Abstract
We hypothesized that the lipid-activated transcription factor, the peroxisome proliferator-activated receptor α (PPARα), plays a pivotal role in the cellular metabolic response to fasting. Short-term starvation caused hepatic steatosis, myocardial lipid accumulation, and hypoglycemia, with an inadequate ketogenic response in adult mice lacking PPARα (PPARα−/−), a phenotype that bears remarkable similarity to that of humans with genetic defects in mitochondrial fatty acid oxidation enzymes. In PPARα+/+ mice, fasting induced the hepatic and cardiac expression of PPARα target genes encoding key mitochondrial (medium-chain acyl-CoA dehydrogenase, carnitine palmitoyltransferase I) and extramitochondrial (acyl-CoA oxidase, cytochrome P450 4A3) enzymes. In striking contrast, the hepatic and cardiac expression of most PPARα target genes was not induced by fasting in PPARα−/− mice. These results define a critical role for PPARα in a transcriptional regulatory response to fasting and identify the PPARα−/− mouse as a potentially useful murine model of inborn and acquired abnormalities of human fatty acid utilization.
Keywords: nuclear hormone receptors, lipid metabolism, transcriptional control, dietary regulation
Starvation triggers a complex array of adaptive metabolic responses. A prominent feature of the energy-metabolic response to fasting includes a switch to reliance on fatty acids and ketones for energy production (1–4) and an augmentation in the capacity for mitochondrial fatty acid oxidation (FAO) in tissues with high oxidative energy demands such as heart and liver (5). The importance of the fasting-inducible capacity for cellular lipid utilization is underscored by the dramatic phenotype of human inborn errors in mitochondrial FAO enzymes (6). Children afflicted with genetically determined enzymatic defects in the FAO pathway typically are asymptomatic under normal feeding conditions. However, short-term fasting, such as that associated with an infectious illness, precipitates a dramatic and often fatal clinical picture characterized by hypoketotic hypoglycemia, liver dysfunction, and cardiomyopathy (6–8). Postmortem studies of FAO enzyme-deficient patients have demonstrated marked intracellular accumulation of neutral lipid in liver and heart. The capacity to oxidize fats is also diminished in several common acquired cardiac diseases including cardiac hypertrophy and myocardial ischemia (9–17). The molecular pathogenesis of target organ dysfunction from inherited and acquired alterations in cellular FAO has not been elucidated.
A previous study in rodents demonstrated that the hepatic expression of genes encoding mitochondrial FAO enzymes is induced, at the transcriptional level, in response to fasting (5). This transcriptional regulatory response likely plays a key role in the fasting-induced augmentation of FAO capacity in liver and other oxidative tissues. The mechanisms involved in the fasting-induced transcriptional activation of FAO enzyme genes are unknown. However, recent studies have identified a role for a nuclear receptor, the peroxisome proliferator-activated receptor α (PPARα), in the transcriptional control of genes encoding mitochondrial FAO enzymes in heart and liver (16, 18–23). PPARα first was identified based on its control of genes encoding peroxisomal FAO enzymes in response to peroxisome proliferators such as fibric acid derivatives (24). PPARα now is known to regulate the transcription of genes encoding peroxisomal, mitochondrial, and certain cytochrome P450 enzymes (18, 24–26) involved in long-chain FAO. Fatty acids and peroxisome proliferators are among the compounds known to activate PPARα (27–29), which binds its cognate elements as a heterodimeric partner with the retinoid X receptor (30). The results of recent studies have indicated that endogenous PPARα ligands are metabolized by peroxisomal (31) and mitochondrial (18) FAO pathways, providing further evidence that fatty acid intermediates activate PPARα in vivo. Accordingly, PPARα as a lipid-activated nuclear receptor is an excellent candidate for mediating the physiologic and dietary control of genes encoding cellular fatty acid utilization enzymes. We sought to test the hypothesis that PPARα is involved in the metabolic response to fasting. We predicted that, as in humans with defective FAO, PPARα−/− mice would be unable to increase the capacity for cellular fatty acid utilization in the context of short-term starvation.
METHODS
Animal Studies.
The PPARα−/− and PPARα+/+ mice were a generous gift of Frank J. Gonzalez (National Cancer Institute, Bethesda, MD) and have been described (32, 33). The PPARα−/− and PPARα+/+ lines were pure-bred on an sv129 background (32). All experiments were performed with mice ranging in age from 12 to 16 weeks (20–30 g). Male and female littermate or age-matched mice were separated into individual cages at the beginning of each fasting experiment. Fasting was initiated at 9:00 a.m. For the gene expression studies, fasting was initiated at 5:00 p.m. The mice were fasted for 24 h or 48 h as indicated in the text. Control mice (fed ad libitum) and fasted mice were kept in identical light/dark cycles. The control mice were allowed free access to standard lab chow (Diet 5053; Purina). At the time of harvest, animals were killed by CO2 inhalation and tissue from liver and cardiac ventricles were rapidly dissected free, snap-frozen in liquid nitrogen, and stored at −80°C until processed for isolation of RNA or lipid extraction. All animal experiments were conducted in strict accordance with the National Institutes of Health guidelines regarding humane treatment for the care and use of laboratory animals and were reviewed and approved by the Animal Studies Committee of Washington University School of Medicine.
Mouse tail vein blood glucose levels were determined by use of a standard clinical blood glucometer (B-Glucose Analyzer; Hemocue, Angelholm, Sweden). Plasma β-hydroxybutyrate levels were determined by the Washington University School of Medicine Diabetes and Research Training Center RIA Core Laboratory. Serum nonesterified fatty acid (NEFA) levels were determined by the Core laboratory of the General Clinical Research Center at Washington University School of Medicine.
Tissue Histology Studies.
After harvest, liver and heart were sliced quickly and snap-frozen in a cryomold for cutting. The sections were stained with oil red O and counterstained with Giemsa.
RNA-Blotting Studies.
Isolation of total RNA and the protocol for Northern blotting have been described (34). 32P-labeled probes were derived from the following cDNAs: mouse medium-chain acyl-CoA dehydrogenase (MCAD; ref. 16); rat liver-form carnitine palmitoyltransferase I (L-CPT I) generated by reverse transcription–PCR (RT-PCR) amplification using primers 5′-TCCCCACTCAAGATGGCAGAGGCT-3′ (sense) and 5′-CTTCCGTGTGGCTCAGGGGTTTAC-3′ (antisense); rat muscle-form CPT I (M-CPT I) generated by RT-PCR using primers 5′-GCGGAAGCACACCAGGCAGTA-3′ (sense) and 5′-ATGTTTGGAAGCTATAGAGCA-3′ (antisense); rat acyl-CoA oxidase (ACO; ref. 33) and rat cytochrome P450 4A3 (CYP 4A3; ref. 33); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ref. 35).
Statistical Methods.
Statistical comparisons were made by using Student’s t test or ANOVA coupled to a Fisher’s test. A statistically significant difference was defined as P < 0.05.
RESULTS
The Hepatic and Cardiac Lipid Homeostatic Response to Fasting Is Altered in PPARα−/− Mice.
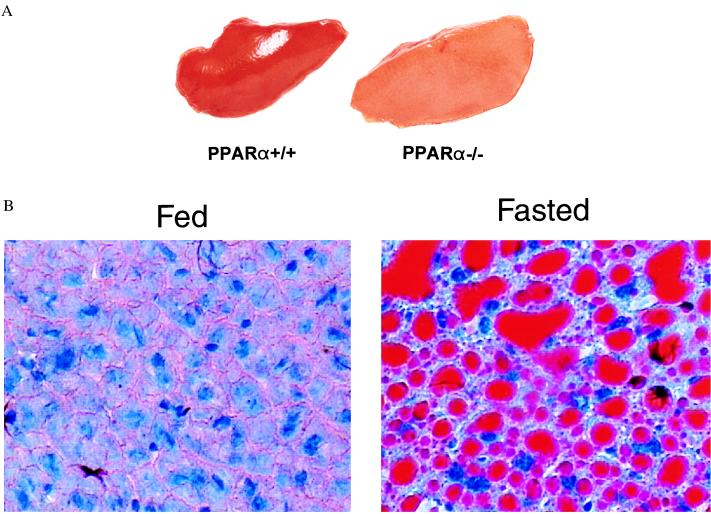
To explore the role of PPARα in the response to fasting, the metabolic phenotype of PPARα−/− mice was investigated after short-term starvation. As described previously, PPARα−/− mice do not exhibit an overt phenotype during the first 6 months of life but have been shown to express reduced levels of FAO enzymes and to lack a peroxisomal, biogenic response to peroxisome proliferators (32, 36). Our studies focused on heart and liver, organs known to increase the capacity for fat oxidation in response to fasting. Age- and strain-matched PPARα−/− and PPARα+/+ animals were subjected to a 48-h fast (see Methods). At the end of the fasting period, hearts and livers were harvested from both groups and characterized. All of the PPARα+/+ animals (n = 29) survived the fast whereas 2/32 PPARα−/− mice died between 24 and 48 h of fasting. The PPARα−/− mice that died were both male. After a 24-h fast, the livers of the PPARα−/− mice that survived the fast were slightly enlarged and appeared significantly paler compared with that of the fasted PPARα+/+ mice (Fig. 1A). Histologic sections of livers from fasted PPARα−/− mice stained with oil red O (to detect neutral lipid) demonstrated marked micro- and macrovesicular lipid intracellular droplet accumulation (Fig. 1B). Examination of liver sections from the fasted PPARα−/− mice revealed that the distribution of lipid droplet accumulation was massive and homogeneous rather than exhibiting a regional pattern (data not shown). TLC analysis of lipid extracted from the livers of the fasted PPARα−/− mice (compared with fed PPARα−/− controls) revealed that the majority of accumulated lipid was in the triglyceride fraction (data not shown). The degree of hepatic lipid accumulation was not significantly different in male compared with female fasted PPARα−/− mice. These results indicate that fasting induces a significant derangement in hepatic lipid balance in the PPARα-null mice.
Figure 1.
Accumulation of intracellular lipid in the liver of fasted PPARα−/− mice. (A) Representative photograph demonstrating the appearance of livers from fasted PPARα+/+ (Left) and PPARα−/− (Right) mice. The livers were harvested rapidly from age-matched PPARα−/− and PPARα+/+ mice after a 24-h fast. The left hepatic lobes are shown. (B) Photomicrographs demonstrating the histologic appearance of the livers of PPARα−/− mice under fed and fasted conditions. Frozen tissue sections were prepared from the livers of PPARα−/− mice after a 24-h fast (FASTED) and age-matched PPARα−/− controls fed ad libitum (FED). The sections were stained with oil red O. The red droplets indicate positive staining for neutral lipid.
The gross appearance and weights of the hearts of fasted PPARα−/− animals was not significantly different than that of fasted PPARα+/+ mice or fed PPARα−/− controls. Histologic analysis of the myocardium of the fasted mice revealed patchy oil red O-positive regions in the PPARα−/− mice but was consistently negative in the PPARα+/+ mice (data not shown), indicating that as in liver, the cellular lipid homeostatic response to fasting is altered in the hearts of PPARα−/− mice.
The levels of circulating NEFA were determined in the PPARα−/− and PPARα+/+ mice. In the fed state, mean serum (±SD) NEFA levels were 938 ± 154 μM in the PPARα+/+ mice and 862 ± 200 μM in the PPARα−/− group (P = not significant). After a 48-h fast, mean NEFA levels were 1,541 ± 343 μM in PPARα−/− mice compared with 838 ± 195 μM in PPARα+/+ mice, a significant difference (P < 0.001). These results suggest that in addition to a reduced capacity for FAO, PPARα−/− mice exhibit abnormally high fasting, circulating free fatty acids, both of which could contribute to the hepatic and cardiac lipid accumulation.
The Glucose Homeostatic Response to Fasting Is Altered in PPARα−/− Mice.
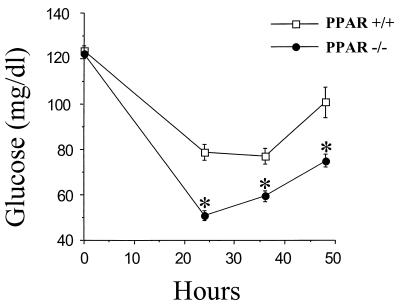
Children with inherited defects in mitochondrial FAO develop severe hypoglycemia during fasting (6, 7). Hypoglycemia occurring in the context of defective mitochondrial FAO likely is due to a combination of glycogen depletion and a blunted gluconeogenic response. To determine whether PPARα−/− mice exhibit a similar abnormality in glucose homeostasis, circulating glucose levels were delineated in age-matched male and female PPARα−/− and PPARα+/+ mice during a 48-h fast. Blood glucose levels in the PPARα+/+ animals exhibited an initial drop during the first 24 h of fasting followed by a rebound increase returning to near normal levels by 48 h. In fasted PPARα−/− mice, the initial fall in mean blood glucose levels was significantly greater than that of the PPARα+/+ mice, dropping from 122 ± 2 mg/dl to 51 ± 2 mg/dl during the first 24 h of fasting (Fig. 2). Compared with the PPARα+/+ group, the blood glucose levels of fasted PPARα−/− mice were significantly lower at all time points measured (24, 36, and 48 h of fasting; Fig. 2). During the second 24 h of fasting, the PPARα−/− mice did exhibit a rebound increase in blood glucose levels. The mean blood glucose levels were not significantly different in male compared with female fasted PPARα+/+ or PPARα−/− mice (data not shown). Thus, the glucose homeostatic response is abnormal in the PPARα−/− mice as manifested by a hypoglycemic response to fasting.
Figure 2.
PPARα−/− mice exhibit a hypoglycemic response to fasting. Mean blood glucose levels (ordinate) of age-matched PPARα−/− and PPARα+/+ mice at various time points during a 48-h fast. Mean values were based on 21 PPARα+/+ mice (6 male; 15 female) and 30 PPARα−/− mice (12 male; 18 female). ∗ indicates a statistically significant difference (P < 0.001; Fisher’s test) compared with the corresponding value obtained in age-matched PPARα+/+ animals.
The Ketogenic Response to Fasting Is Blunted in PPARα−/− Mice.
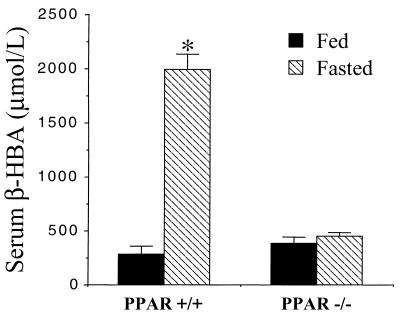
During a period of fasting, substrate for the production of ketone bodies is derived from acetyl-CoA moieties produced mainly by mitochondrial β-oxidation. Accordingly, a characteristic feature of the fasting-induced hypoglycemia associated with defective capacity for mitochondrial FAO is an inadequate ketogenic response (6). Indeed, hypoketotic hypoglycemia is a well described clue to establishing the diagnosis of human genetic defects in FAO (6). To explore the possibility that the reduced capacity for mitochondrial FAO in PPARα−/− mice would result in an inadequate ketogenic response to fasting, plasma β-hydroxybutyrate levels were measured in PPARα−/− and PPARα+/+ mice after a 48-h fast. Mean fasting plasma ketone levels were markedly induced in fasted PPARα+/+ mice (Fig. 3). In striking contrast, fasting did not increase the plasma ketone levels in PPARα−/− mice despite development of hypoglycemia (Fig. 3). These were no significant gender differences in the plasma ketone levels in the control and fasted PPARα+/+ and PPARα−/− mice (data not shown).
Figure 3.
The ketogenic response to fasting is blunted in PPARα−/− mice. Bars = mean ± SE serum β-hydroxybutyrate (β-HBA) levels determined in age-matched PPARα−/− and PPARα+/+ mice after a 48-h fast compared with the fed ad libitum state. ∗ denotes a significant difference (P < 0.05) compared with the value of the fed PPARα+/+ group. Values are based on at least six mice per group and two independent experiments.
The Fasting-Induced Activation of Hepatic and Cardiac PPARα Target Gene Expression Is Markedly Blunted in PPARα−/− Mice.
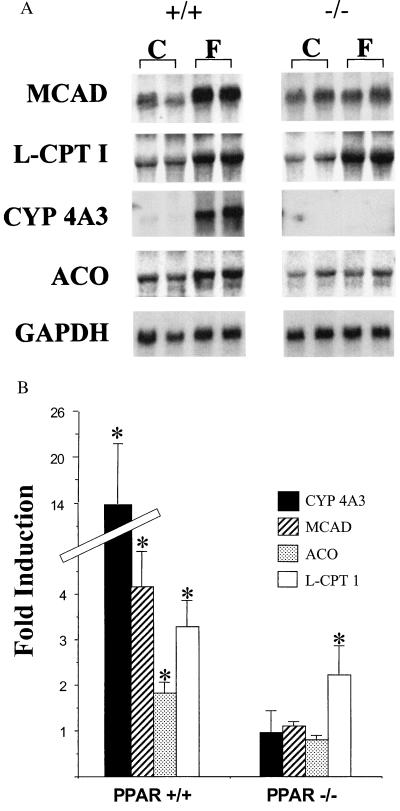
The expression of genes encoding mitochondrial FAO enzymes is increased in the liver of fasted rats (5). Given that in PPARα−/− mice the cellular lipid metabolic response to fasting is altered, we hypothesized that PPARα serves to activate the transcription of hepatic and cardiac FAO enzyme genes in response to fasting. To explore this possibility, the expression of PPARα target genes encoding mitochondrial [MCAD; liver (L-) or muscle (M-), CPT I], peroxisomal (ACO), and cytochrome P450 (CYP 4A3) FAO enzymes was characterized in the livers and hearts of control (fed) and 24-h-fasted PPARα−/− and PPARα+/+ mice. Fasting induced the steady-state levels of mRNA encoding MCAD, L-CPT I, and ACO 2- to 4-fold in liver of the PPARα+/+ mice (Fig. 4). As shown recently by others (37), fasting induced the hepatic expression of the CYP 4A3 gene 15- to 20-fold in PPARα+/+ mice (Fig. 4). In striking contrast, the fasting-mediated induction of MCAD, ACO, and CYP 4A3 gene expression was abolished in PPARα−/− mice. The fasting response of the L-CPT I gene, however, was preserved in PPARα−/− mice.
Figure 4.
The fasting-induced hepatic expression of most PPARα target genes encoding cellular FAO enzymes is abolished in PPARα−/− mice. (A) Representative autoradiographs of Northern blot analyses performed with total RNA (15 μg/lane) isolated from the livers of fed control (C) and 24-h-fasted (F) littermate PPARα+/+ (+/+) and PPARα−/− (−/−) mice. The cDNA probes are denoted on the left (abbreviations defined in the text). The signal for GAPDH is shown as a control for loading and RNA integrity. (B) Bars represent mean fasting fold induction compared with the values of fed littermate controls based on phosphorimager analysis of Northern blots described in A. All values were normalized to the signal for GAPDH. ∗ denotes a significantly (P < 0.05) higher mean signal intensity compared with the corresponding fed control values. The values are based on a minimum of nine animals per group and four independent experiments.
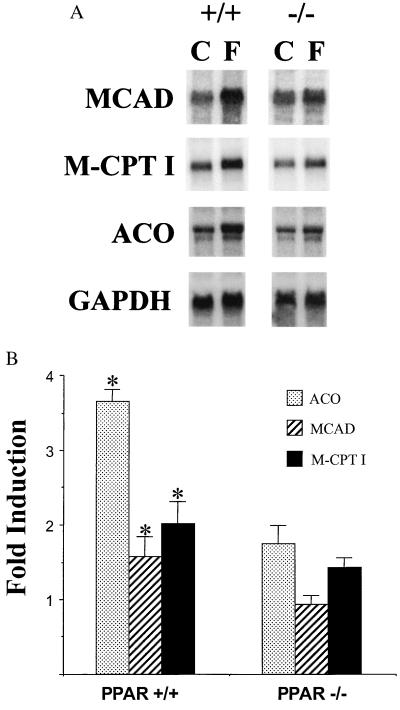
In the hearts of PPARα+/+ mice, fasting caused a modest but significant induction of the expression of PPARα target genes encoding M-CPT I, MCAD, and ACO (1.5- to 3.5-fold; Fig. 5). The fasting response of the PPARα target genes (M-CPT I, MCAD, and ACO) was abolished in the hearts of PPARα−/− mice (Fig. 5). Taken together, these data indicate that PPARα is necessary for the fasting-induced expression of target genes involved in hepatic and cardiac mitochondrial and extramitochondrial fatty acid utilization. Moreover, these results suggest that the observed cellular lipid imbalance in PPARα−/− mice is a result, at least in part, of an inadequate capacity to meet increased demands for cellular FAO in heart and liver.
Figure 5.
The fasting-induced cardiac expression of PPARα target genes is altered in PPARα−/− mice. (A) Representative autoradiographs of Northern blot analyses performed with total RNA (15 μg/lane) isolated from the hearts of fed control (C) and 24-h-fasted (F) littermate PPARα+/+ (+/+) and PPARα−/− (−/−) mice. The cDNA probes are denoted on the left. The signal for GAPDH is shown as a control for loading and RNA integrity. (B) Bars represent mean fasting fold induction compared with fed littermate controls after normalization to the GAPDH signal based on phosphorimager analysis of Northern blots described in A. Asterisks represent a significantly (P < 0.05) higher mean signal intensity compared with corresponding fed control values. The values are based on a minimum of six animals per group and three independent experiments.
DISCUSSION
Our results identify a critical role for PPARα in the gene transcriptional regulatory response to short-term starvation. PPARα is shown to be required for the fasting-activated expression of target genes involved in mitochondrial and extramitochondrial FAO in liver and heart, tissues known to increase FAO rates during short-term starvation. Our findings are consistent with a recent report by Kroetz et al. (37), demonstrating that PPARα mediates the induced hepatic expression of CYP 4A genes in response to starvation and in the diabetic state. Accordingly, PPARα is one of only a few transcription factors known to be involved in the cellular response to fasting. Recently, the transcription factor ADD1/SREBP was shown to confer fasting-mediated transcriptional down-regulation of the gene encoding fatty acid synthase (FAS), an enzyme involved in lipid synthesis (38). Taken together, these results indicate that distinct transcriptional regulatory pathways are involved in two key metabolic responses to fasting: induction of cellular fatty acid utilization and reduction of lipid synthesis.
The dramatic fasting phenotype of the PPARα−/− mice reflects a defective cellular lipid homeostatic response. The fasting-induced accumulation of intracellular lipid demonstrated here is consistent with the known reduction in the hepatic and cardiac expression of PPARα target genes encoding FAO enzymes (36). We propose that a reduced capacity for fatty acid utilization coupled with a loss of the fasting-induced expression of several key FAO enzyme genes results in a mismatch between cellular lipid import and utilization, leading to cellular lipid accumulation in the PPARα−/− mice. In support of this proposed mechanism, we demonstrated recently that in the context of a perturbation of mitochondrial long-chain fatty acid import caused by pharmacologic inhibition of CPT I, PPARα−/− mice develop marked hepatic and cardiac lipid accumulation (33). In this report, we also found abnormally high fasting, circulating NEFA levels in PPARα−/− mice. Our data do not allow us to determine whether elevated-fasting serum NEFA levels in PPARα−/− mice contribute to the observed hepatic and cardiac lipid accumulation.
The phenotype of the fasted PPARα−/− mice unveils an important role for PPARα in glucose and energy homeostasis. In contrast to extramitochondrial FAO pathways, mitochondrial β-oxidation is an important source of ATP and acetyl-CoA, particularly in the fasting state. Acetyl-CoA derived from the mitochondrial FAO cycle is a precursor for ketogenesis. In PPARα−/− mice, in which the fasting-mediated induction of mitochondrial FAO enzyme expression is altered, the capacity for ATP and ketone production is limited. As predicted, the PPARα−/− mice exhibit a severely blunted ketogenic response to fasting despite the development of hypoglycemia. Interestingly, hypoketotic hypoglycemia is a diagnostic hallmark of inherited defects in mitochondrial FAO enzymes in humans (6). The hypoglycemic response of the fasted PPARα−/− mice likely reflects both a depletion of hepatic glycogen and a reduced capacity for gluconeogenesis because of a decrease in the intracellular levels of acetyl-CoA, a potent activator of the gluconeogenic pathway (39, 40).
Inborn errors in mitochondrial β-oxidation enzymes are an important cause of inherited episodic hypoglycemia, hepatic dysfunction, cardiomyopathy, and skeletal myopathy during childhood (6–8). The clinical manifestations of genetic defects in FAO enzymes are “stress”-induced; affected children are usually asymptomatic until faced with a dietary or physiologic condition that dictates an increased reliance on the oxidation of fats for energy. For example, during a period of fasting, affected children often develop a clinical episode or “crisis” characterized by the precipitous onset of symptoms related to multiorgan toxicity. The pathogenesis and pathophysiology of end-organ abnormalities that occur in children with inborn errors in FAO pathway enzymes are poorly understood but likely involve energy starvation and the effects of potentially toxic lipid intermediates. PPARα−/− mice exhibit fasting-induced lipid accumulation in liver and heart in association with hypoketotic hypoglycemia, a phenotype that bears a striking resemblance to that of humans with genetic defects in mitochondrial FAO enzymes. Recently, mice null for the mitochondrial FAO enzyme, long-chain acyl-CoA dehydrogenase (LCAD), also were shown to have a metabolic phenotype similar to that of humans with inherited defects in FAO (41). PPARα- and LCAD-deficient mice should be useful for studies aimed at characterizing the pathogenesis of this important group of inherited childhood disorders as well as several common acquired human cardiovascular diseases known to be associated with reduced capacity for mitochondrial FAO such as pressure overload-induced cardiac hypertrophy and myocardial ischemia (9–14).
We recently have demonstrated that PPARα−/− mice exhibit a gender-influenced response to pharmacologic inhibition of CPT I (33). The metabolic abnormalities of the etomoxir-treated PPARα−/− mice were shown to be more severe in males compared with females. In addition, gender-related serum and hepatic lipid abnormalities recently were reported in aged PPARα−/− mice (42). In contrast, the fasting-induced phenotype of the PPARα−/− mice described here was not influenced by gender. It is possible that CPT I inhibition induces a more severe perturbation in hepatic and cardiac metabolism compared with that of fasting, such that the gender effect was manifest only in the context of the former. Alternatively, CPT I inhibition may induce a distinct gender-influenced metabolic pathway that is not activated by fasting.
In summary, our results identify PPARα as a key factor in the cellular metabolic response to fasting. We propose that in response to fasting and other physiologic conditions known to cause a mismatch between cellular lipid uptake and utilization, PPARα is activated by an endogenous lipid ligand to induce the orchestrated expression of genes involved in FAO and, thus, increase the capacity for cellular fatty acid utilization—a key metabolic response that serves to maintain cellular lipid balance in tissues such as liver that increase fatty acid uptake and utilization in the fasting state. Our results also indicate that PPARα−/− mice should be useful in the investigation of the pathogenesis and treatment of inherited and acquired human FAO disorders.
Acknowledgments
We thank Kelly Hall for assistance with preparation of the manuscript. This work was supported by National Institutes of Health Grants DK45416 and HL58493 and American Heart Association Grants EIA 95001150 and GIA 9750199N.
ABBREVIATIONS
- PPAR
peroxisome proliferator-activated receptor
- FAO
fatty acid oxidation
- NEFA
nonesterified free fatty acids
- MCAD
medium-chain acyl-CoA dehydrogenase
- CPT I
carnitine palmitoyltransferase I
- ACO
acyl-CoA oxidase
- CYP
cytochrome P450
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Seitz H J, Müller M J, Krone W, Tarnowski W. Arch Biochem Biophys. 1977;183:647–663. doi: 10.1016/0003-9861(77)90399-x. [DOI] [PubMed] [Google Scholar]
- 2.Christiansen R Z. Biochim Biophys Acta. 1977;488:249–262. doi: 10.1016/0005-2760(77)90182-5. [DOI] [PubMed] [Google Scholar]
- 3.Parvin R, Pande S V. J Biol Chem. 1979;254:5423–5429. [PubMed] [Google Scholar]
- 4.Bergseth S, Lund H, Poisson J-P, Bremer J, Davis-Van Thienen W, Davis E J. Biochim Biophys Acta. 1986;876:551–558. doi: 10.1016/0005-2760(86)90043-3. [DOI] [PubMed] [Google Scholar]
- 5.Nagao M, Parimoo B, Tanaka K. J Biol Chem. 1993;268:24114–24124. [PubMed] [Google Scholar]
- 6.Roe C R, Coates P M. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A I, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1501–1533. [Google Scholar]
- 7.Kelly D P, Hale D E, Rutledge S L, Ogden M L, Whelan A J, Zhang Z, Strauss A W. J Inherit Metab Dis. 1992;15:171–180. doi: 10.1007/BF01799626. [DOI] [PubMed] [Google Scholar]
- 8.Kelly D P, Strauss A W. N Engl J Med. 1994;330:913–919. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- 9.Wittels B, Spann J F., Jr J Clin Invest. 1968;47:1787–1794. doi: 10.1172/JCI105868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop S P, Altschuld R A. Am J Physiol. 1970;218:153–159. doi: 10.1152/ajplegacy.1970.218.1.153. [DOI] [PubMed] [Google Scholar]
- 11.Taegtmeyer H, Overturf M L. Hypertension. 1988;11:416–426. doi: 10.1161/01.hyp.11.5.416. [DOI] [PubMed] [Google Scholar]
- 12.Christe M D, Rodgers R L. J Mol Cell Cardiol. 1994;26:1371–1375. doi: 10.1006/jmcc.1994.1155. [DOI] [PubMed] [Google Scholar]
- 13.Takeyama D, Kagaya Y, Yamane Y, Shiba N, Chida M, Takahashi T, Ido T, Ishide N, Takishima T. Cardiovasc Res. 1995;29:763–767. [PubMed] [Google Scholar]
- 14.Massie B M, Schaefer S, Garcia J, McKirnan M D, Schwartz G G, Wisneski J A, Weiner M W, White F C. Circulation. 1995;91:1814–1823. doi: 10.1161/01.cir.91.6.1814. [DOI] [PubMed] [Google Scholar]
- 15.Sack M N, Rader T A, Park S, Bastin J, McCune S A, Kelly D P. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 16.Sack M N, Disch D L, Rockman H A, Kelly D P. Proc Natl Acad Sci USA. 1997;94:6438–6443. doi: 10.1073/pnas.94.12.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sack M N, Kelly D P. Int J Mol Med. 1998;1:17–24. doi: 10.3892/ijmm.1.1.17. [DOI] [PubMed] [Google Scholar]
- 18.Gulick T, Cresci S, Caira T, Moore D D, Kelly D P. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Disch D L, Rader T A, Cresci S, Leone T C, Barger P M, Vega R, Wood P A, Kelly D P. Mol Cell Biol. 1996;16:4043–4051. doi: 10.1128/mcb.16.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandt J, Djouadi F, Kelly D P. J Biol Chem. 1998;273:23786–23793. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 21.Djouadi, F., Brandt, J., Weinheimer, C. J., Leone, T. C., Gonzalez, F. J. & Kelly, D. P. (1999) Prostaglandins Leukotrienes Essent. Fatty Acids, in press. [DOI] [PubMed]
- 22.Mascaró C, Acosta E, Ortiz J A, Marrero P F, Hegardt F G, Haro D. J Biol Chem. 1998;273:8560–8563. doi: 10.1074/jbc.273.15.8560. [DOI] [PubMed] [Google Scholar]
- 23.Yu G-S, Lu Y-C, Gulick T. J Biol Chem. 1998;273:32901–32909. doi: 10.1074/jbc.273.49.32901. [DOI] [PubMed] [Google Scholar]
- 24.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 25.Muerhoff A S, Griffin K J, Johnson E F. J Biol Chem. 1992;267:19051–19053. [PubMed] [Google Scholar]
- 26.Aldridge T C, Tugwood J D, Green S. Biochem J. 1995;306:473–479. doi: 10.1042/bj3060473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Göttlicher M, Widmark E, Li Q, Gustafsson J-A. Proc Natl Acad Sci USA. 1992;89:4653–4657. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Proc Natl Acad Sci USA. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Nature (London) 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan C-Y, Pan J, Usuda N, Yeldandi A V, Rao M S, Reddy J K. J Biol Chem. 1998;273:15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 32.Lee S S T, Pineau T, Drago J, Lee E J, Owens J W, Kroetz D L, Fernandez-Salguero P M, Westphal H, Gonzalez F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Djouadi F, Weinheimer C, Saffitz J E, Pitchford C, Bastin J, Gonzalez F J, Kelly D P. J Clin Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly D P, Gordon J I, Alpers R, Strauss A W. J Biol Chem. 1989;264:18921–18925. [PubMed] [Google Scholar]
- 35.Levy F H, Kelly D P. Am J Physiol. 1997;272:C457–C465. doi: 10.1152/ajpcell.1997.272.2.C457. [DOI] [PubMed] [Google Scholar]
- 36.Aoyama T, Peters J M, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez F J. J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 37.Kroetz D L, Yook P, Costet P, Bianchi P, Pineau T. J Biol Chem. 1998;273:31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 38.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerich J, Haymond M, Rizza R, Verdonk C, Miles J. In: The Regulation of Carbohydrate Formation and Utilization in Mammals. Veneziale C, editor. Baltimore: Univ. Park Press; 1981. pp. 419–457. [Google Scholar]
- 40.Fanelli C, Calderone S, Epifano L, De Vincenzo F, Modarelli F, Pampanelli S, Perriello G, De Feo P, Brunetti P, Gerich J E, et al. J Clin Invest. 1993;92:1617–1622. doi: 10.1172/JCI116746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurtz D M, Rinaldo P, Rhead W J, Tian L Q, Millington D S, Vockley J, Hamm D A, Brix A E, Lindsey J R, Pinkert C A, et al. Proc Natl Acad Sci USA. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costet P, Legendre C, Moré J, Edgar A, Galtier P, Pineau T. J Biol Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]