Abstract
A somatic mutation in the X linked PIGA gene is responsible for the deficiency of glycosyl phosphatidylinositol (GPI)-anchored proteins on blood cells from patients with paroxysmal nocturnal hemoglobinuria. No inherited form of GPI-anchor deficiency has been described. Because conventional Piga gene knockout is associated with high embryonic lethality in chimeric mice, we used the Cre/loxP system. We generated mice in which two loxP sites flank part of Piga exon 2. After crossbreeding with female mice of the EIIa-cre strain, the floxed allele undergoes Cre-mediated recombination with high efficiency during early embryonic development. Because of X chromosome inactivation, female offspring are mosaic for cells that express or lack GPI-linked proteins. Analysis of mosaic mice showed that in heart, lung, kidney, brain, and liver, mainly wild-type Piga is active, suggesting that these tissues require GPI-linked proteins. The salient exceptions were spleen, thymus, and red blood cells, which had almost equal numbers of cells expressing the wild-type or the recombined allele, implying that GPI-linked proteins are not essential for the derivation of these tissues. PIGA(−) cells had no growth advantage, suggesting that other factors are needed for their clonal dominance in patients with paroxysmal nocturnal hemoglobinuria.
Keywords: Cre, loxP, glycosyl phosphatidylinositol, Xp22, paroxysmal nocturnal hemoglobinuria
The gene PIGA (phosphatidylinositol glycan class A) encodes a subunit of the α1–6-N-acetylglucosaminyltransferase complex, an enzyme essential for the biosynthesis of glycosyl phosphatidylinositol (GPI) anchors (1, 2). In paroxysmal nocturnal hemoglobinuria (PNH), an acquired hemolytic anemia, a somatic mutation in the PIGA gene causes a proportion of blood cells to be deficient in all GPI-linked molecules (3, 4). PIGA maps to the X chromosome (4, 5). Its location on the X chromosome explains why a single mutation is sufficient to abrogate the expression of GPI-linked proteins. Because of X chromosome inactivation in female cells, both male and female cells contain one single active Piga gene. In PNH, the mutation is thought to occur in a hematopoietic stem cell, because GPI-deficient cells are found in all blood cell lineages (for review, see ref. 6). Although the mutations account for the deficiency of GPI-linked proteins on the affected blood cells, it is not clear that mutations of the PIGA gene cause the clonal expansion that enables GPI-anchor-deficient blood cells to become the dominant blood cell population in patients with PNH.
GPI-linked proteins are found in almost every tissue and serve many different functions (7). An inherited form of complete GPI-anchor deficiency has not been reported and is probably not compatible with life. Conforming to the evolutionary conservation of synteny of the mammalian X chromosome (“Ohno’s law;” ref. 8), the murine Piga maps to X-F3/4. Piga gene inactivation in murine embryonic stem (ES) cells followed by blastocyst injection is associated with a high rate of early embryonic lethality and low chimerism in surviving animals (9, 10). Female mice heterozygous for a mutant Piga gene have never been obtained. To study the consequences of a nonfunctional Piga gene and to address the issue of a maternally inherited Piga mutation, we generated mice carrying a Piga mutation using Cre/loxP-controlled DNA recombination (11). High efficiency of Piga gene recombination was obtained by targeting Piga gene inactivation directly to the preimplantation female embryo. Because of X inactivation, newborn female mice are mosaic, with cells that express or lack GPI-linked proteins. To assess the importance of PIGA in different organs, we determined the relative contribution of cells expressing or lacking GPI-linked proteins. Female mice that had high efficiency of Piga gene recombination enabled us to further investigate the possibility of an inherited Piga gene mutation.
MATERIALS AND METHODS
Production of the lox-Piga-lacZ Mice.
The production of the lox-Piga-lacZ mice has been described in detail previously (12). In brief: two loxP sites were introduced by homologous recombination into the Piga locus of 129SV-derived ES cells CJ7, flanking 662 bp of exon 2 and 1.6 kb of the adjoining intron 2. In addition, the coding region of the lacZ gene was inserted into intron 2 in such a way that in the lox-Piga-lacZ configuration (floxed Piga gene), LacZ is not expressed and does not interfere the function of PIGA. However, after Cre-mediated Piga recombination (lox-ΔPiga-lacZ), lacZ will find itself in-frame 3′ of the translation start of the Piga gene and therefore will be driven by the endogenous Piga promoter. The structure of wild-type (wt) Piga, lox-Piga-lacZ, and Lox-ΔPiga-lacZ are shown in Fig. 1A. ES cells carrying the lox-Piga-lacZ gene were injected into C57BL/6 blastocysts to generate chimeric mice. Chimeric males were crossbred with C57BL/6 female mice. Lox-Piga-lacZ showed the expected X linked transmission pattern. Hemizygous lox-Piga-lacZ males were obtained in the N2 generation.
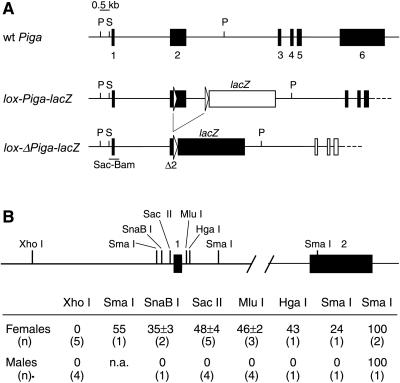
Figure 1.
(A) Genomic structure of wt Piga, lox-Piga-lacZ, and lox-Piga-lacZ after recombination (lox-ΔPiga-lacZ). Two loxP sites (open arrows) and the coding region for lacZ (lacZ-box) were introduced into the Piga locus by homologous recombination in murine ES cells (12). In the lox-Piga-lacZ configuration, PIGA function is not impaired (filled boxes), and LacZ is not expressed (open lacZ box). However, after Cre-mediated excision of the DNA sequences between the two lox sites Piga becomes inactivate (open Piga boxes) and LacZ falls under the endogenous Piga promoter and is expressed (filled lacZ box). Restriction sites used in the methylation assay are shown. P, PstI; S, SacII. Genomic DNA was digested with PstI (P) to obtain restriction fragments suitable for Southern blotting. Each PstI fragment has a different size depending on whether it is derived from the wt (7.0 kb), the lox-Piga-lacZ (10.4 kb), or the lox-ΔPiga-lacZ (8.1 kb) gene. To determine the extent of methylation DNA was subsequently digested with the methylation-sensitive restriction endonuclease SacII (S). If the 5′ SacII site is unmethylated, each fragment is shortened by 0.5 kb. The Sac-Bam DNA probe used for hybridization is shown. (B) The diagram shows the promoter region and intron 1 of the Piga gene with the endonuclease cleavage sites that are subject to methylation depending on the inactivation status of the gene. Lower shows the proportions of DNA that is methylated in wt mice. Values were defined by the use of a phosphoimager (see Materials and Methods) and are shown as the mean and SD of all measured samples. n.a., not analyzed.
FVB/NJ mice homozygous for the EIIa-cre transgene (EIIa-cre(+/+) mice) were generously provided by Heiner Westphal (National Institutes of Health, Bethesda, MD; ref. 13). Lox-Piga-lacZ mice were crossbred with EIIa-cre(+/+) mice as indicated. The lox-Piga-lacZ/EIIa-cre offspring therefore had a mixed genetic background.
DNA Preparation and Amplification.
DNA was isolated by the SDS/proteinase K procedure (14). Genotyping was performed by using PCR analysis. wt Piga and lox-Piga-lacZ were amplified with the forward primer “−13 + 8” (5′-GGACCACCTCAGCATGGCCAA-3′) and the reverse primer “Piga 600 rev” (5′-TATTTCAGGATTCAGTGCTGC-3′). The loss of the SmaI restriction site in exon 2 was used to assess the presence of the 5′ loxP site. Lox-ΔPiga-lacZ was amplified with the forward primer “−13 + 8” and the reverse primer “lacZ 500 rev” (5′-CGACAGTATCGGCCTCAGGAAGA-3′), yielding a 382-bp fragment. The EIIa-cre transgene was amplified by using the forward primer “cre 5′” (5′-CCAATTTACTGACCGTACACC-3′) and the reverse primer “cre 3′” (5′-TTACGTATATCCTGGCAGCG-3′), yielding a 476-bp fragment.
Southern Blot Analysis.
Genomic DNA (30 μg) was digested with restriction endonucleases. To obtain suitable restriction fragments, double or triple digests were performed. Digests of the methylation-sensitive enzymes SacII, MluI, or SmaI were performed in conjunction with PstI. To measure the extent of methylation at the SnaBI and HgaI site, DNA was first digested with PstI and EcoRI. The XhoI site was examined by using a XhoI monodigest. Digested DNA was resolved on 0.7% agarose gels at 38 V for 60–72 hours and transferred to Hybond-N nylon membranes (Amersham Pharmacia) by using Southern blotting. The filters were hybridized to a SacII–BamHI DNA probe including exon 1 (Sac-Bam). The band intensity ratios were defined with a phosphoimager (Molecular Imager System GS-525, Bio-Rad) by using molecular analyst version 2.1.2 software (Bio-Rad).
Analysis of GPI-Anchored Membrane Proteins.
GPI-linked surface proteins on erythrocyte membranes were measured by using flow cytometry (FACscan, Becton Dickinson) with FITC-conjugated mAbs against CD24 (M1/69, PharMingen) (10).
RESULTS
Production of Mice with High Level of Piga Gene Recombination.
To obtain high efficiency of Cre-mediated Piga gene recombination, we took advantage of the expression pattern of the EIIa promoter. Expression from the EIIa promoter in the absence of its natural transcriptional activator, the E1A gene product, is restricted to oocytes and preimplantation embryos (15). Therefore, we predicted that Cre-mediated Piga gene recombination in the early embryo will be most efficient if EIIa-cre is maternally derived. Accordingly, we crossbred hemizygous lox-Piga-lacZ(+) males with homozygous EIIa-cre(+/+) females. Thirty-one mice were born. Nineteen were males, which, because of the breeding strategy, did not inherit a lox-Piga-lacZ gene. Twelve offspring were females, and all carried a lox-Piga-lacZ and an EIIa-cre gene. The efficiency of Cre-mediated Piga gene recombination in female offspring varied. Three had, in virtually all cells, a recombined lox-Piga-lacZ gene (highly recombined animals), as verified by the absence of a lox-Piga-lacZ allele determined by PCR of tail DNA and Southern blot analysis (see below). In the remaining nine females, the extent of Cre-mediated lox-Piga-lacZ recombination varied between 0 and 95% (partially recombined mice). Thus, the site-specific DNA recombination approach proved successful in generating female mice that had high-efficiency Cre-mediated Piga gene recombination. However, all three highly recombined animals had phenotypic abnormalities, including oro-facial malformations with cleft palate. They were unable to suckle and to feed properly. Unlikely to survive, the animals were sacrificed for the analysis. Partially recombined females, however, survived to adulthood.
Methylation Analysis to Study Transcriptional Activity at the Piga Locus.
Cre-mediated Piga gene inactivation in our mice abrogates the expression of GPI-linked surface proteins and simultaneously leads to the expression of LacZ under the Piga promoter. β-d-Galactosidase activity is therefore a marker for cells that express the lox-ΔPiga-lacZ gene (G. Cattoretti and M.B., unpublished results). However, all PIGA(−) cells may not be detected by activity of β-d-galactosidase because Piga promoter activity may be too low to express detectable levels of enzyme. To estimate the number of PIGA(−) cells, we therefore determined the transcriptional activity of the Piga alleles on a molecular level and used methylation-sensitive restriction sites to probe for the methylation status at the Piga locus. Cytosine methylation of the promoter region is associated with transcriptional silencing of genes subject to X chromosome inactivation (16, 17). We therefore analyzed the Piga promoter region for CpG dinucleotides that are differentially methylated on the active and inactive X chromosome. In pilot studies, we identified six restriction endonuclease cleavage sites that are essentially unmethylated on the active, but methylated to various extent on the inactive, X chromosome (see Fig. 1B). Digestion with the corresponding methylation-sensitive restriction endonuclease produced restriction fragments that distinguish wt Piga, lox-Piga-lacZ, and lox-ΔPiga-lacZ by Southern blotting. Cleavage at the SacII restriction site 5′ of exon 1 (5′ SacII) was 100% complete in DNA obtained from wt males (n = 4), indicating that the SacII cleavage site is unmethylated on the single active male X chromosome. In wt females, DNA cleavage at this site was 46.8 ± 2% (n = 4), suggesting that the SacII restriction site on the inactive X chromosome is almost always methylated. We determined the degree of methylation at the 5′ SacII restriction site to assess transcriptional activity of the Piga alleles.
Methylation of the 5′ SacII site in EIIa-cre (+/+) and lox-Piga-lacZ mice.
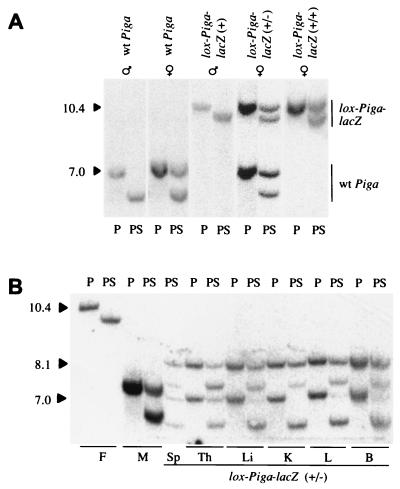
First, we verified the methylation pattern of 5′ SacII in both lox-Piga-lacZ and EIIa-cre animals. Southern blot analysis of genomic DNA isolated from hemizygous lox-Piga-lacZ(+) males (n = 6) and from males that carry the EIIa-cre transgene (n = 2) confirmed that the 5′ SacII site is 100% unmethylated in both mouse strains (see also Fig. 2A). Likewise, genomic DNA obtained from homozygous EIIa-cre(+/+) females (n = 2) and DNA (n = 17) from various tissues of homozygous lox-Piga-lacZ(+/+) females (n = 2) was digested only to 47 ± 12% and 45 ± 7% respectively, which is in agreement with an almost complete methylation of 5′ SacII on the inactive X chromosome (Fig. 2A).
Figure 2.
Southern blot analysis to determine the transcriptional activity of the Piga alleles. (A) Southern blot analysis of DNA obtained from wt animals and mice hemi-, hetero-, or homozygous for the lox-Piga-lacZ gene. (B) Southern blot analysis of DNA isolated from different organs of a female mouse with a lox-ΔPiga-lacZ gene in almost all cells (mouse E, Fig. 3B). DNA was digested with PstI alone (P) or with PstI and SacII (PS). Southern blots were hybridized with the Sac-Bam DNA probe. The 10.4-kb fragment corresponds to lox-Piga-lacZ, the 8.1-kb fragment to lox-ΔPiga-lacZ, and the 7.0-kb fragment to the wt Piga gene. Digestion of the nonmethylated portion with SacII shortens each restriction fragment by 0.5 kb. Sp, spleen; Th, thymus; Li, liver; K, kidney; L, lung; B, brain. F (father) displays DNA of a male lox-Piga-lacZ(+) mouse, M (mother) shows DNA of a female EIIa-cre(+/+) mouse.
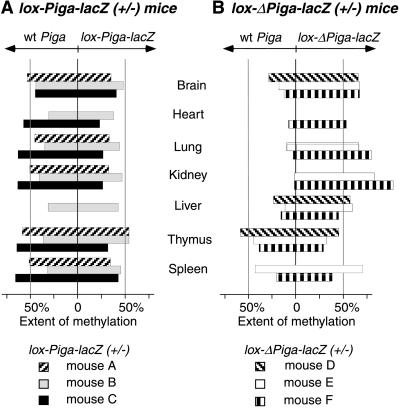
Similarly, DNA (n = 21) obtained from different organs of heterozygous lox-Piga-lacZ(+/−) mice (n = 3) demonstrated that the 5′ SacII site of the wt Piga was methylated to 49.5 ± 12% and the lox-Piga-lacZ was methylated 37 ± 11%. The balanced proportion of methylated and unmethylated wt Piga suggests that half of the wt Piga is on the active and half is on the inactive X chromosome, which represents the expected balanced pattern of X chromosome inactivation. The extent of methylation of the lox-Piga-lacZ allele was somewhat lower than predicted by the degree of methylation of the wt Piga (see Fig. 3A). A similar difference in methylation was found at the MluI site (data not shown), a methylation-sensitive restriction site 3′ of Piga exon 1 (see Fig. 1B). This result suggests that in heterozygous lox-Piga-lacZ(+/−) mice, random X inactivation occurs normally but that on the inactive X chromosome, methylation of the lox-Piga-lacZ gene might not be complete.
Figure 3.
Diagram summarizing methylation at the 5′ SacII restriction site in different tissues. (A) Methylation of wt and lox-Piga-lacZ in 3 mice heterozygous for lox-Piga-lacZ. (B) Methylation of wt and lox-ΔPiga-lacZ in 3 mice that had high levels of Piga gene recombination. The Southern blot analysis of mouse E is shown in Fig. 2B. DNA samples from the heart of mouse A and E and from the liver of mouse A and C were degraded. DNA from heart, lung, kidney, and spleen of mouse D were lost accidentally.
Methylation Analysis in Female Mice with High Level of Piga Gene Recombination.
In three female mice, the absence of the 10.4-kb fragment derived from lox-Piga-lacZ and the appearance of a new 8.1-kb restriction fragment derived from lox-ΔPiga-lacZ suggested that the lox-Piga-lacZ gene had recombined in virtually all cells (see Fig. 2B).
Methylation analysis was performed on DNA from different tissues. A representative example is shown in Fig. 2B. Fig. 3B summarizes the extent of methylation of the Piga alleles in various tissues. The wt Piga gene was essentially unmethylated in kidney, lung, and heart and to a lesser extent also in the brain and liver. This result indicates that these tissues preferentially express the wt Piga gene. In contrast, DNA isolated from thymus and spleen revealed an almost equal proportion of cells expressing either the wt or the recombined Piga gene, suggesting that a functional PIGA protein is nonessential in these tissues.
The methylation of lox-Piga-lacZ in heterozygous animals and the methylation of lox-ΔPiga-lacZ in mosaic animals were lower than expected. We do not know the cause of the incomplete methylation of lox-Piga-lacZ and lox-ΔPiga-lacZ. A possible explanation might be that insertion of the foreign prokaryotic DNA sequences, loxP in exon 2 and/or lacZ in intron 2, interfered with methylation of the Piga locus. Alternatively, homologous recombination with unmethylated plasmid DNA [replacement vector used encompassed the entire 5′ Piga region (12)] might have introduced an altered methylation pattern into ES cells. Persistence of altered methylation during mammalian embryonic development has been described (18), and heritable changes in DNA methylation have been reported in plants (19). The altered methylation of lox-ΔPiga-lacZ does not interfere with our intent to determine the proportion of PIGA(−) cells, because the expression of GPI-linked proteins on the cell surface depends on the expression of wt PIGA (20).
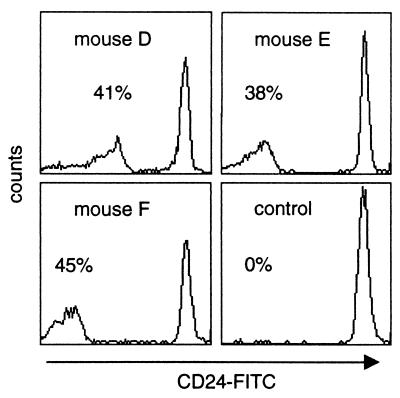
DNA isolated from bone marrow cells was not sufficient for Southern blot analysis. To evaluate the contribution of PIGA(−) cells to hematopoiesis, we therefore determined the proportion of PIGA(−) red blood cells in peripheral blood by using flow cytometry and monoclonal antibody toward CD24 (heat stable antigen), a GPI-linked protein highly expressed on normal mouse red blood cells. The proportion of CD24(−) red blood cells in our three highly recombined animals were 41%, 38%, and 45% (Fig. 4), suggesting that GPI-linked proteins are dispensable for differentiation and maturation of red blood cells.
Figure 4.
Proportion of PIGA-deficient red blood cells in mice with a lox-ΔPiga-lacZ gene in almost every cell. Flow cytometric analysis of peripheral red blood cells obtained from mice D–F and from an age-matched lox-Piga-lacZ control mouse were stained with FITC-conjugated mAb against the GPI-linked antigen CD24 and analyzed by flow cytometry.
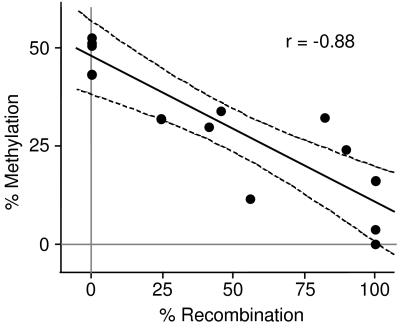
Methylation analysis of tail DNA obtained from animals that did not have a recombined lox-Piga-lacZ in all cells (i.e., partially recombined) showed a negative correlation between the extent of lox-Piga-lacZ recombination and extent of methylation in the wt Piga locus (Fig. 5). This result further supports our finding that, with the exception of hematopoiesis, the growth of cells expressing GPI-linked proteins is favored.
Figure 5.
Negative correlation of the level of DNA methylation and the extent of lox-Piga-lacZ recombination. The plot displays the percentage of methylation at the 5′ SacII site of the wt gene (y axis) as a function of the amount of recombination of the lox-Piga-lacZ gene (x axis). Shown are the regression line and the 95% confidence interval. r, correlation coefficient.
Inheritance of a Piga Gene Mutation.
To test the mode of inheritance of a mutated Piga gene, we bred mosaic female mice that survived to adulthood (partially recombined mice) with C57BL/6 males carrying a wt Piga gene. Of 28 pups (16 females and 12 males), none inherited a lox-ΔPiga-lacZ gene. Interestingly, however, none of the offspring carried a lox-Piga-lacZ gene. This result suggests that Cre-mediated recombination of the lox-Piga-lacZ gene occurred to a 100% in the maturing oocyte, and that mice with an inherited Piga mutation are embryonic lethal.
DISCUSSION
A somatic mutation in the X linked PIGA gene is responsible for the deficiency of GPI-linked proteins on the surface of blood cells in patients with PNH. An inherited form of the disease has not been described. To study the importance of PIGA in different organs and to address the question of a heritable Piga mutation, we generated female mice that had high efficiency of Cre-mediated Piga gene recombination during early embryonic development. The fact that Piga maps to the X chromosome (21) and is subject to X chromosome inactivation (12) was to our advantage because (i) in males as well as in females, a single Piga gene recombination is sufficient to abrogate the expression of GPI-linked proteins (provided that in females the recombined Piga gene is on the active X chromosome); (ii) X inactivation in female mice that had high efficiency of Cre-mediated Piga gene recombination generates a mosaicism with cells that express or lack GPI-linked proteins; and (iii) the methylation status of the locus can be used to estimate the transcriptional activity.
In contrast to previous attempts using blastocyst injection of PIGA(−) ES cells (9, 10), using the Cre/loxP system proved to be successful in the generation of female mice that carry a Piga-null mutation in virtually all cells. In addition, in highly recombined female mice all cells are genetically identical, whereas in chimeric mice PIGA(−) ES cells are derived from strain 129 and the host blastocysts belonged to strain C57BL/6 (9, 10). Differences in the genetic background may have influenced the growth of PIGA(−) cells in chimeric animals (22, 23). In contrast, in mosaic animals, the proportion of PIGA(+) to PIGA(−) cells reflects directly the consequences of a nonfunctional Piga gene on cell proliferation and survival (24).
Methylation of the 5′ Region of Piga Is Associated with Transcriptional Activity.
During mammalian development, one of the two X chromosomes in female embryos is randomly inactivated in somatic cells to achieve dosage compensation (25). X inactivation proceeds in different schedules in different somatic tissues commencing at embryonic day 6.5 and is largely completed by embryonic day 10.5 (26). In somatic cell hybrid experiments, we have previously shown that the Piga mutation behaves recessively (20). Cells heterozygous for lox-ΔPiga-lacZ will therefore not express the PIGA(−) phenotype until after X chromosome inactivation. In the newborn, a predominant expression of wt Piga is therefore found in tissues in which PIGA is functionally important at the time of and after X chromosome inactivation. In contrast, predominant expression of the mutant Piga gene will occur if the mutation confers a proliferative advantage.
Analysis using 5-methylcytosine modification-sensitive endonucleases revealed that the 5′ region of the Piga gene is essentially unmethylated on the active X chromosome. On the inactive X chromosome, the 5′ region of the Piga gene is methylated, but the extent of methylation varies within the examined DNA region (see Fig. 1B). To assess the transcriptional activity of the Piga alleles, we therefore chose to measure the level of methylation at the endonuclease restriction site SacII 5′ of exon 1, which is not methylated on the active, but is almost 100% methylated on the inactive, X chromosome.
Mosaic analysis in our mice showed that in tissues such as heart, lung, kidney, and brain, the wt Piga gene is expressed predominantly, suggesting that these tissues require the expression of GPI-linked proteins. Cells that lack the expression of GPI-linked proteins most likely were lost either by a growth disadvantage or because of cell death. The salient exception to this finding were thymus, spleen, and red blood cells (see Figs. 3B and 4), which had an approximately equal contribution of PIGA(+) and PIGA(−) cells. This result suggests that for the derivation of hematopoietic cells, GPI-linked proteins are dispensable. The latter is in agreement with our previous finding showing erythroid and myeloid differentiation in embryoid bodies obtained from PIGA(−) ES cells (10).
A predominant expression of lox-ΔPiga-lacZ was never observed. This result indicates that a Piga mutation does not provide a proliferative advantage to the mutant cell, suggesting that a second factor is required in addition to the PIGA mutation to cause the clonal dominance of PIGA(−) blood cells in patients with PNH.
Inherited Piga Mutations?
The fact that we were able to obtain female mice that carry in virtually all cells a mutated Piga gene raises the interesting issue of whether a heritable form of PNH exists. Because of X inactivation followed by cellular selection, female mice with high levels of Piga gene recombination were born alive. The biased male/female ratio of 1.5 suggests fetal wastage of highly recombined animals not rescued by the relative growth advantage of PIGA(+) cells.
An inherited Piga mutation would be expected to follow a male lethal, female dominant inheritance pattern, with a varied phenotype in females depending on the proportion of cells expressing the mutant Piga gene. We tested the mode of inheritance by breeding mosaic females that survived to adulthood and had some degree of lox-Piga-lacZ recombination (partially recombined) with male mice carrying a wt Piga gene. Not one of the offspring inherited a lox-ΔPiga-lacZ gene, but surprisingly, none inherited either the lox-Piga-lacZ gene. This indicates that Cre-mediated recombination of the maternal lox-Piga-lacZ gene occurred 100% in the maturing oocyte and that a maternally inherited Piga mutation is embryonic lethal. The latter result seems at first sight in apparent contrast to our female offspring, which had high efficiency of Cre-mediated Piga gene inactivation. However, in our offspring, the lox-Piga-lacZ is paternally derived. In the embryo proper, X chromosome inactivation occurs at random. In contrast, in the trophoectoderm and in the primitive endoderm of the implanting embryo, the paternally derived X chromosome is preferentially inactivated (27). It is therefore conceivable that PIGA is essential in these tissues. In fact, previous studies in PIGA(−) embryoid bodies suggested that PIGA might be required for proper endoderm formation (10).
Our animal model suggests that in mice, and possibly also in humans, a genetic lesion that involves the Piga gene and abolishes PIGA function may not be heritable. However, this does not exclude the possibility of sporadic mutations that, if occurring during early embryogenesis, may almost exclusively be found in females and thus mimic an X linked dominant disease with prenatal lethality in males and a variable phenotype in females. In fact, recently, Ogata et al.(28) reported a female infant mosaic for an interstitial deletion within Xp22 spanning the critical region of the gene responsible for microphthalmia with linear skin defect and the PIGA gene as determined by using microsatellite analysis (28). Findings in our lox-ΔPiga-lacZ mosaic mice suggest that (i) in most tissues, the normal X chromosome will be active, (ii) immunostaining of blood cells with antibodies toward GPI-linked proteins would probably be useful to test for a functional PIGA gene in X linked dominant male lethal disorders mapping to Xp22, and (iii) women mosaic for cells carrying a PIGA gene mutation are not at risk to pass on the PIGA gene mutation.
Acknowledgments
We thank Lucio Luzzatto for his help in starting this project and for his unparalleled generosity, which allowed us to continue this work at Washington University, St. Louis. We thank David Schlessinger for his expertise and advice. We are grateful to Dan Link and Alvaro Pereira for stimulating discussions and David Wilson, Philip W. Majerus, and Philip J. Mason for critically reading the manuscript. EIIa-cre transgenic mice were kindly provided by Heiner Westphal. This work was supported by Grant RO1-HL-56678-01 from the National Institute of Health, the McDonnell Foundation, the Howard Hughes Medical Institute, Barnes–Jewish Hospital Foundation, and the Edward Mallinckrodt Foundation. P.K. is supported by the Theodor und Ida Herzog-Egli-Stiftung and the Fritz-Rohrer-Fonds, Switzerland. M.B. receives the Junior Faculty Award of the American Society of Hematology.
ABBREVIATIONS
- PIGA
phosphatidylinositol glycan class A
- PIGA refers to the human gene
Piga to the murine gene, and PIGA to the gene product of either species
- ES
embryonic stem
- PNH
paroxysmal nocturnal hemoglobinuria
- GPI
glycosyl phosphatidylinositol
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Miyata T, Takeda J, Iida Y, Yamada N, Inoue N, Takahashi M, Maeda K, Kitani T, Kinoshita T. Science. 1993;259:1318–1320. doi: 10.1126/science.7680492. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe R, Inoue N, Westfall B, Taron C H, Orlean P, Takeda J, Kinoshita T. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessler M, Mason P J, Hillmen P, Miyata T, Yamada N, Takeda J, Luzzatto L, Kinoshita T. EMBO J. 1994;13:110–117. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda J, Miyata T, Kawagoe K, Iida Y, Endo Y, Fujita T, Takahashi M, Kitani T, Kinoshita T. Cell. 1993;73:703–711. doi: 10.1016/0092-8674(93)90250-t. [DOI] [PubMed] [Google Scholar]
- 5.Bessler M, Hillmen P, Longo L, Luzzatto L, Mason P J. Hum Mol Genet. 1994;3:751–757. doi: 10.1093/hmg/3.5.751. [DOI] [PubMed] [Google Scholar]
- 6.Bessler M, Hillmen P. Semin Hematol. 1998;35:149–167. [PubMed] [Google Scholar]
- 7.Ferguson M A J. Biochem Soc Transact. 1992;20:243–256. doi: 10.1042/bst0200243. [DOI] [PubMed] [Google Scholar]
- 8.Ohno S. In: Chromosomes and Sex-linked Genes. Ohno S, editor. Berlin: Springer; 1967. pp. 46–69. [Google Scholar]
- 9.Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, Kinoshita T, Takeda J. Blood. 1996;87:3600–3606. [PubMed] [Google Scholar]
- 10.Rosti V, Tremml G, Soares V, Pandolfi P P, Luzzatto L, Bessler M. J Clin Invest. 1997;100:1028–1036. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer B, Henderson N. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tremml, G., Dominguez, C., Rosti, V., Zang, Z., Pandolfi, P. P., Keller, P. & Bessler, M. (1999) Blood, in press. [PubMed]
- 13.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.14–9.19. [Google Scholar]
- 15.Dooley T P, Miranda M, Jones N C, DePamphilis M L. Development. 1989;107:945–956. doi: 10.1242/dev.107.4.945. [DOI] [PubMed] [Google Scholar]
- 16.Nan X, Cross S, Bird A. Novartis Found Symp. 1998;214:6–16. doi: 10.1002/9780470515501.ch2. [DOI] [PubMed] [Google Scholar]
- 17.Razin A. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses J J, Reik W, Feil R. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 19.Jeddeloh J A, Bender J, Richards E J. Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillmen P, Bessler M, Bungey J, Luzzatto L. Somatic Cell Mol Genet. 1993;19:123–129. doi: 10.1007/BF01233528. [DOI] [PubMed] [Google Scholar]
- 21.Kawagoe K, Takeda J, Endo Y, Kinoshita T. Genomics. 1994;23:566–574. doi: 10.1006/geno.1994.1544. [DOI] [PubMed] [Google Scholar]
- 22.De Haan G, Nijhof W, Van Zant G. Blood. 1997;89:1543–1550. [PubMed] [Google Scholar]
- 23.Van Zant G, Thompson B P, Chen J J. Exp Hematol. 1991;19:941–949. [PubMed] [Google Scholar]
- 24.Gartler S M, Andina R J. Adv Hum Genet. 1976;7:99–140. doi: 10.1007/978-1-4757-0659-8_3. [DOI] [PubMed] [Google Scholar]
- 25.Lyon M F. Nature (London) 1974;250:651–653. doi: 10.1038/250651a0. [DOI] [PubMed] [Google Scholar]
- 26.Tan S S, Williams E A, Tam P P. Nat Genet. 1993;3:170–174. doi: 10.1038/ng0293-170. [DOI] [PubMed] [Google Scholar]
- 27.Takagi N, Sasaki M. Nature (London) 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 28.Ogata T, Wakui K, Muroya K, Ohashi H, Matsuo N, Brown D M, Ishii T, Fukushima Y. Hum Genet. 1998;103:51–56. doi: 10.1007/s004390050782. [DOI] [PubMed] [Google Scholar]