Abstract
JC virus (JCV) is a polyoma virus that commonly infects humans. We have found T antigen DNA sequences of JCV in the mucosa of normal human colons, colorectal cancers, colorectal cancer xenografts raised in nude mice, and in the human colon cancer cell line SW480. A larger number of viral copies is present in cancer cells than in non-neoplastic colon cells, and sequence microheterogeneity occurs within individual colonic mucosal specimens. The improved yield of detection after treatment with topoisomerase I suggests that the viral DNA is negatively supercoiled in the human tissues. These results indicate that JCV DNA can be found in colonic tissues, which raises the possibility that this virus may play a role in the chromosomal instability observed in colorectal carcinogenesis.
Human cancers often are characterized by aneuploidy and widespread chromosomal rearrangements that result in the excessive activity of certain growth-stimulating genes and the deletion of other growth-limiting (tumor suppressor) genes. These genomic deletions are the result of an active process called chromosomal instability (1), which can be detected at the earliest stages of multistage carcinogenesis of colorectal tumors (2). The mechanism that permits the accumulation of this extreme degree of chromosomal disorder in cancer currently is unexplained.
Aneuploid lymphocytes termed rogue cells have been encountered in short-term lymphocyte cultures of people from locations throughout the world (3, 4). Experimental and epidemiological evidence suggest rogue cells may be the result of infection by the very widespread JC polyoma virus, a DNA virus with a supercoiled 5.13-kb genome that shows a high degree of homology with the well-known, fibroblast-transforming simian virus 40 (SV40) (5, 6). Significant antibody titers to JC virus (JCV) capsid protein are encountered in 70–80% of adult populations throughout the world (7–13). Rogue cells are so badly damaged it is unlikely they would often survive mitosis. Consequently, these cells are assumed to exist only transiently. However, the data suggest that there also may be a significant increase in simple chromosome damage in the cultured lymphocytes of people exhibiting rogue cells (14).
The possible wider significance of the JCV depends on what cells or tissues it can infect in addition to the lymphocytoid series. The virus lytically infects oligodendrocytes, the principal target cells in humans. Its activation from latency in people with immunosuppressive conditions, particularly AIDS, is responsible for the demyelinating central nervous system disorder, progressive multifocal leukoencephalopathy. JCV DNA also has been isolated from an unusual oligoastrocytoma, although no clear association between JCV and glial tumors has been shown. In Japan, viral DNA has been recovered from 45% of urine samples obtained from older persons. The virus is presumed latent in the kidney as well as in lymphoid tissues such as bone marrow (15–18).
In this paper, we report the detection of JC viral DNA fragments from a fourth tissue, very different from those in which the virus previously has been reported, namely, normal and malignant colorectal epithelium. That this viral DNA presence is not the result of the occurrence in the tissue of transitory, infected cells of the lymphocytoid line is strongly suggested by the fact that viral nucleotide sequences also were detected in five of 10 colon cancer xenografts and in the colon cancer cell line SW480. Portions of this work have been reported previously as abstracts at national meetings (19, 20).
MATERIALS AND METHODS
Human Tissue Specimens.
For this study, we used matched pairs of colorectal cancer and normal, adjacent mucosa from surgical resection specimens. DNA was isolated from each specimen (21) and used as a template for the PCR to amplify DNA sequences coding the amino terminus of the JCV T antigen. The PCR product was a 520-bp target derived from the Mad1 strain sequence (22) (Fig. 1). Specimens of surgically resected human colorectal tissues from patients with cancer were obtained from the University of Michigan School of Medicine Department of Pathology after surgical resection, under Institution Review Board approval, and after a variable period of ischemic time (usually 30–60 min), were slowly frozen and stored at −80°C. The surgical samples were thawed and homogenized in Trizol (GIBCO) by using a 1.5-ml plastic tube (Kontes) and a disposable pestle for each specimen. Care was taken to prevent carryover between specimens. The DNA-containing fraction was digested overnight in proteinase K (Boehringer Mannheim), followed by extraction with phenol/chloroform, and quantitated by spectrophotometry (OD260).
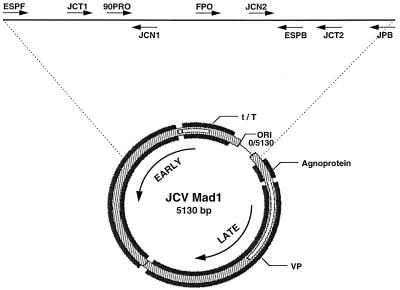
Figure 1.
Schematic map of the JCV genome. Target sequences in the T antigen gene and adjacent regions were selected for amplification as described. The target sequences from nucleotide positions 4383–272 (crossing the origin of viral replication, designated 0/5130) are highlighted by the dotted line, and the nine oligonucleotide probes used to amplify the viral sequences or to hybridize the PCR products are indicated at the top. The positions of the primers and oligoprobes are as follows: ESPF, 4383–4405; JCT1, 4481–4500; 90PRO, 4521–4550; JCN1, 4573–4554; FPO, 4721–4743; JCN2, 4881–4900; ESPB, 4949–4927; JCT2, 5000–4981; JPB, 272–247. The positions of the large T and small t antigens are indicated, and the arrows indicate the direction of transcription of the early (T/t antigens) and late (agnoprotein and capsid or VP) genes.
Demonstration of JCV DNA Sequences in Human Colonic Tissues.
The Mad1 viral sequence in a plasmid vector was used as a positive template control, and template blanks (with no DNA, but containing all other reagents) were run as negative controls in each gel to exclude the possibility of contamination of the reagents or equipment with each PCR amplification. The results also were confirmed by using fresh reagents in two physically distinct laboratories that had never previously been exposed to any of the reagents, human tissues, or PCR products. Additional JCV DNA targets of 120 bp and 93 bp located at either end of the initial 520-bp T antigen nucleotide sequence were amplified by PCR in confirmatory experiments.
Considerable effort was expended to optimize the PCR conditions for these studies (L.L., A.E.R., G.M., and C.R.B., unpublished work). The PCR targets used to detect three JCV T antigen regions were based on published JCV sequences, as illustrated in Fig. 1. Except as indicated, PCRs were run in 50-μl volumes by using the “hot start” technique (with Ampliwax, Perkin–Elmer), with 50 pmol of each primer, 1–2.5 units of recombinant Taq polymerase (GIBCO), a standard buffer (50 mM KCl/10 mM Tris⋅HCl/2 mM MgCl2), 200 μM of each dNTP, and 500–1,000 ng of template DNA. The initial denaturation occurred at 94°C for 3 min and a final extension of 72°C for 10 min. Oligonucleotides were used as PCR primers as well as radiolabeled oligoprobes, and the positions described below correspond to the positions on the Mad1 isolate.
The 520-bp target was amplified by using the primers JCT1 (5′-AATAGTGGTTTACCTTAAAG, complementary to positions 4481–4500 on the sense strand) and JCT2 (5′-TGAATAGGGAGGAATCCATG, complementary to positions 5000–4981 on the antisense strand), by using 1.5 mM MgCl2 in this reaction, for 40 cycles at 92°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec. PCR products were electrophoresed on agarose gels, blotted onto Hybond N+ membranes (Amersham Pharmacia), and hybridized with the oligoprobes 90PRO (5′-TGTCTCCAAGAACTTTCTCCCAGCAATGAA, at positions 4521–4550) or FPO (5′-CATTCCTTGCAATAAAGGGTATC, at positions 4721–4743). After 40 cycles of PCR with the JCT1/JCT2 primers, nested PCR was performed by using 1–2% of the first-round product for another 30 cycles, at 62°C for 1 min for the annealing/extension reaction, with primers 90PRO and ESPB (5′-CTGCATGGGGGAACATTCCTGTC, corresponding to positions 4949–4927). This procedure amplifies a 429-bp target sequence. Similarly, by using primers FPO and ESPB, a 229-bp amplicon was generated. The 93-bp target was amplified by using primers JCT1 and JCN1 (5′-GTGTGACTTAACCCAAGAAG, at positions 4573–4554), for 60 cycles at 92°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec. The oligoprobe 90PRO was used to hybridize the Southern transfer of the PCR products. A 120-bp target was amplified by using primers JCT2 and JCN2 (5′-TATCAGGGTGGAGTTCTTTG, corresponding to positions 4881–4900). This reaction was run in a 10-μl volume with 1 pmol of each primer for 20 cycles (92°C for 30 sec and 62°C for 30 sec), followed by the addition of fresh reagents for a standard 100-μl reaction, cycled another 30 times by annealing/extension at 62°C for 1 min. The oligoprobe ESPB was used for hybridization of the Southern blot of the PCR products.
JCV in Cultured Human Colorectal Cancer Cell Lines.
Colon cancer cell lines SW480, HCT116, HT29, and LoVo were obtained from the American Type Culture Collection (ATCC), grown under standard conditions as recommended by ATCC. The DNA was extracted and JCV sequences were sought by PCR, as described above.
DNA from Xenografts of Human Colon Cancers.
DNA samples extracted from 10 primary human colon cancers explanted and raised s.c. as xenografts in nude mice were generously provided by Bert Vogelstein and Kenneth Kinzler (Johns Hopkins University, Baltimore). These were provided at a concentration of 20 ng/ml DNA and were not further purified or diluted with any reagents from our laboratory before PCR. Similar controls were run with each experiment, as described above.
Topoisomerase I Treatment of DNA.
DNA specimens (500 ng) were treated with 0.5 units of topoisomerase I (GIBCO/BRL) at 37°C for 45 min before preparation for PCR amplification, in a modification of the published method of Keller (23) (L.L., A.E.R., G.M., and C.R.B. unpublished work).
Quantitation of JCV DNA.
A modification of the method of Tornatore et al. (24) was used to establish a semiquantitative PCR-based technique for estimating the JCV copy number. JCV-containing plasmids were serially diluted in 1-μg aliquots of human placental DNA (HumanCOT, GIBCO) and subjected to PCR amplification of the 120-bp target described above, by using primers JCT2 and JCN2, denaturing at 94°C for 2 min, annealing and extending at 62°C for 60 sec over 45 cycles by using Taq polymerase (GIBCO) and standard buffers (50 mM KCl, 10 mM Tris⋅HCl, and 2.0 mM MgCl2). The PCR products were electrophoresed in 2.5% agarose (GIBCO) and stained with ethidium bromide (EtdBr). Serial dilutions of the plasmid suspension provided a standard reference for the relative ability to amplify the JCV sequences. Forty-five cycles of amplification were found to be sufficient to permit the quantification of a difference in DNA copy number between normal colon and tumor samples.
Heteroduplex Analysis.
Heteroduplex analysis (25) was performed by nested PCR, first by amplifying a long (1,020 bp) sequence from the JCV genome (using primers ESPF: 5′-GTATTCCACCAGGATTCCCATTC from positions 4383–4405 and JPB: 5′-AGCTGGTGACAAGCCAAAACAGCTCT from positions 272–247) that included promoter sequences for viral transcription. The first round of amplification was carried out by using 35 cycles at 92°C for 30 sec and 62°C for 2 min. One percent of the PCR product served as template for nested PCR of the 520-bp amplicon by using primers JCT1 and JCT2, this time incorporating [α-32P]dCTP into the PCR products. Gel electrophoresis was carried out by denaturing the PCR products at 94°C for 2 min, cooling slowly in the thermal cycler for 60 min, and electrophoresing through a 6% polyacrylamide gel (30:1 acrylamide/bis), followed by autoradiography. Duplexes were created by mixing the PCR products of the samples with similarly radiolabeled PCR products obtained by using the Mad1 plasmid as a template.
Cloning and Sequencing of JCV DNA.
The 520-bp PCR products were cloned into pUC18 (SureClone, Amersham Pharmacia) according to the manufacturer’s instructions, grown in INVα (Invitrogen), and reamplified, and the PCR products were sequenced from both ends of the PCR products by using original and nested primers with an ABI 310 Genetic Analyzer (Perkin–Elmer).
Sequencing the JCV Regulatory Region.
Sequencing of the regulatory region downstream of the origin of replication was performed to further confirm the identity of the virus as distinct from BK virus or SV40, by using PCR primers spanning the T antigen (5060–5079) and regulatory region (297–317). Clones were derived from the amplified bands of selected samples in a Bluescript vector and sequenced as described (26).
RESULTS
In a preliminary study of a group of DNA samples extracted from 23 pairs of normal colorectal epithelium and adjacent cancers, we ran a total of 198 amplifications of the 520-bp target (an average of approximately four PCRs per DNA sample), and 16 of these (8%) were positive. Overall, 12 of the samples (26%) yielded at least one positive amplification of JCV by PCR (Table 1). The PCR products were documented as authentic JCV sequences by Southern blot analysis as illustrated in Fig. 2. The intensity of the Southern blot bands varied compared with the more uniform intensity of the EtdBr-stained bands. This finding raised the possibility of some sequence variation in the PCR products, which would result in variable degrees of hybridization with the probe (see below).
Table 1.
Amplification of the 520-bp fragment (bp 4481-5000 of the JCV T antigen sequence) from colorectal tissues
| Method of amplification | Number of samples investigated | Samples containing JCV DNA (% positive) |
|---|---|---|
| PCR | 46 | 12 (26%) |
| (23 normal-cancer pairs) | ||
| TISPA | 54 | 48 (89%) |
| (25 normal-cancer pairs and four unpaired cancers) |
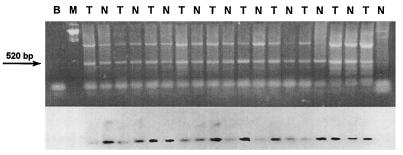
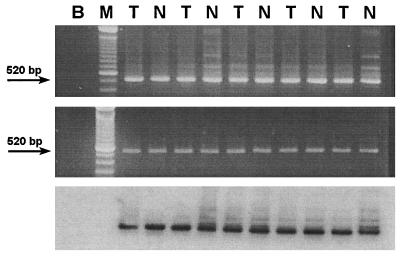
Figure 2.
Detection of the JCV T Ag sequence in human colorectal tissues. The 520-bp fragment (spanning JCV nucleotides 4481–5000, by using the primers JCT1 and JCT2) was amplified from DNA extracted from matched samples of normal colon (N) and colorectal cancers (T). (Upper) An EtdBr-stained 2% agarose gel demonstrating the 520-bp amplicons. (Lower) A Southern blot of the gel hybridized with a radiolabeled 416-bp probe for the JCV T antigen sequence obtained from the plasmid pMITC digested with HindIII. B indicates a PCR control lane (blank template plus all other reagents), and M refers to the molecular weight marker lane.
The relatively low yield from amplification of the 520-bp sequence led to the hypothesis that the physical configuration of the viral DNA may be important in permitting its efficient amplification. Therefore, primers for the previously described 120- and 93-bp targets were developed. These smaller PCR targets were more readily amplified than the longer sequence (data not presented). The improved yield of positives obtained by amplifying shorter target sequences led us to a consideration of developing technical improvements in methodology for identifying JCV DNA by PCR.
SV40 contains a supercoiled DNA genome, a feature that produced unexpected biochemical characteristics when it was first studied in 1963 (27). We hypothesized that a supercoiled topology of the highly homologous JCV DNA might limit the efficiency of PCR amplification and used topoisomerase I treatment to determine whether the relaxation of supercoiling would improve our yield (which we refer to as topoisomerase I-sensitive polyomavirus amplification, or TISPA). In a series of 54 new DNA samples from normal colons and cancer specimens, TISPA was found to enhance the sensitivity of detection, resulting in the amplification of the 520-bp JCV sequence from 89% of the samples (see Table 1 and Fig. 3). In 22 instances of paired samples, both the normal and tumor tissues were positive. There were six failures to amplify JCV sequences from a colonic tissue, which were distributed as follows: 0 normal negative/tumor negative; two normal negative/tumor positive; one normal positive/tumor negative; and three tumors were negative in which there was no normal tissue for comparison. We suggest that supercoiling of the viral DNA may inhibit efficient amplification and that intact supercoiled viral DNA may result in inconsistent amplification, and it remains possible that other factors may induce sufficient relaxation of supercoiled DNA to improve its detectability by using PCR (unpublished work). Because the positive control (JCV cloned in a plasmid vector) was readily amplifiable without TISPA, the enhanced sensitivity after TISPA for the DNA samples extracted from tissues may be related to a small viral copy number in the human colonic mucosal specimens.
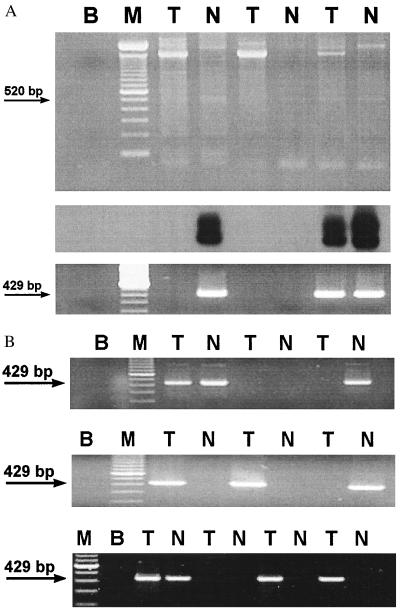
Figure 3.
Effect of topoisomerase I relaxation on the amplification of JCV DNA. (A) Three matched normal-tumor pairs of DNA were selected that did not yield a 520-bp PCR product with JCT1/JCT2 primers (data not shown). After treatment with topoisomerase I, the 520-bp fragment was successfully amplified from three of the six as shown on the gel. (Top) An EtdBr-stained 2% agarose gel separation of PCR products obtained from topoisomerase I-treated templates. (Middle) A Southern blot of the same gel hybridized with radiolabeled oligoprobe FPO, confirming that three of the faintly identifiable bands on the original gel contained the authentic viral sequence. (Bottom) An EtdBr-stained 2% agarose gel of a 429-bp nested PCR product generated after 1 μl of the initial reaction was transferred to a fresh tube and amplified with the primers 90PRO/ESPB for 28 more cycles. (B) Twenty samples of colonic DNA (10 matched normal-cancer pairs), all negative for JCV sequences after one round of PCR, were treated with topoisomerase I followed by nested PCR for the 429-bp amplicon as described above. Half of the initially negative samples were found to harbor JCV sequences on three independent experiments, presented as EtdBr-stained 2% agarose gels. T, tumors; N, normal samples; M, molecular size marker; B, control (blank).
The demonstration of JCV nucleotide sequences in the DNA from the surgically excised tissues did not exclude the possibility that the virus was present in nonepithelial cells contaminating the specimens. Colon cancer cell lines were investigated for the presence of JCV DNA. DNA isolated from the colon cancer cell line SW480 also was found to harbor JCV, whereas DNA from the colon cancer cell lines HCT116, HT29, and LoVo did not, in spite of numerous attempts at amplification, which included the use of nested PCR and TISPA (Table 2).
Table 2.
JCV DNA sequences in cultured human colorectal cancer cell lines and nude mouse xenografts
| DNA source | TISPA |
|---|---|
| HCT116 | Negative |
| LoVo | Negative |
| HT29 | Negative |
| SW480 | Positive* |
| Human xenografts | 5/10 positive* |
All TISPA bands confirmed to be JCV by Southern analysis.
To further test whether JCV was present in epithelial cells or caused by the presence of contaminating lymphocytes (or other cell types), DNA was extracted from 10 colon cancer xenografts. Each of these was derived from a primary colorectal cancer specimen that had been grown s.c. in a nude mouse. This technique permits selection and clonal expansion of neoplastic cells and eliminates non-neoplastic cell types that characteristically are present in a primary tumor. JCV sequences were amplified from five of these DNA samples (Table 2). These findings indicated that neoplastic colorectal epithelium is a source of JCV DNA and excluded the interpretation that infiltrating lymphocytes or other contaminating cells were responsible for these results.
We hypothesized that not every normal epithelial cell in the colon would be infected by JCV. However, if the virus were involved mechanistically in the clonal expansion of neoplastic tissues, there would be more copies of the virus in cancer cells than in normal colonic cells. To estimate viral sequence copy number, we adapted the semiquantitative PCR technique of Tornatore et al. (24). With both the 520-bp and 120-bp fragments as target sequences from the T antigen, DNA specimens from normal colon samples amplified as if they contained at least 0.01 JCV viral copies/human genome equivalent, whereas the data from the tumors suggested the presence of at least 1 order of magnitude more copies compared with the matched normal colonic tissues from the same patients (Fig. 4). These results are consistent with clonal origin and expansion of virally infected epithelial cells in neoplastic tissues. The low viral copy number per cell could reflect that only rare colonic epithelial cells are infected or could be an underestimate reflecting inefficient amplification of the viral target sequence compared with genomic human DNA.
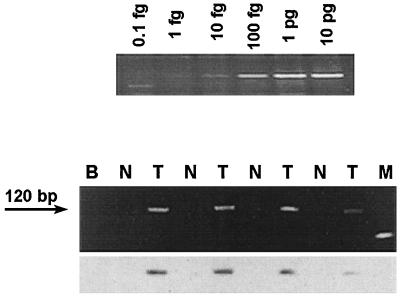
Figure 4.
Semiquantitative amplification of JCV T Ag sequence from colorectal tissue samples. (Upper) The 120-bp PCR product (spanning JCV nucleotides 4881–5000) obtained by amplifying the pMad1 (JCV sequence) plasmid. Concentrations of the plasmid DNA initially were determined by A260, and then diluted in 1 μg of human placental DNA (GIBCO), which provided an estimate of the minimal amount of template required to detect viral sequences (assuming similar efficiencies of amplification of plasmid DNA and authentic viral DNA). By using serial dilutions of template DNA to determine the point of extinction of the PCR product, the viral content in tumor specimens was estimated to be ≈0.01 viral copies/human genome equivalent. (Lower) An EtdBr-stained 2% agarose gel, and immediately below it, the corresponding Southern blot hybridized with the radiolabeled oligoprobe ESPB to confirm the sequence. Paired samples of template DNA from normal colon (N) and tumor specimens (T) from individual patients were diluted in parallel before amplification to determine the point at which the viral sequence could no longer be detected. Extinction of the PCR product from the normal samples, although still detectable from tumor samples, indicates that the tumor specimens contained a greater amount of amplifiable JCV than the matched normal colonic epithelium.
The PCR product derived from each of these tissues was identical in size to the control amplicon obtained from the Mad1 JCV sequence that had been cloned into a plasmid vector. We hypothesized that the DNA recovered from a longstanding viral infection could, because of the multiple rounds of replication, be characterized by diverse DNA sequences. In contrast, if our findings reflected laboratory contamination, most or all of our PCR products would have identical sequences. To deal with this concern, heteroduplex analysis was performed by amplifying all of the JCV sequences present in the DNA preparations by nested PCR and identifying imperfectly annealed, double-stranded heteroduplexes in nondenaturing polyacrylamide gels (25). Our results indicated that multiple viral sequences (i.e., numerous heteroduplex bands that migrate more slowly than the perfectly annealed homoduplexes in the gel) were present in individual samples from both normal and tumor specimens (Fig. 5). To confirm this finding, PCR products were extracted from agarose gels, cloned, and sequenced in both directions through two different segments (bp 4628–4679 and 4831–4864) from 15 different samples of normal colon or colorectal cancer. Sequence microheterogeneity was observed between different tissue samples and between matched normal and tumor specimens (Table 3).
Figure 5.
Heterogeneity of JCV T Ag sequences in human colorectal tissues. (Top) An EtdBr-stained 2% agarose gel demonstrating the 520-bp amplicons indicated by the arrow. A number of slowly migrating heteroduplex bands are visible in many of the lanes (T indicates a tumor specimen, N indicates a normal colonic specimen), suggesting the presence of imperfectly matched complementary sequences that form heteroduplexes that do not migrate with perfect homoduplexes. (Middle) An EtdBr-stained 2% agarose gel after the heteroduplexes were resolved by performing one additional denaturing, hybridization, and extension cycle with excess JCT1/JCT2 primers. The disappearance of the heteroduplex bands indicates that the extra bands in the top gel represented hybridization between imperfect matches of 520-bp amplicons, because the one additional amplification cycle permits copying of a perfect match for each DNA strand. If the slowly migrating bands had represented nonspecific amplification, these would not have resolved into the 520-bp band with additional amplification. (Bottom) A Southern blot derived from an electroblot of the middle gel containing resolved duplexes, hybridized with the 32P-radiolabeled oligonucletide probe 90PRO, confirming the identity of the 520-bp band.
Table 3.
Variations JCV sequences
| Tissue | Clone | Position | Variation | Predicted change
|
|
|---|---|---|---|---|---|
| Large T | Small t | ||||
| 124, normal | #2 | 4844 | G insertion | Truncated | Truncated |
| #12 | 4922 | C → T | R → K31 | R → K31 | |
| 124, tumor | #1 | 4643 | T → C | None | K → R124 |
| #5 | 4811 | A → T | V → D68 | V → D68 | |
| 4924 | C → A | M → I30 | M → I30 | ||
| #3 | 4900 | G → C | C → W38 | C → W38 | |
| 150, normal | #19 | 4903 | T → C | None | None |
| #2 | 4919 | T → G | K → T32 | K → T32 | |
| 150, tumor | #10 | 4643 | T → C | None | K → R124 |
| #7 | 4891 | G → T | None | None | |
| 151, normal | #2 | 4667 | A → G | None | L → P126 |
| #4 | None | ||||
| #10 | None | ||||
| 151, tumor | #5 | 4709 | G → A | None | A → V102 |
| 4718 | G → T | None | P → H99 | ||
| #11 | 4635 | T → C | None | D → G91 | |
| 4718 | G → T | None | P → H99 | ||
| 4742 | T → C | None | R → G127 | ||
| #12 | None | ||||
| #18 | 4718 | G → T | None | P → H99 | |
| 153, tumor | #19 | 4820 | C → T | G → D65 | G → D65 |
| 4840 | T → C | None | None | ||
| 164, tumor | #7 | 4649 | T → C | ||
| 4817 | A → G | ||||
| 4903–4908 | Deleted T | Truncated | Truncated | ||
| #8 | 4810 | A → T | |||
| 4903–4908 | Deleted T | Truncated | Truncated | ||
| #2 | 4903–4908 | Deleted T | Truncated | Truncated | |
| #9 | 4903–4908 | Deleted T | Truncated | Truncated | |
| SW480 | #4 | 4745 | G → A | None | None |
| 4834 | T → C | S → F90 | S → F90 | ||
In these samples, TISPA was performed by using degenerate primers to amplify other possible JCV quasispecies present in the human colon (L.L., A.E.R., and C.R.B., unpublished work). Nucleotide position, variation, and the predicted change are in reference to the Mad1 strain. DNA sequence variations all were verified by bidirectional sequence analysis.
To further confirm the identity of the DNA sequences found in the normal and cancer tissues, nucleotide templates from both the normal colon and cancer tissue from one patient (#724) were amplified by using a new set of primers targeted to the JCV regulatory region. This part of the viral genome contains the nucleotide sequences that regulate DNA replication and early and late transcription. The regulatory region sequences that occur in 98-bp tandem repeats (as found in brain tissue from progressive multifocal leukoencephalopathy patients) were used as “signature” sequences because they differ substantially from BK virus (BKV) and SV40. The structure of the viral regulatory region also has been identified in other tissues, including tonsils, tonsillar stromal cells, and lymphocytes (26). DNA sequences in this region are heterogeneous, presumably resulting from deletions and rearrangements that occur during multiple rounds of viral replication. JCV-specific bands from the regulatory region were amplified from the DNA of samples 124N (normal colon) and 124T (tumor), based on sequence-specific Southern blot hybridization. Eight clones were derived in a Bluescript vector from the amplified bands of each of the two samples and sequenced as described (26). The predominant sequence rearrangements in the regulatory region were similar to Mad1. However, there were several clones that showed deletions and rearrangements in comparison with the Mad1 sequence, but none that were similar to either BKV or SV40 (data not shown).
DISCUSSION
Polyomaviruses are widespread among animal species, the two best known being the polyomavirus of mice and the SV40 of the African green monkey (5–13). In humans, the two most extensively studied are JCV and the BK virus, which are equally frequent in humans. Both viruses have approximately 70% DNA sequence homology with SV40, whose human cell transforming properties have been widely studied. This observation has led to extensive speculation concerning the oncogenic potentialities of the two human viruses. However, the reports of an association in humans between polyomavirus infection (including SV40) and tumorigenesis have been variable and controversial (28–45). Thus, one must remain circumspect about the identification of the present virus until it has been completely sequenced. Nonetheless, our data strongly indicate the presence of JCV. Specifically, we have found T antigen DNA sequences characteristic of JCV in normal and malignant colorectal epithelium.
The transforming properties of SV40 depend on the T antigen produced by the virus. T antigen functions both as a DNA helicase and as a component in many protein–protein interactions, including interactions with p53 and the retinoblastoma family of proteins. Our studies, which link both complex rogue cells and simple chromosome damage with JCV (3, 4, 14), suggest this virus also may have the helicase activity of SV40. Other studies indicate that JCV T antigen functional domains are similar to those in the SV40 T antigen (5, 6, 46–48). We and others have found evidence for chromosomal instability in the earliest stages of colorectal neoplasia, as chromosomal deletions may be found in even tiny, benign colorectal adenomas (2). Whatever mechanism is responsible for the appearance of this type of genomic instability, it must occur very early in the process and account for both the unbalanced mitoses and the evasion of cell cycle checkpoint control. An infection that introduced the T antigen could explain the abrupt onset of chromosomal instability in multistep carcinogenesis, and we now have evidence that JCV can be present in human colons.
Recently, Lengauer et al. (1) reported that aneuploid colorectal tumors without microsatellite instability exhibit a striking defect in chromosome segregation, resulting in gains or losses in excess of 10−2 per chromosome per cell generation; this chromosomal instability appeared to be a dominantly inherited somatic trait. Our studies included three of the chromosomally unstable cell lines studied by these investigators (SW480, HT29, and LoVo), as well as one line with microsatellite instability (HCT116). If the presence of JCV, T antigen DNA sequences were the sole cause of this chromosomal instability, then, on the basis of the present findings, one might expect to find JCV T antigen DNA sequences in the chromosomally unstable lines but not those with microsatellite instability. In keeping with the hypothesis, no JCV T antigen DNA sequences were recovered from HCT116. Only SW480 among the three aneuploid lines was positive for JCV T antigen DNA sequences. However, the pertinent viral DNA sequences, having induced the instability, subsequently may have been lost from the tumor genome and may not be necessary for neoplastic behavior after the genomic instability has permitted critical genetic losses at tumor suppressor loci.
In summary, we have found evidence that human polyomavirus JCV DNA can be found in colonic tissues of patients with colorectal cancers. The detection of the viral DNA was improved substantially by the addition of topoisomerase I treatment to the preparative regimen, suggesting that copies of the virus exist in its supercoiled, episomal form. The viral load appears to be low in the normal colon and at least 10-fold higher in the DNA extracted from cancers. There is sequence microheterogeneity in the JCV DNA, both in the T antigen and regulatory domains. The presence of JCV sequences in human colon cancer xenografts and one colon cancer cell line supports our conclusion that the virus infects colonic epithelium, rather than a contaminating cell, such as lymphocytes. Taken in the context of the known biological properties of T antigen and recent epidemiological findings that link JCV to aneuploid “rogue” cells, we propose that infection with JCV is a prime candidate for a role in chromosomal instability, a phenomenon commonly found in colorectal cancers that is currently without a mechanistic explanation.
Acknowledgments
We thank Dr. Luigi Ricciardiello for assistance in the generation of the graphics in the manuscript. This work was supported in part by the Research Service of the Department of Veterans Affairs, The University of Michigan Comprehensive Cancer Center (CA46592), The University of California San Diego Cancer Center, and grants from the National Cancer Institute R01 CA26803 (to J.V.N.) and R01 CA72851 (to C.R.B.).
ABBREVIATIONS
- JVC
JC virus
- SV40
simian virus 40
- EtdBr
ethidium bromide
- TISPA
topoisomerase I-sensitive polyomavirus amplification
References
- 1.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 2.Boland C R, Sato J, Appelman H D, Bresalier R S, Feinberg A P. Nat Med. 1995;1:902–909. doi: 10.1038/nm0995-902. [DOI] [PubMed] [Google Scholar]
- 3.Neel J V, Major E O, Awa A A, Glover T, Burgess A, Traub R, Curfman B, Satoh C. Proc Natl Acad Sci USA. 1996;93:2690–2695. doi: 10.1073/pnas.93.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazutka J R, Neel J V, Major E O, Dedonyte V, Mierauskine J, Slapsyte G, Kesminiene A. Cancer Lett. 1996;109:177–183. doi: 10.1016/s0304-3835(96)04448-5. [DOI] [PubMed] [Google Scholar]
- 5.Fanning E, Knippers R E. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 6.Dorries K. Adv Virus Res. 1997;48:205–261. doi: 10.1016/s0065-3527(08)60289-4. [DOI] [PubMed] [Google Scholar]
- 7.Walker D L, Frisque R J. In: The Papovaviridae: The Polyomaviruses. Salzman N P, editor. Vol. 1. New York: Plenum; 1986. pp. 327–377. [Google Scholar]
- 8.Brown P, Tsai T, Gajdusek D C. Am J Epidemiol. 1975;102:331–340. doi: 10.1093/oxfordjournals.aje.a112169. [DOI] [PubMed] [Google Scholar]
- 9.Taguchi F, Kajioka J, Miyamura T F. Microbiol Immunol. 1982;26:1057–1064. doi: 10.1111/j.1348-0421.1982.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 10.Walker D L, Padgett B L, ZuRhein G M, Albert A E, Marsh R F. Science. 1973;181:674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- 11.London W T, Houff S A, Madden D L, Fuccillo D A, Gravell M, Wallen W C, Palmer A E, Sever J L, Padgett B L, Walker D L, et al. Science. 1978;201:1246–1249. doi: 10.1126/science.211583. [DOI] [PubMed] [Google Scholar]
- 12.Frisque R J, Rifkin D B, Walker D L. J Virol. 1980;35:265–269. doi: 10.1128/jvi.35.1.265-269.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padgett B L, Walker D L. Prog Med Virol. 1976;22:1–35. [PubMed] [Google Scholar]
- 14.Neel J V. Am J Hum Genet. 1998;63:489–497. doi: 10.1086/301954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinnell B W, Padgett B L, Walker D L. J Infect Dis. 1983;147:669–675. doi: 10.1093/infdis/147.4.669. [DOI] [PubMed] [Google Scholar]
- 16.Major E O, Amemiya K, Tornatore C S, Houff S A, Berger J R. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monaco M C, Atwood W J, Gravell M, Tornatore C S, Major E O. J Virol. 1996;70:7004–7014. doi: 10.1128/jvi.70.10.7004-7012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallia G L, Houff S A, Major E O, Khalili K. J Infect Dis. 1997;176:1603–1609. doi: 10.1086/514161. [DOI] [PubMed] [Google Scholar]
- 19.Laghi L, Chauhan D P, Marra G, Major E O, Neel J V, Boland C R. Gastroenterology. 1996;110:A548. (abstr.). [Google Scholar]
- 20.Laghi L, Randolph A, Chauhan D P, Marra G, Neel J V, Boland C R. Gastroenterology. 1997;112:A598. (abstr.). [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Mannual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Frisque R J, Bream G L, Cannella M T. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller W. Proc Natl Acad Sci USA. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tornatore C, Berger J R, Houff S A, Curfman B, Meyers K, Winfield D, Major E O. Ann Neurol. 1992;31:454–462. doi: 10.1002/ana.410310426. [DOI] [PubMed] [Google Scholar]
- 25.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 26.Monaco M C, Jensen P N, Hou J, Durham L, Major E O. J Virol. 1998;72:9918–9923. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weil R, Vinograd J. Proc Natl Acad Sci USA. 1963;50:730–738. doi: 10.1073/pnas.50.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbone M, Rizzo P, Grimley P M, Procopio A, Mew D J, Shridhar V, de Bartolomeis A, Esposito V, Giuliano M T, Steinberg S M, et al. Nat Med. 1997;3:908–912. doi: 10.1038/nm0897-908. [DOI] [PubMed] [Google Scholar]
- 29.De Luca A, Baldi A, Esposito V, Howard C M, Bagella L, Rizzo P, Caputi M, Pass H I, Giordano G G, Baldi F, et al. Nat Med. 1997;3:913–916. doi: 10.1038/nm0897-913. [DOI] [PubMed] [Google Scholar]
- 30.Monini P, Rotola A, de Lellis L, Corallini A, Secchiero P, Albini A, Benelli R, Parravicini C, Barbanti-Brodano G, Cassai E. Int J Cancer. 1996;66:717–722. doi: 10.1002/(SICI)1097-0215(19960611)66:6<717::AID-IJC1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Carbone M, Rizzo P, Procopio A, Giuliano M, Pass H I, Gebhardt M C, Mangham C, Hansen M, Malkin D F, Bushart G, et al. Oncogene. 1996;13:527–535. [PubMed] [Google Scholar]
- 32.Martini F, Iaccheri L, Lazzarin L, Carinci P, Corallini A, Gerosa M, Iuzzolino P, Barbanti-Brodano G, Tognon M. Cancer Res. 1996;56:4820–4825. [PubMed] [Google Scholar]
- 33.Lednicky J A, Garcea R L, Bergsagel D J, Butel J S. Virology. 1995;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- 34.De Mattei M, Martini F, Corallini A, Gerosa M, Scotlandi K, Carinci P, Barbanti-Brodano G, Tognon M. Int J Cancer. 1995;61:756–760. doi: 10.1002/ijc.2910610603. [DOI] [PubMed] [Google Scholar]
- 35.Carbone M, Pass H I, Rizzo P, Marinetti M, Di Muzio M, Mew D J, Levine A S, Procopio A. Oncogene. 1994;9:1781–1790. [PubMed] [Google Scholar]
- 36.Bergsagel D J, Finegold M J, Butel J S, Kupsky W J, Garcea R L. N Engl J Med. 1992;326:988–993. doi: 10.1056/NEJM199204093261504. [DOI] [PubMed] [Google Scholar]
- 37.Corallini A, Pagnani M, Viadana P, Silini E, Mottes M, Milanesi G, Gerna G, Vettor R, Trapella G, Silvani V. Int J Cancer. 1987;39:60–67. doi: 10.1002/ijc.2910390111. [DOI] [PubMed] [Google Scholar]
- 38.Dorries K, Loeber G, Meixensberger J. Virology. 1987;160:268–270. doi: 10.1016/0042-6822(87)90071-7. [DOI] [PubMed] [Google Scholar]
- 39.Brinster R L, Chen H Y, Messing A, van Dyke T, Levine A J, Palmiter R D. Cell. 1984;37:367–379. doi: 10.1016/0092-8674(84)90367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strickler H D, Goedert J J, Fleming M, Travis W D, Williams A E, Rabkin C S, Daniel R W, Shah K V. Cancer Epidemiol Biomarkers Prev. 1996;5:473–475. [PubMed] [Google Scholar]
- 41.Arthur R R, Grossman S A, Ronnett B M, Bigner S H, Vogelstein B, Shah K V. J Neurooncol. 1994;20:55–58. doi: 10.1007/BF01057961. [DOI] [PubMed] [Google Scholar]
- 42.Israel M A, Martin M A, Takemoto K K, Howley P M, Aaronson S A, Solomon D, Khoury G. Virology. 1978;90:187–196. doi: 10.1016/0042-6822(78)90302-1. [DOI] [PubMed] [Google Scholar]
- 43.Wold W S, Mackey J K, Brackmann K H, Takemori N, Rigden P, Green M. Proc Natl Acad Sci USA. 1978;75:454–458. doi: 10.1073/pnas.75.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pepper C, Jasani B, Navabi H, Wynford-Thomas D, Gibbs A R. Thorax. 1996;51:1074–1076. doi: 10.1136/thx.51.11.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butel J S, Lednicky J A. J Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 46.Dyson N, Buchkovich K, Whyte P, Harlow E. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 47.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swenson J J, Trowbridge P W, Frisque R J. J Neurovirol. 1996;2:78–86. doi: 10.3109/13550289609146541. [DOI] [PubMed] [Google Scholar]