigens in the context with a monomorphic CD1d (6, 7), whose mode of antigen recognition is entirely distinct from that of conventional T cells that recognize peptide antigens in association with the MHC, which is polymorphic in nature. Therefore, NKT cells seem to constitute a novel immune system mainly against glycolipid antigens. Moreover, Vα24JαQ T cells, a human counterpart for Vα14 NKT cells, have been identified (1) and are shown to recognize CD1d molecules (8, 9). The conservation of the invariant Vα14 NKT cell receptor and of the recognition of CD1d ligand through evolution implies a critical role of Vα14 NKT cells in a host-defense system.
A great deal of effort has been made to identify the molecules essential for the development of Vα14 NKT cells. In particular, studies with various genetically manipulated mice have revealed that NKT cell development depends on the molecules, including β2-microglobulin (β2m) (10–12), CD1d (13–15), pTα (16), RAG-1 and 2 (17), TCF-1 (18), and Jα281 (4). These results suggest that Vα14 NKT cells are differentiated through selection events mediated by various cell surface receptors, such as mature or immature forms of NKT cell antigen receptors and cytokine receptors during the interaction with CD1d-expressing stroma cells. However, neither precursor populations nor precise molecular requirement for the development of Vα14 NKT cells has been clarified.
Such emerging views on a novel subset of lymphocytes prompted us to search for their precursors and molecular requirements for NKT cell differentiation. Here, we have identified a precursor of mature Vα14 NKT cells and characterized the molecular requirements for differentiation. In addition, granulocyte/macrophage colony-stimulating factor (GM-CSF) is found to be a critical molecule for the initiation of the Vα14 NKT receptor gene rearrangement during the development of these lymphocytes.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from Japan SLC (Hamamatsu, Japan). Vβ8.2 transgenic (Vβ8.2tg) mice, RAG-1−/− mice, RAG-1−/− Vα14tgVβ8.2tg mice, and β2m-deficient (β2m−/−) mice were described elsewhere (5, 6, 12). RAG-1−/− Vβ8.2tg mice were produced by crossing Vβ8.2tg mice to RAG-1−/− mice. Common β (βc)-deficient mice, which were generated originally by Nishinakamura et al. (19), were kindly provided by H. Nakauchi (Tsukuba University). RAG-1−/− mice were used as a source of feeders. All animals were maintained under specific pathogen-free conditions.
Antibodies.
mAbs against Fcγ receptors II and III (FcγRII/III) (2.4G2), TCRαβ (H57–597), NK1.1 (PK136), CD3ɛ (145–2C11), Vβ8 (F23.1), Gr1 (RB6–8C5), TER119 (TER-119), CD19 (1D3), anti-Thy-1 (53–2.1), and anti-B220 (RA3–6B2) were obtained from PharMingen except for anti-Vα14 mAb (5). Cychrome-streptavidin (PharMingen) was used for the detection of biotinylated antibodies.
Flow Cytometric Analysis.
Freshly prepared or cultured cells (1 × 106) were stained as described (5). The specificity of staining was confirmed by using isotype-matched irrelevant mAbs or preincubating cells with an excess amount of the unlabeled mAb before staining with the fluorochrome-conjugated staining mAb (cold blocking). Stained cells were analyzed on an Epics XL flow cytometer (Coulter). Data were live-gated by size and lack of propidium iodide uptake. In Fig. 1C, 1 × 106 cells were incubated with 0.3 unit of phosphatidylinositol-specific phospholipase C (PIPLC; Boehringer Mannheim) in 1% BSA-supplemented RPMI 1640 medium at 37°C, in 5% CO2 for 1 hr, washed, stained, and then analyzed in a way similar to that described above.
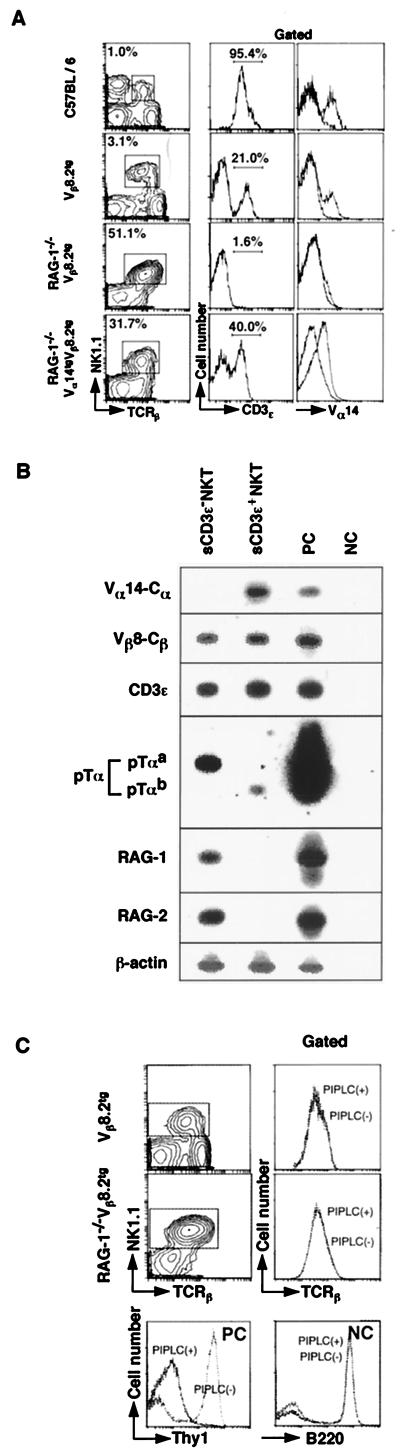
Figure 1.
Characterization of sCD3ɛ− NKT cells by flow cytometric analysis and RT-PCR. (A) Spleen cells from C57BL/6, Vβ8.2tg, RAG-1−/− Vβ8.2tg, and RAG-1−/− Vα14tgVβ8.2tg mice were stained with antibodies against TCRβ, NK1.1, and either CD3ɛ or Vα14. The profiles of CD3ɛ and Vα14 expression on the gated NKT cells (TCRβ+NK1.1+ cells) are shown. The percentage of gated cells is shown in for each profile. (B) RT-PCR analysis of sCD3ɛ− NKT and sCD3ɛ+ NKT cells. BM cells from Vβ8.2tg mice were fractionated into CD3ɛ− and CD3ɛ+ NKT cells by a cell sorter, and RT-PCR was carried out. PC, C57BL/6 thymocytes; NC, mock. (C) Effect of PIPLC on surface expression of TCRβ on TCRβ+NK1.1+ cells of Vβ8.2tg and RAG-1−/−Vβ8.2tg mice. Vβ8.2tg and RAG-1−/− Vβ8.2tg spleen cells were treated with or without PIPLC and then stained with anti-TCRβ and NK1.1 mAbs. The gated NKT cells were evaluated for surface TCRβ expression. For positive control (PC), C57BL/6 spleen cells with or without PIPLC were examined for surface expression of Thy-1.2. For negative control (NC), B220 expression was assessed.
Cell Purification.
CD3−NK1.1+ and CD3+NK1.1+ cells were sorted electrically on an Epics Elite cytometer (Coulter). NK1.1+ cells were enriched by positive selection by using MACS system (Miltenyi Biotec, Auburn, CA), and then CD3− and CD3+ cells were sorted on an Epics Elite cytometer. Sorted cells were generally 97–99% pure, as determined by postsort analysis. For reverse transcription–PCR (RT-PCR) analysis, cells routinely were double-sorted and the purity was more than 99.5%. For bone marrow (BM) cell preparation, cells were incubated with biotin-labeled anti-Gr1, anti-TER119, and anti-CD19 and then mixed with streptavidin-conjugated magnetic beads (PerSeptive Biosystems, Framingham, MA) to eliminate myeloid, erythroid, and B cells. Negatively selected cells were stained and sorted as above.
Cytokines.
Simian IL-15 and murine GM-CSF (both from Genzyme) were used at the concentrations of 200 and 20 ng/ml, respectively.
Cell Culture.
The sorted CD3−NK1.1+ cells (2 × 104) mixed with BM cells (2 × 106) from RAG-1−/− or β2m−/− mice were cultured in 24-well plates in 1.0 ml RPMI 1640 medium (GIBCO/BRL) supplemented with 10% FBS in the presence of indicated cytokines at 37°C for 5 days. Where indicated, sorted spleen cells (1 × 106) were cultured in the presence of indicated cytokines without additional feeder cells for 12, 24, and 36 hr in 96-well, round-bottomed plates.
RT-PCR.
PCR amplifications except for pTα, GM-CSFRα, and βc were performed under the conditions as described (3, 5). PCR primers for pTα, GM-CSFRα, and βc were 5′-CTGGTGGTTTGCCTGGTCCTC-3′ and 5′-GTGGGTGGGAGGCAGGGACTT-3′ for pTα, 5′-CTCGACCTGGGCATCCTT-3′ and 5′-CACGTCGTCGGACACCTTGT-3′ for GMCSFRα, and 5′-ATGCTCTCCGGTGGTGAA-3′ and 5′-TGGCGCAGTATGAGGTGTCT-3′ for βc. DNA was amplified for 35 cycles at annealing temperatures at 60°C for pTα, 68°C for GM-CSFRα, and 65°C for βc. The radioactivities of DNA blots of PCR products were measured by a BioImage Analyzer (Fuji) (3, 5). The probes for the detection were 5′-GCAGAAGCAGTTTGAAGAGGA-3′ for pTα, 5′-GCGGGCGACACGAGGATGAAG-3′ for GM-CSFRα, and 5′-CCGGGGGCCAGTGTCTACACC-3′ for βc.
Genomic PCR.
Detection of coding and signal sequences of Vα14 as well as RAG-2 genes was performed as described (20). Nuclear DNA was amplified by nested PCR for circular DNA detection. PCR was carried out for 30 cycles and then followed by 35 cycles of amplification by using the second set of primers. The primer sequences and the detection probes have been published (20).
Competitive PCR.
The competitor DNAs were constructed by PCR by using a competitor DNA construction kit following the manufacturer’s instructions (Takara Shuzo, Kyoto). After the amounts of input DNA were equalized (600 ng per lane for freshly isolated cells, comparable to about 105 cells, 300 ng per lane for cultured cells for circular Vα14-Jα281, and 300 ng per lane for genomic Vα14-Jα281 detection), the mixture of the DNA and defined copy numbers of competitors was subjected to nested PCR amplifications under the same conditions as above. The ratio of a longer competitive product to a shorter target DNA product was used to estimate the amounts of target DNAs (363 bp vs. 310 bp for genomic Vα14-Jα281, and 348 bp vs. 288 bp for circular Vα14-Jα281).
RESULTS
Phenotypic Characterization of TCRβ+ NKT Cells in the TCR Vβ8.2tg Spleen Cells.
The goal of this study is to determine the molecular requirement for the differentiation of a novel subset of lymphocytes, Vα14 NKT cells. C57BL/6 splenocytes contained 1–2% of mature NKT cells that coexpressed NK1.1 and intermediate levels of TCRβ and CD3ɛ on the cell surface (Fig. 1A, first row). However, in the spleen cells of mice with transgenic Vβ8.2, which is used preferentially by Vα14 NKT cells, all of NK1.1+ cells expressed TCRβ, and about 80% of them were negative for surface CD3ɛ (sCD3ɛ) expression (Fig. 1A, second row). In RAG-1−/− Vβ8.2tg mice, where endogenous TCRα gene rearrangement does not take place, no significant number of sCD3ɛ+ NKT cells was detected, although there was a dramatic increase in percentages of TCRβ+NK1.1+ cells (3.1 vs. 51.1%) (Fig. 1A, third row), indicating the essential requirement of α-chain gene rearrangements for the generation of sCD3ɛ+ NKT cells. In fact, the introduction of the rearranged invariant Vα14 gene into RAG-1−/− Vβ8.2tg mice resulted in increased numbers of sCD3ɛ+ NKT cells (1.6–40%) and also in the significant expression of Vα14 on NKT cells (Fig. 1A, fourth row). From these results, we have anticipated that a unique subset of NKT cells such as sCD3ɛ− Vα14− NK1.1+ TCRβ+ NKT cells (sCD3ɛ− NKT cells) contains a precursor population of mature sCD3ɛ+ Vα14 NKT cells. The pre-NKT cell population, indeed, is present in wild mice, although the size of the population is relatively small compared with that in Vβ8.2tg mice (approximately 0.05% in C57BL/6 vs. more than 2% in Vβ8.2tg mice as shown in Fig. 1A). It might be a result of the Vβ8.2tg situation, in which precursor cell differentiation is accelerated by the forced expression of the rearranged NKT antigen receptor, Vβ8.2 β-chain. The precise mechanism for surface expression of the TCRβ chains without CD3 complex remains unclear. At least the TCRβ chains on sCD3ɛ− NKT cells seemed not to be anchored via glycosyl-phosphatidylinositol (GPI), because they were highly resistant to the digestion with PIPLC (Fig. 1C). Phenotypic characterization of sCD3ɛ− NKT cells revealed that they express CD44, Thy-1 (40.0%), and IL-2 receptor β (IL-2Rβ) on their surface, but not CD3ɛ, CD4, CD8, c-kit, or CD25 (data not shown). This phenotype is reminiscent of immature c-kit−, CD44+, CD25−, CD3−, and CD4−CD8− double-negative thymocytes, which differentiate to CD3+ TCRαβ+ double-negative T cells after short-term culture (21).
To elucidate further molecular properties of sCD3ɛ− NKT cells, both sCD3ɛ− and sCD3ɛ+ NKT cells were purified from BM cells of Vβ8.2tg mice and were subjected to RT-PCR analysis (Fig. 1B). The rearranged Vα14 TCR transcripts were detected in sCD3ɛ+ NKT cells, but not in sCD3ɛ− NKT cells (Fig. 1B, top blot). Despite their undetectable sCD3ɛ expression, considerable amounts of CD3ɛ transcripts were detected in the sCD3ɛ− NKT cells. As expected, sCD3ɛ− NKT cells expressed pTα transcript as well as that of RAG-1 and RAG-2. Interestingly, NKT cells appeared to express an alternative spliced form of pTα (pTαb), which recently has been identified in “β-only cells” (CD4+, CD8−, TCRβ+, CD3−) in the periphery of TCRα-deficient mice (22). These data confirmed the previous results and suggested further that the sCD3ɛ− NKT cell subpopulation contained pre-NKT-like progenitors that expressed pre-TCR and other machineries for TCRα gene rearrangement.
Differentiation of sCD3ɛ− Pre-NKT Cells into sCD3ɛ+ Mature NKT Cells in Vitro.
Consequently, it was examined whether the sCD3ɛ− NKT cells differentiate into mature sCD3ɛ+ NKT cells in vitro in the presence of various cytokines reported to induce NKT cell growth, including IL-3, IL-7, GM-CSF, IL-12, IL-2, and IL-15. Although data are not shown, singular use of IL-2 or IL-15 induced cell growth, and a combination of IL-15 and GM-CSF generated sCD3ɛ+ NKT cells in the culture. To determine further whether such sCD3ɛ+ mature Vα14 NKT cells were generated de novo from sCD3ɛ− pre-NKT cells, sCD3ɛ− NKT cells were purified from the Vβ8.2tg BM cells (Fig. 2A) and subjected to a short-term culture with BM feeder cells from RAG-1−/− or β2m−/− mice in the presence of IL-15 and GM-CSF. As shown in Fig. 2B, neither IL-15 alone nor the combined use of IL-2 and GM-CSF supported the development of mature sCD3ɛ+ Vα14 NKT cells, whereas significant numbers of sCD3ɛ+ Vα14 NKT cells were generated in the presence of both IL-15 and GM-CSF. Importantly, expression of β2m or its associated class I-like molecules on BM stroma cells was found to be required because the development of mature Vα14 NKT cells was hampered severely when β2m−/− BM cells were used (Fig. 2B, bottom row).
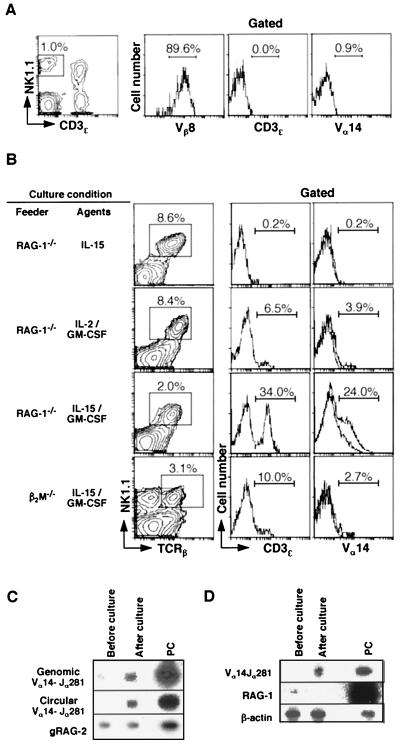
Figure 2.
Differentiation of sCD3− NKT cells into sCD3+ mature Vα14 NKT cells in vitro. (A) Flow cytometric profiles of purified sCD3ɛ− NKT cells. sCD3ɛ− NKT cells purified from the Vβ8.2tg mouse BM were assessed for their expression of Vβ8, CD3ɛ, and Vα14. (B) Generation of sCD3ɛ+ mature Vα14 NKT cells from sCD3ɛ− pre-NKT cells in vitro. Sorted sCD3ɛ− NKT cells (2 × 104) from Vβ8.2tg BM cells were cocultured for 5 days with RAG-1−/− or β2m−/− BM feeder cells (2.0 × 106) in the presence of indicated cytokines. The cells were stained with mAbs against TCRβ, NK1.1, and either CD3ɛ or Vα14 TCR. The percentages of CD3ɛ+ and Vα14+ cells are indicated in each profile. (C) Genomic PCR for the detection of coding (genomic Vα14-Jα281) and signal (circular Vα14-Jα281) joints of Vα14-Jα281 gene rearrangement after culture. Cells harvested before (at the onset) and after culture were examined for gene rearrangement by detection of coding and signal joints generated from Vα14-Jα281 gene recombination. RAG-2 (gRAG-2) was used as input DNA control. (D) RT-PCR analysis of mRNAs for Vα14 TCR and RAG-1 using the same samples as in A. β-Actin was used as a control. PC, C57BL/6 thymocytes.
The differentiation of Vα14 NKT cells was confirmed further at a molecular level. The starting population of sCD3ɛ− pre-NKT cells showed no significant Vα14-Jα281 gene rearrangement at the level of circular DNA and genomic DNA (Fig. 2C, first lane). After 5-day culture with IL-15 and GM-CSF, significant amounts of both circular DNA generated by Vα14-Jα281 gene rearrangement (circular Vα14-Jα281) and rearranged genomic DNA became detectable, suggesting ongoing Vα14-Jα281 gene rearrangement (Fig. 2C, second lane). These results excluded a possibility that mature Vα14 NKT cells detected were consequences of a simple expansion of a minor contaminant of mature NKT cells in the sorted cell samples.
Further RT-PCR analysis revealed that the rearranged invariant Vα14 gene indeed was transcribed (Fig. 2D), indicating that mature Vα14 NKT cells were generated from pre-NKT cells in situ. In addition, the RAG-1 gene expression was down-regulated and disappeared after the culture. Based on both phenotypic and molecular evidence, we concluded that sCD3ɛ− pre-NKT cells contained precursor cells that differentiated in vitro into mature Vα14 NKT cells with BM stroma cells in the presence of both IL-15 and GM-CSF.
Induction of Vα14 Gene Rearrangement in sCD3ɛ− pre-NKT Cells by GM-CSF.
Our present study clearly showed that IL-15 and GM-CSF induced differentiation of sCD3ɛ− pre-NKT cells to mature sCD3ɛ+ Vα14 NKT cells (Fig. 2). To examine the possibility that either IL-15 or GM-CSF directly induced Vα14 gene rearrangement in pre-NKT cells, the purified sCD3ɛ− pre-NKT cells were cultured without feeder cells for a very short period (12, 24, and 36 hr) in the presence of either IL-15 or GM-CSF and analyzed by the detection of the signal sequence generated by Vα14-Jα281 gene rearrangement. Surprisingly, in the presence of GM-CSF alone, but not IL-15, newly generated signal sequences were detected 24 hr after the onset of culture and maintained at least by the 36-hr time point (Fig. 3A Left). No signal sequence was detected in the cultured cells from RAG-1−/− Vβ8.2tg mice (Fig. 3A Right). Subsequently, we assessed the frequency of the generation of circular Vα14-Jα281 in this culture by using competitive PCR. As shown in Fig. 3B, the specificity of the signal sequences in the culture was confirmed by the competitive-PCR analysis; also, the frequency of the signal sequence was estimated to be one of several thousand input cells. This result supports the random Vα14-Jα281 gene assembly, given that all input sCD3ɛ− NKT cells equally have precursor potential. It cannot be ruled out that the NKT precursor population still may be a mixture of heterogeneous cell populations and that only a part of them are true precursors of Vα14 NKT cells. If this is the case, the frequency of Vα14-Jα281 gene rearrangement would be higher than the above estimation.
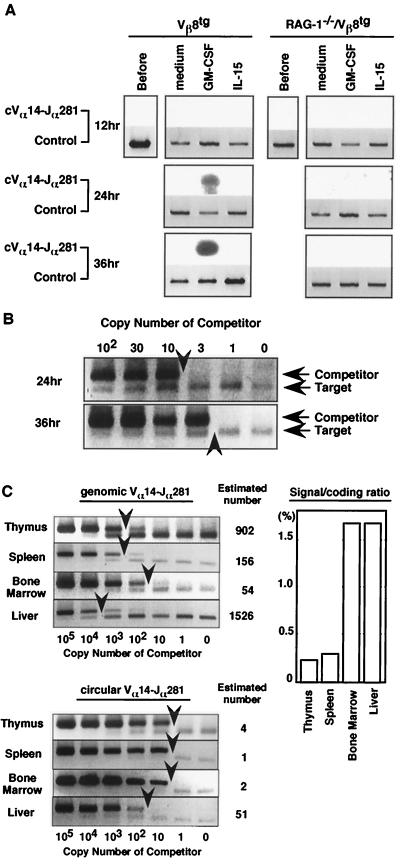
Figure 3.
Induction of Vα14 gene rearrangement events in sCD3ɛ− pre-NKT cells by GM-CSF. (A) Genomic PCR analysis of circular Vα14-Jα281 extrachromosomal DNA. The sCD3ɛ−NKT cells collected were cultured and harvested at time points indicated. Similar results were obtained in four independent experiments. RAG-2 DNA (Control) was used as input DNA control. (B) Competitive-PCR analysis of the amounts of circular Vα14-Jα281 DNA in sCD3ɛ− NKT cells. The copy numbers of circular Vα14-Jα281 DNA at 24- and 36-hr time points after culture were estimated by competitive PCR. The upper band represents amplified competitor DNA, and the lower one represents target DNA. Arrowheads indicate target DNA amounts estimated by comparing densities of competitor and target DNA bands. (C) Competitive-PCR analysis of genomic and circular Vα14-Jα281 DNA in freshly isolated tissues. Arrowheads indicate estimated DNA amounts. Numbers along the right indicate copy numbers of genomic and circular DNA estimated by measuring densities of competitor (upper band) and target DNA bands (lower band). The ratio of signal sequence to coding joint in each organ is shown in the graph (Right).
To evaluate further the rearrangement events detected in the culture, we compared the frequency of Vα14 gene rearrangements in the culture with that in various lymphoid organs in situ. Consequently, genomic DNA was prepared from thymus, BM, spleen, and liver cells and subjected to competitive PCR. As shown in Fig. 3C, the frequency of the signal sequence in various tissues was estimated to be in the range of about 1/105 in spleen to 1/2,000 in liver. Because the frequency of the Vα14-Jα281-mediated signal sequences was around one in several thousand starting cells in the culture (Fig. 3B), it is in the range equivalent to that in normal liver tissue. The frequency of the coding sequence in each organ also was examined by competitive PCR. The ratio of the signal sequence to the coding sequence resulting from Vα14-Jα281 gene rearrangement was the highest in the liver (1.7%) and BM (1.7%) (Fig. 3C Right), suggesting preferential, ongoing gene recombination in these tissues.
Subsequently, we confirmed the expression of GM-CSF receptor in the pre-NKT cell population (Fig. 4A). The sCD3ɛ− NKT cells from Vβ8.2tg mice and TCRβ− NK1.1+ (NK) cells from C57BL/6 mice were purified and analyzed for GM-CSFR transcripts. In sCD3ɛ− NKT cells, both GM-CSFRα and βc mRNAs were easily detectable, whereas NK cells in wild-type C57BL/6 mice expressed barely detectable levels of GM-CSFRα and no significant level of βc mRNAs. These results indicated that sCD3ɛ− pre-NKT cells expressed GM-CSFR mRNA (Fig. 4A). These observations also ensured that sCD3ɛ− pre-NKT cells possessed a precursor potential to differentiate into mature NKT cells by GM-CSFR-mediated Vα14 gene rearrangement.
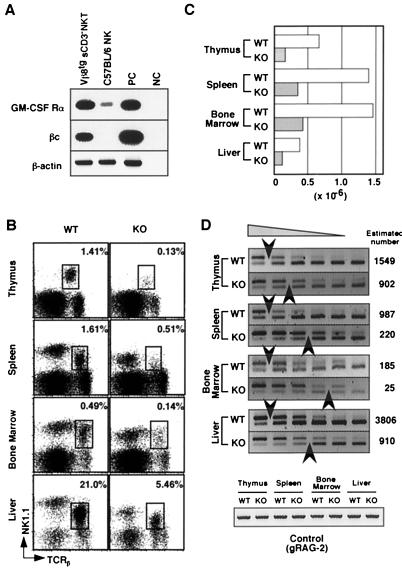
Figure 4.
Expression of GM-CSF receptor in sCD3ɛ− NKT cells and Vα14 NKT cell development in βc-deficient mice. (A) Total RNA was prepared from sorted Vβ8.2tg sCD3ɛ− NKT and NK cells from C57BL/6 spleen, and the expressions of GM-CSFRα and βc were assessed by RT-PCR. β-Actin was used as input cDNA control. (B) TCRβ/NK1.1 profiles of NKT cells in wild-type (WT) and βc-deficient (KO) mice. The percentages of gated NKT cells are shown. Cells pooled from three animals were analyzed for each group. (C) Absolute numbers of NKT cells in each organ were calculated from flow cytometric profiles. (D) Competitive-PCR analysis of Vα14-Jα281 gene rearrangement in WT and KO mice. Genomic DNA (300 ng) were PCR-amplified with primers specific to Vα14 and Jα281 gene segments in the presence of a defined number of competitors serially diluted by 2-fold. The number of competitors in the first lane is 2,500 copies for thymus, 1,250 for spleen, 313 for BM, and 5,000 for liver. Arrowheads indicate estimated target DNA amounts as in Fig. 3B. The upper band represents amplified competitor DNA and the lower one represents the target DNA.
Finally, we addressed the role of GM-CSFR by using βc-deficient mice, in which GM-CSFR-mediated signaling pathway was abrogated because of lack of a signaling-receptor subunit, βc chain (19). In βc-deficient mice, both percentages and absolute numbers of NKT cells in all lymphoid tissues examined were reduced significantly compared with wild-type controls (Fig. 4 B and C), whereas the numbers of other cell types, including T, B, and NK cells, were similar (data not shown). Furthermore, the βc-deficient mice showed 2- to 10-fold decreased numbers of cells with rearranged Vα14-Jα281 TCRα as assessed by competitive PCR (Fig. 4D). These results indicated strongly that GM-CSF was crucial for the development of Vα14 NKT cells in vivo.
DISCUSSION
In this report, we have identified a unique pre-NKT cell population (Vβ8.2+, pTα+, sCD3ɛ−, and Vα14−) and demonstrated that the pre-NKT cells can differentiate into mature NKT cells in vitro (Vβ8.2+, sCD3ɛ+, and Vα14+). The pre-NKT cells are revealed to express transcripts relevant to immature developing T lineage cells, such as RAG-1, RAG-2, pTα, and cytoplasmic CD3ɛ. The expression of pTα in sCD3ɛ− pre-NKT cells is in good agreement with the observation that NKT cells are absent in pTα-deficient mice (16). These results suggest that pre-TCR-dependent selection (β-selection) and subsequent proliferation are required for NKT cell development. Although the physiological significance remains to be clarified, NKT cells appear to express an alternative-spliced form of pTα, “pTbα,” which is different from that of pre-T cells and has been identified recently in “β-only cells” in the periphery of TCR Cα-deficient mice (22). In addition to the characteristic phenotype of pre-NKT cells, GM-CSFR is expressed in sCD3ɛ− pre-NKT cells at a level comparable to myeloid cells in BM (Fig. 4A), although GM-CSFR is known to be expressed on myeloid cells but not on conventional T cells (23). Moreover, newly generated signal sequence and rearranged Vα14-Jα281 genomic DNA are detectable after culture with GM-CSF alone without feeder cells, indicating the de novo generation of mature phenotype of Vα14 NKT cells. Therefore, it is likely that GM-CSF directly acts on sCD3ɛ− pre-NKT cells and may induce TCRα gene rearrangement (Fig. 3A) (although the possibility cannot be completely excluded that minor contaminants in sorted samples, but not sCD3ɛ−NKT cells, are direct targets of GM-CSF, and that Vα14 NKT cell development is facilitated by such GM-CSF-responsive contaminants).
One intriguing explanation of the effect of GM-CSF is that GM-CSF allows the recombination machinery accessible to TCRα gene loci. In some pre-B cell lines, certain humoral factors, such as bacterial lipopolysaccharide and interferon γ, are demonstrated to induce germ-line transcription and rearrangement of Igκ locus by converting the inactive Igκ locus to an accessible recombinase substrate (24, 25). Similarly, germ-line transcripts of Jγ-Cγ appear to be induced by IL-3, implying that IL-3 allowed the TCRγ loci to open (26). Furthermore, other cytokines, such as IL-4, erythropoietin, and GM-CSF, are reported to induce transcription from germ-line TCRγ loci (26). Based on the above findings, GM-CSF might make TCRα gene loci accessible for preexisting recombination enzymes and result in Vα14-Jα281 gene assembly in sCD3ɛ− pre-NKT cells.
The βc-deficient mice show significantly reduced numbers of Vα14 NKT cells (Fig. 4 B and C). These results are in good agreement with in vitro findings that GM-CSF is crucial for Vα14 NKT cell development and TCRα gene rearrangements (Figs. 2 and 3 A and B). This is also confirmed by the molecular analysis that the frequency of Vα14-Jα281 gene rearrangement is reduced significantly. It is interesting, however, that the gene rearrangements are not blocked completely (Fig. 4D), suggesting that the Vα14-Jα281 rearrangements are compensated to some extent by other redundant factors. There exists a functional redundancy between GM-CSF and IL-3 (27). Thus, Vα14 NKT cells in βc-mutant mice might be generated by IL-3. In fact, IL-3-mediated signaling can be transduced through an additional IL-3β-subunit, βIL-3, in βc-mutant mice (28). In any event, our results suggest that βc-mediated signals are critical for Vα14 NKT cell development in vivo.
Finally, we propose a model of the differentiation scenario of Vα14NKT cells from the sCD3ɛ− pre-NKT cells as follows. (i) The first step before Vα14 gene rearrangement is the step at which the precursors can be maintained or expanded to some extent solely by IL-15, but not GM-CSF (unpublished data). (ii) The Vα14 gene rearrangement step is the step at which GM-CSF alone, but not IL-15, could facilitate Vα14 gene rearrangement. This observation also is supported by the fact that decreased but significant numbers of NKT cells develop in IL-15 receptor α-deficient mice (29). In this step, stroma cells are dispensable, as shown in Fig. 3A. (iii) The positive selection step is the step at which NKT cells, after successful NKT cell receptor (Vα14Jα281 TCRα) formation, are positively selected by CD1/β2m molecules on stromal cells, similar to positive selection of conventional T cells in the thymus.
Acknowledgments
We thank Drs. Toshitada Takemori and Haruhiko Koseki for advice and helpful discussions. This work was supported, in part, by a Grant-in-Aid for Scientific Research on Priority Areas (06282103) from the Ministry of Education, Culture, and Science of Japan and a research grant from Taisho Pharmaceutical.
ABBREVIATIONS
- NK
natural killer
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- RT-PCR
reverse transcription–PCR
- PIPLC
phosphatidylinositol-specific phospholipase C
- sCD3
surface CD3
- TCR
T cell receptor
- tg
transgenic
- BM
bone marrow
- βc
common β
- β2m
β2-microglobulin
References
- 1.Lantz O, Bendelac A. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makino Y, Kanno R, Ito T, Higashino K, Taniguchi M. Int Immunol. 1995;7:1157–1161. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 3.Makino Y, Kanno R, Koseki H, Taniguchi M. Proc Natl Acad Sci USA. 1996;25:6516–6520. doi: 10.1073/pnas.93.13.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi M, Koseki H, Tokuhisa T, Masuda K, Sato H, Kondo E, Kawano T, Cui J, Perkes A, Koyasu S, et al. Proc Natl Acad Sci USA. 1996;93:11025–11028. doi: 10.1073/pnas.93.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 7.Burdin N, Brossay L, Koezuka Y, Smiley S T, Grusby M J, Gui M, Taniguchi M, Hayakawa K, Kronenberg M. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 8.Exley M, Garcia J, Balk S P, Porcelli S. J Exp Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bendelac A, Killeen N, Littman D R, Schwartz R H. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 11.Ohteki T, MacDonald H R. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi Y, Koseki H, Zijlstra M, Taniguchi M. Proc Natl Acad Sci USA. 1995;92:1200–1204. doi: 10.1073/pnas.92.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y H, Chiu N M, Mandal M, Wang N, Wang C R. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 14.Mendiratta S K, Martin W D, Hong S, Boesteanu A, Joyce S, Van Kaer L. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 15.Smiley S T, Kaplan M H, Grusby M J. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 16.Di Santo J P, Rodewald H R. Curr Opin Immunol. 1998;10:196–207. doi: 10.1016/s0952-7915(98)80249-5. [DOI] [PubMed] [Google Scholar]
- 17.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 18.Ohteki T, MacDonald H R. J Exp Med. 1996;183:1277–1282. doi: 10.1084/jem.183.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishinakamura R, Nakayama N, Hirabayashi Y, Inoue T, Aud D, McNeil T, Azuma S, Yoshida S, Toyoda Y, Arai K, et al. Immunity. 1995;2:211–222. doi: 10.1016/1074-7613(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Makino Y, Yamagata N, Sasho T, Adachi Y, Kanno R, Koseki H, Kanno M, Taniguchi M. J Exp Med. 1993;177:1399–1408. doi: 10.1084/jem.177.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey D I, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 22.Barber D F, Passoni L, Wen L, Geng L, Hayday A C. J Immunol. 1998;161:11–16. [PubMed] [Google Scholar]
- 23.Park L S, Friend D, Gillis S, Urdal D L. J Biol Chem. 1986;261:4177–4183. [PubMed] [Google Scholar]
- 24.Scherer D C, Brockman J A, Bendall H H, Zhang G M, Ballard D W, Oltz E M. Immunity. 1996;5:563–574. doi: 10.1016/s1074-7613(00)80271-x. [DOI] [PubMed] [Google Scholar]
- 25.O’Brien D P, Oltz E M, Van Ness B G. Mol Cell Biol. 1997;17:3477–3487. doi: 10.1128/mcb.17.7.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstein Y, Morishita K, Cleveland J L, Ihle J N. J Exp Med. 1989;169:2059–2071. doi: 10.1084/jem.169.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishinakamura R, Miyajima A, Mee P J, Tybulewicz V L, Murray R. Blood. 1996;88:2458–2464. [PubMed] [Google Scholar]
- 28.Gorman D M, Itoh N, Kitamura T, Schreurs J, Yonehara S, Yahara I, Arai K, Miyajima A. Proc Natl Acad Sci USA. 1990;87:5459–5463. doi: 10.1073/pnas.87.14.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodolce J P, Boone D L, Chai S, Swain R E, Dassopoulos T, Trettin S, Ma A. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]