Abstract
Background
DNA viruses have a wide range of genome sizes (5 kb up to 1.2 Mb, compared to 0.16 Mb to 1.5 Mb for obligate parasitic bacteria) that do not correlate with their virulence or the taxonomic distribution of their hosts. The reasons for such large variation are unclear. According to the traditional view of viruses as gifted "gene pickpockets", large viral genome sizes could originate from numerous gene acquisitions from their hosts. We investigated this hypothesis by studying 67 large DNA viruses with genome sizes larger than 150 kb, including the recently characterized giant mimivirus. Given that horizontally transferred DNA often have anomalous nucleotide compositions differing from the rest of the genome, we conducted a detailed analysis of the inter- and intra-genome compositional properties of these viruses. We then interpreted their compositional heterogeneity in terms of possible causes, including strand asymmetry, gene function/expression, and horizontal transfer.
Results
We first show that the global nucleotide composition and nucleotide word usage of viral genomes are species-specific and distinct from those of their hosts. Next, we identified compositionally anomalous (cA) genes in viral genomes, using a method based on Bayesian inference. The proportion of cA genes is highly variable across viruses and does not exhibit a significant correlation with genome size. The vast majority of the cA genes were of unknown function, lacking homologs in the databases. For genes with known homologs, we found a substantial enrichment of cA genes in specific functional classes for some of the viruses. No significant association was found between cA genes and compositional strand asymmetry. A possible exogenous origin for a small fraction of the cA genes could be confirmed by phylogenetic reconstruction.
Conclusion
At odds with the traditional dogma, our results argue against frequent genetic transfers to large DNA viruses from their modern hosts. The large genome sizes of these viruses are not simply explained by an increased propensity to acquire foreign genes. This study also confirms that the anomalous nucleotide compositions of the cA genes is sometimes linked to particular biological functions or expression patterns, possibly leading to an overestimation of recent horizontal gene transfers.
Background
During the last decade the study of virus evolution has been neglected to the point where 'virus evolution' most often refers to studies more akin to population genetics, such as the worldwide scrutiny of new polymorphisms appearing daily in the H5N1 avian flu virus [1], than to the fundamental question of where viruses come from [2-4]. Phylogenetic studies on viruses have long been considered unfeasible for two main reasons: 1) their reputed propensity to randomly acquire genetic material from their host or the environment and 2) their reputed very high sequence divergence rate. The generality of this vision, probably inherited from the study of RNA viruses (in particular retroviruses), now deserves to be revisited for DNA viruses in light of the increasing amount of available genomic sequence data, and the recent characterization of some giant viruses [5-8].
When analyzing DNA virus genome sequences on a global scale [9] one is immediately struck by their tremendous variation in size. Even if viral DNA genomes are expected to be larger than viral RNA genomes due to the improved accuracy of the replication system, it is not as easy to understand how DNA viruses with apparently similar "fitness" (as judged from their virulence and burst sizes) may have come to exhibit sizes ranging from a few kilobases up to more than a megabase [5,6].
Even more intriguing is the fact that such a genome size variation is commonly found among viruses infecting the same or similar hosts, for prokaryotic viruses (e.g. bacteriophage ranging from 30 kb up to nearly 670 kb for bacteriophage G [10]), as well as animal viruses [from less than 5 kb (polyomaviruses) to 360 kb (poxviruses)]. Currently the largest known eukaryotic DNA viruses are plankton parasite Emiliania huxleyi virus 86 (407 kb) [7] and the amoeba-infecting mimivirus (1.2 Mb) [5].
Finally, unlike the situation for eukaryotic cellular organisms, the increase in viral genome size is not correlated with either accumulation of "junk" DNA (e.g. low complexity sequences or non-coding regions), invasion of mobile elements, gene duplication or repeat expansion [5,7,11].
In the present work, we now examine to which extent horizontal gene transfer (HGT) from host might account for the exceptional genome size of several families of large double stranded DNA viruses (LDVs) with genomes exceeding 150 kb in size.
These viruses are found in a wide variety of viral families including those classified in Nucleo-Cytoplasmic Large DNA viruses [12] (Asfarviridae, Poxviridae, Phycodnaviridae, Iridoviridae, Mimiviridae) as well as herpesviruses, nimaviruses, baculoviruses, and bacteriophages. At the time of this study, 67 LDV genomes (>150 kb) were available in public databases (Table 1). Each of these genomes encodes hundreds of predicted protein-coding genes. With this increasing body of data, it has now become possible to analyze different structural and functional aspects of those LDV genomes. Note that polydnaviruses were not analyzed due to their anomalous low gene density, atypical chromosomal organization and life style [13].
Table 1.
Proportions of cA genes in the 67 large DNA viruses.
| Classification | Virus | Genome size (bp) | Genomic G+C (%) | Number of analyzed CDS | Number of cA genes | Proportions of cA genes (%) | Co-localizationa |
| Caudovirales | Enterobacteria phage RB43 | 180500 | 43.2 | 177 | 15 | 8.47 | - |
| Enterobacteria phage T4 | 168903 | 35.3 | 175 | 8 | 4.57 | - | |
| Pseudomonas phage phiEL | 211215 | 49.33 | 178 | 18 | 10.11 | - | |
| Pseudomonas phage phiKZ | 280334 | 36.83 | 278 | 27 | 9.71 | - | |
| Mycobacterium phage Bxz1 | 156102 | 64.77 | 147 | 27 | 18.37 | - | |
| Enterobacteria phage RB69 | 167560 | 37.66 | 160 | 11 | 6.88 | * | |
| Enterobacteria phage RB49 | 164018 | 40.44 | 160 | 12 | 7.5 | - | |
| Vibrio phage KVP40 | 244835 | 42.6 | 256 | 18 | 7.03 | - | |
| Aeromonas phage 44RR2.8t | 173591 | 43.88 | 173 | 7 | 4.05 | - | |
| Aeromonas phage Aeh1 | 233234 | 42.78 | 228 | 13 | 5.7 | - | |
| Bacteriophage S-PM2 | 196280 | 37.82 | 135 | 28 | 20.74 | * | |
| Cyanophage P-SSM2 | 252401 | 35.51 | 212 | 33 | 15.57 | * | |
| Cyanophage P-SSM4 | 178249 | 36.74 | 121 | 20 | 16.53 | ** | |
| Aeromonas phage 31 | 172963 | 43.91 | 173 | 10 | 5.78 | - | |
| Bacteriophage c-st | 185683 | 26.28 | 151 | 8 | 5.3 | - | |
| Alphaherpesvirinae | Cercopithecine herpesvirus 16 | 156487 | 76.09 | 71 | 18 | 25.35 | - |
| Equid herpesvirus 1 | 150224 | 56.67 | 79 | 12 | 15.19 | - | |
| Cercopithecine herpesvirus 1 | 156789 | 74.46 | 71 | 23 | 32.39 | - | |
| Human herpesvirus 2 | 154746 | 70.39 | 72 | 21 | 29.17 | * | |
| Human herpesvirus 1 | 152261 | 68.28 | 72 | 24 | 33.33 | - | |
| Psittacid herpesvirus 1 | 163025 | 60.95 | 71 | 8 | 11.27 | - | |
| Gallid herpesvirus 2 | 174077 | 43.89 | 106 | 19 | 17.92 | * | |
| Gallid herpesvirus 3 | 164270 | 53.61 | 92 | 19 | 20.65 | *** | |
| Cercopithecine herpesvirus 2 | 150715 | 75.97 | 71 | 22 | 30.99 | * | |
| Meleagrid herpesvirus 1 | 159160 | 47.56 | 85 | 8 | 9.41 | *** | |
| Betaherpesvirinae | Pongine herpesvirus 4 | 241087 | 61.7 | 155 | 67 | 43.23 | *** |
| Human herpesvirus 6B | 162114 | 42.77 | 91 | 6 | 6.59 | - | |
| Murid herpesvirus 1 | 230278 | 58.73 | 156 | 52 | 33.33 | *** | |
| Human herpesvirus 5 strain AD169 | 230287 | 57.19 | 145 | 53 | 36.55 | *** | |
| Human herpesvirus 6 | 159321 | 42.44 | 107 | 11 | 10.28 | - | |
| Human herpesvirus 7 | 153080 | 36.22 | 78 | 8 | 10.26 | - | |
| Cercopithecine herpesvirus 8 | 221454 | 49.14 | 223 | 80 | 35.87 | - | |
| Murid herpesvirus 2 | 230138 | 61.01 | 165 | 87 | 52.73 | *** | |
| Human herpesvirus 5 strain Merlin | 235645 | 57.48 | 153 | 56 | 36.6 | *** | |
| Tupaiid herpesvirus 1 | 195859 | 66.61 | 144 | 59 | 40.97 | *** | |
| Gammaherpesvirinae | Human herpesvirus 4 | 171823 | 59.5 | 90 | 37 | 41.11 | * |
| Equid herpesvirus 2 | 184427 | 57.5 | 74 | 28 | 37.84 | - | |
| Cercopithecine herpesvirus 15 | 171096 | 61.94 | 77 | 22 | 28.57 | * | |
| Herpesvirinae Uncl. | Ostreid herpesvirus 1 | 207439 | 38.73 | 121 | 7 | 5.79 | - |
| Chordopoxvirinae | Monkeypox virus | 196858 | 33.09 | 162 | 3 | 1.85 | - |
| Camelpox virus | 205719 | 33.17 | 171 | 6 | 3.51 | ** | |
| Cowpox virus | 224499 | 33.4 | 189 | 2 | 1.06 | - | |
| Myxoma virus | 161773 | 43.56 | 147 | 5 | 3.4 | - | |
| Rabbit fibroma virus | 159857 | 39.53 | 138 | 4 | 2.9 | - | |
| Ectromelia virus | 209771 | 33.18 | 154 | 6 | 3.9 | - | |
| Variola virus | 185578 | 32.73 | 152 | 6 | 3.95 | - | |
| Molluscum contagiosum virus | 190289 | 63.36 | 139 | 43 | 30.94 | *** | |
| Canarypox virus | 359853 | 30.37 | 291 | 14 | 4.81 | - | |
| Fowlpox virus | 288539 | 30.89 | 230 | 12 | 5.22 | - | |
| Lumpy skin disease virus | 150773 | 25.91 | 136 | 4 | 2.94 | - | |
| Vaccinia virus | 194711 | 33.34 | 179 | 3 | 1.68 | - | |
| Entomopoxvirinae | M. sanguinipes entomopoxvirus | 236120 | 18.27 | 208 | 15 | 7.21 | - |
| A. moorei entomopoxvirus 'L' | 232392 | 17.78 | 218 | 16 | 7.34 | - | |
| Poxvirinae Uncl. | Mule deer poxvirus | 166259 | 26.16 | 148 | 2 | 1.35 | - |
| Asfarviridae | African swine fever virus | 170101 | 38.95 | 129 | 22 | 17.05 | ** |
| Baculovirviridae | M. configurata NPV-A | 155060 | 41.68 | 146 | 7 | 4.79 | - |
| M. configurata NPV-B | 158482 | 40.04 | 146 | 7 | 4.79 | - | |
| L. dispar MNPV | 161046 | 57.47 | 133 | 34 | 25.56 | - | |
| Xestia c-nigrum granulovirus | 178733 | 40.68 | 145 | 8 | 5.52 | - | |
| Iridoviridae | Lymphocystis disease virus | 186250 | 27.23 | 127 | 7 | 5.51 | - |
| Invertebrate iridescent virus 6 | 212482 | 28.63 | 178 | 15 | 8.43 | - | |
| Mimiviridae | A. polyphaga mimivirus | 1181404 | 27.96 | 910 | 83 | 9.12 | - |
| Phycodnaviridae | E. huxleyi virus 86 | 407339 | 40.18 | 416 | 44 | 10.58 | - |
| P. bursaria Chlorella virus 1 | 330743 | 39.97 | 426 | 117 | 27.46 | - | |
| E. siliculosus virus 1 | 335593 | 51.73 | 209 | 25 | 11.96 | - | |
| Nimaviridae | Shrimp white spot syndrome virus | 305107 | 41.01 | 245 | 96 | 39.18 | - |
| Unclassified Virus | H. zea virus 1 | 228089 | 41.86 | 104 | 25 | 24.04 | * |
a. '-' no significance; * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001.
Nucleotide composition is one of the specific properties of viral and cellular genomes [14-16]. The mimivirus genome is A+T rich (72%), and exhibits rather homogenous nucleotide compositions along the chromosome [5]. In contrast, some betaherpesviruses exhibit a clear bimodal heterogeneity in G+C composition along their genomes [17]. Different factors can shape compositional heterogeneity within and across genomes [18], including mutational biases, physical constraints on DNA molecules, functional requirements at the level of transcription [19] and translation [20-22], and genetic exchanges with other genomes by horizontal transfer [23-26]. Many of the initial surveys of LDV genomes characterized nucleotide compositional properties of individual genomes. However, there are few studies systematically addressing their compositional properties in a comparative way [27].
Here we analyzed the nucleotide compositional properties of 67 LDV genomes. We first compared global nucleotide compositions across these viruses and their hosts. We next identified compositionally anomalous (cA) genes in the viral genomes, examined their correlation with strand asymmetry, a possible cause of compositional biases, and described their functional and physical (i.e. chromosomal co-localization) properties. Finally, we investigated potential exogenous origins of the cA genes through phylogenetic tree reconstruction.
Results
Global compositional bias differs across LDVs and their hosts
G+C content is a simple measure of genomic nucleotide composition, and it has been shown to be species-specific for prokaryotes [15]. LDVs also present a large variation in global G+C content across viral families (27%–76%; Additional file 1). Large variations in G+C content are also observed within a viral family or subfamily (Additional file 2), for instance, the lumpy skin disease virus (26%) and the molluscum contagiosum virus (64%) belonging to the same chordopoxvirus subfamily. Large variations in G+C content were previously noted for herpesviruses [28].
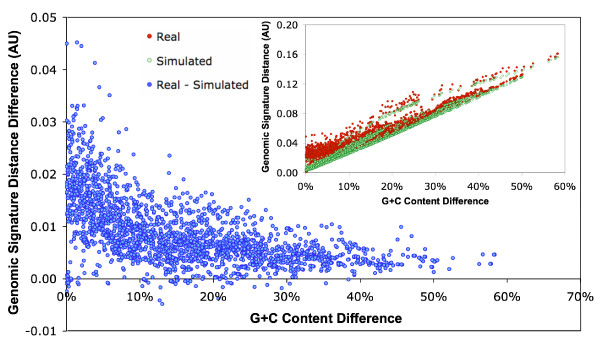
Nucleotide word frequency is a finer indicator of genomic specificity [29]. We computed the frequencies of tetra-nucleotide words in the LDV genomes (i.e. genomic signatures [30]) and genomic signature distances for all possible pairs of the 67 LDVs (see Methods). The genomic signature distance was strongly correlated with the difference in G+C content (R2 = 0.92) as a consequence of the important role of G+C content in shaping nucleotide word frequencies (Fig. 1). However, we observed a large variation in the genomic signature distance for similar differences in G+C contents. This variation suggests the existence of higher order compositional biases that are independent of G+C content. We shuffled every LDV genome sequence to erase tetra-nucleotide word biases that were independent of base compositions, and re-computed the genomic signature distances for all the pairs of the 67 randomized sequences. We observed a significant deviation in the genomic signature distances between the real and the randomized data (t-test, p < 0.001; Fig. 1). Thus LDVs maintain species-specific global compositional biases at both single and tetra-nucleotide levels, as previously observed for cellular organisms [30] and for a few viral families [28]. In other words, these LDVs have a certain level of homogeneity in nucleotide composition within their genomes, as Mrázek and Karlin previously suggested for eighteen large DNA viruses [27].
Figure 1.
Difference in genomic signature distance between real and randomized data for all possible pairs of 67 LDVs. The inset figure shows the genomic signature distances between all possible pairs of 67 LDVs for both real and randomized data.
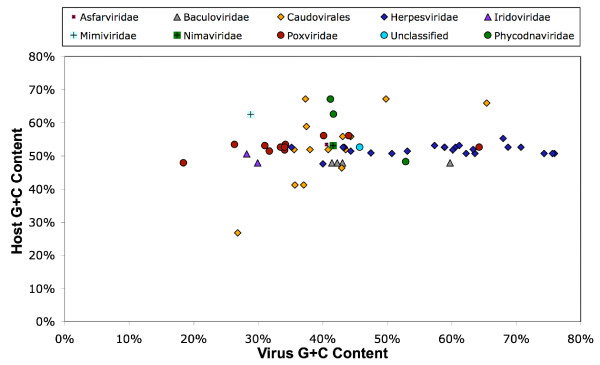
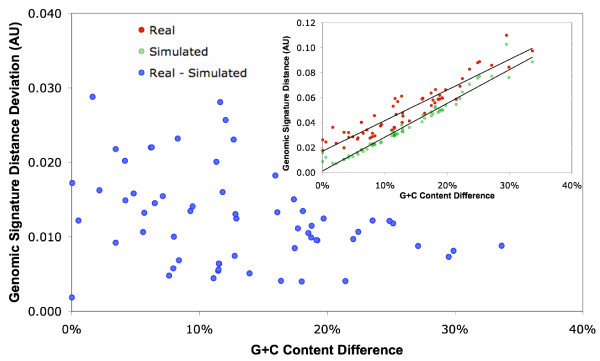
Viruses replicate intracellularly in their hosts, using the host nucleotide pool. One may expect that the global nucleotide compositional biases in viral genomes are similar to those of their hosts [31,32]. Host genomes are also potential sources of genes for viruses, as viral genomes are known to incorporate host genes. Thus the comparison of the nucleotide compositional properties between LDVs and their hosts is of particular interest. We compared nucleotide compositional biases between the LDVs and their hosts (i.e. modern hosts or their close relatives). We observed a clear lack of correlation between the G+C content of the coding regions, (G+C)CDS, of the LDV genomes, and the (G+C)CDS of the cellular host genomic sequences (Fig. 2). A genomic signature analysis also revealed distinct tetra-nucleotide word preferences between LDVs and their hosts independent of their base composition differences (Fig. 3). After a base-by-base random shuffling of the analyzed sequences, we observed a significant deviation between the real and the randomized data in the genomic signature distances for the LDV-host pairs (t-test, p < 0.001; Fig. 3). Thus the species-specific global compositional biases of LDVs are distinct from that of their hosts. This invalidates the concept that viruses tend to adapt their global DNA signatures to the host machinery. Furthermore, HGTs from their modern hosts are not as frequent, if any, as they could have influences on global nucleotide compositions of LDV genomes.
Figure 2.
Comparison of G+C content between LDVs and their hosts.
Figure 3.
Difference in genomic signature distance between real and randomized data for LDV-host pairs. The inset figure shows the genomic signature distances between the LDV-host pairs for both real and randomized data.
Compositionally anomalous genes in LDVs
We next analyzed compositional heterogeneities across different genes encoded in individual LDV genomes. Genes with nucleotide compositions substantially deviating from the average in a genome tend to be under distinct selection pressures or have particular evolutionary histories. We denote such genes as compositionally anomalous (cA) genes. To identify cA genes with a robust statistical support, we used a method [26] based on Markov modeling and Bayesian inference originally developed for the identification of horizontally transferred genes within prokaryotic genomes. Composition-based approaches have a clear limitation in detecting HGTs; they detect only a subset of HGTs (i.e. recent horizontal gene acquisitions by a recipient genome from a donor genome with nucleotide compositions that are different from the recipient genome). Our aim here is to examine the nature of cA genes in LDVs in the light of previous observations of cA genes for prokaryotic genomes.
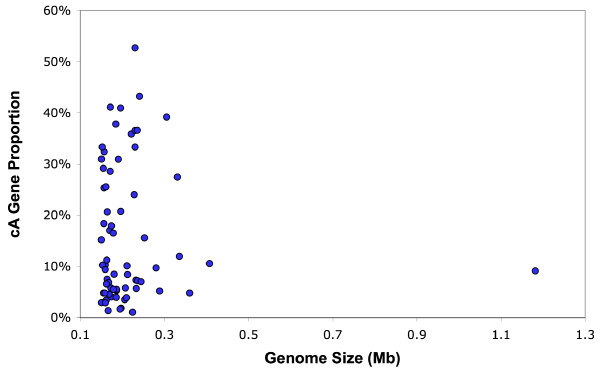
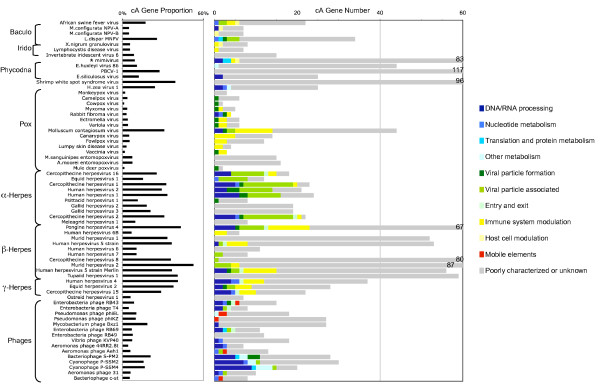
We identified cA genes in all the analyzed LDV genomes. In many LDV genomes, the cA genes were more A+T rich than the remaining genes (for 43 out of 67 LDVs; binomial test, p < 0.05). The proportion of cA genes per genome was highly variable across the 67 LDVs, ranging from 1% to 53% (Table 1, Fig. 4). In contrast, the proportion obtained by the same method is less variable for prokaryotes (0% to 25%) [9]. Herpesviruses exhibited the widest range of cA gene proportion (6% to 53%). All but two of the analyzed NCLDVs, including mimivirus, had a relatively low level of cA proportion (1%–17%). Two exceptions were Paramecium bursaria chlorella virus 1 (28%) and molluscum contagiosum virus (31%). Phages showed a relatively low proportion (4% to 21%). The cA gene proportions for other viruses were 5% to 26% for four baculoviruses, 39% for nimavirus and 24% for Heliothis zea virus 1. The very high cA proportions in some betaherpesviruses appear to be linked with genomic islands harboring betaherpesvirus-specific genes, which we will discuss below (in the "Co-localization of cA genes" section).
Figure 4.
Proportions of the cA genes detected in the 67 LDV genomes, compared with the genome sizes.
Nakamura et al. reported a significant positive correlation between the cA gene proportion and the total number of genes in a genome for prokaryotes [26]; larger prokaryotic genomes tend to more rapidly acquire foreign genes (of "anomalous nucleotide compositions") than smaller ones. In contrast, we found no such correlation for the 67 LDVs (R2 = 0.03) (Additional file 3).
We observed a weak but significant positive correlation (R2 = 0.53, p < 0.005) between the proportion of cA genes and genomic G+C content (Additional file 4). This correlation was not due to a bias induced by a given viral family or subfamily. For example, when we excluded herpesviruses, many of which exhibit both a high cA gene proportion and a high G+C content, the correlation was reduced but still remained significant (R2 = 0.24, p ≤ 0.005). Such a significant correlation was observed also within a single subfamily (i.e. alphaherpesviruses, R2 = 0.66, p ≤ 0.005). A potential variation in gene prediction quality (which could be dependent on genomic G+C compositions) does not appear to explain these correlations (Monier et al., unpublished data). We also examined a possible relationship between the cA gene proportion and chromosome topology. There was no difference in the cA gene proportions between linear LDV genomes (15.5% on average) and circular LDV genomes (17.3% on average; t-test, p = 0.63).
Our global genomic signature analysis revealed remarkably different nucleotide compositions between LDVs and their hosts. To investigate further the discrepancy of nucleotide compositions, we examined if the identified cA genes are enriched in "host-like" sequences in terms of their nucleotide compositions. We determined the genomic signature distances (di-nucleotides for word length) from individual cA genes to the viral genome where they originate as well as to the host genomic sequences. As shown in the Additional file 5, we observed a marked enrichment of genes with host-like signatures among the identified cA genes (19%) relative to non-cA genes (3.3%). The host-like signatures in these cA genes could be due to a large variation in their nucleotide compositions, being unrelated to possible host origins for some of the cA genes. To test this hypothesis, we performed the same analysis by randomizing the pairings between viruses and their hosts. A comparable fraction (23% versus 19%) of the cA genes indeed exhibited smaller distances to the genomic signatures of randomly chosen hosts than to the viral genomes where they originate. Thus the enrichment of genes with host-like signatures in cA genes may not suggest their horizontal acquisitions from hosts.
Correlation between cA genes and replication-associated strand asymmetry
Genome sequences of many prokaryotes and vertebrates [33,34] show strong strand asymmetry in nucleotide composition between the leading and the lagging strands. For most bacteria, compositional strand asymmetry is characterized by an excess of G and T bases in the leading strands relative to the lagging strands [33,35,36]. A substantial part of compositional strand asymmetry is independent of gene distribution in the two distinct DNA strands, and is probably due to mutational biases linked to asymmetric mechanisms of DNA replication [37,38]. Compositional strand asymmetry spanning large genomic segments has been described also for some large DNA viruses, including mimivirus [5,39] and several herpesviruses [40,41]. Such replication-associated strand bias can potentially result in two classes of genes with distinct nucleotide compositions, depending on which strand they are located. We thus examined if there is a correlation between the distribution of cA genes and compositional strand asymmetry.
We first generated cumulative GT-excess plots [42] for all of the 67 LDV genomes to assess their compositional strand asymmetry. LDVs presented various GT-excess patterns. Several LDVs, mostly phages, showed strong global strand asymmetry with a monotonous increase (or decrease) of the cumulative GT-excess curve along their entire genome. Some LDVs including mimivirus exhibited a few peaks in their GT-excess profiles. For other LDVs, the GT-excess curves locally fluctuated with no long genomic segments exhibiting a consistent compositional asymmetry. We selected fifteen LDVs presenting long (>10 kb) genomic segments with uniform compositional asymmetry (Additional file 6). After identifying the genomic coordinates where the sign of the nucleotide-skew (G+T versus C+A) changes, we split the genomic sequences into sub-strands. Those sub-strands were classified into two classes: class I consists of sub-strands having a positive nucleotide-skew (i.e. G+T% > C+A%), and class II with a negative skew (i.e. G+T% < C+A%). Genes were then classified into either class I or II according to the sub-strand they originated from. For instance, the plus strand (according to the GenBank entry) of the Pseudomonas phage phiKZ genome is G and T rich along its entire length, thus being classified as class I. This class I strand contains 229 genes (≥ 300 nt), of which 22 were detected as cA genes. The complementary strand is classified as a class II and contains 49 genes, of which 5 were cA genes. No significant correlation was found between the distribution of cA genes and the compositional strand asymmetry (Fisher's exact test, p-value = 0.99). The mimivirus genome shows a clear switch of nucleotide-skew (G+T versus C+A) at position 380,000 nt as previously noted [5]. Thus a part of the plus strand (from 0 to 380,000 nt) and a part of the complementary strand (from 380,000 to 1,118,404 nt) constitute the class I strands. Class II is represented by the remaining strands (Additional file 6). Again, no significant correlation was found between the distribution of cA genes and the compositional strand asymmetry (p-value = 0.63). Of the fifteen LDVs that we analyzed, only three showed a significant correlation between cA gene distribution and strand asymmetries (p-values < 0.05). The three viruses are the fowlpox virus and two strains of the human herpesvirus 5 (the wild type strain merlin and the laboratory strain AD169). These results suggest that replication-associated compositional strand asymmetry accounts for only a small part of the nucleotide compositional biases of the cA genes observed in LDVs.
Co-localization of cA genes
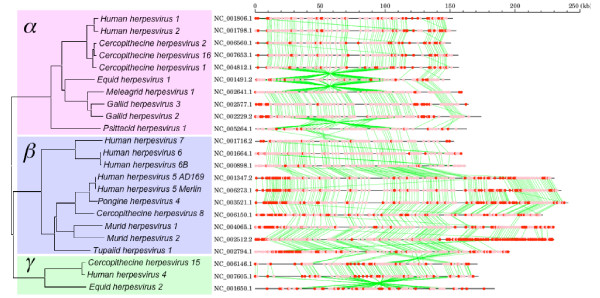
In prokaryotes, horizontally transferred genes are often clustered in the chromosome and make genomic islands [26,43,44]. We investigated co-localization of the cA genes along the chromosomes of the LDVs using a Monte-Carlo simulation (see Methods). None of the genomes of entomopoxviruses, baculoviruses, iridoviruses, mimivirus, nimavirus or phycodnaviruses exhibited a significant propensity for cA genes to be clustered along the genome. Of the 67 LDVs, only 12 presented a significant frequency of pairs of co-localized cA genes (p ≤ 0.01; Table 1). Using the same Monte-Carlo simulation, we identified significantly large sized cA gene islands (ranging from 3 up to 32 neighboring cA genes) in 11 LDV genomes (p ≤ 0.01; Additional file 7). Those were 6 betaherpesviruses, 1 alphaherpesvirus, 2 chordopoxviruses, African swine fever virus, and the cyanophage P-SSM4. Betaherpesviruses showing cA islands of significant sizes include murid herpesvirus 2 with an extravagant 53% of cA gene proportion. The abundance of cA gene islands in betaherpesviruses parallels the mosaicity in G+C composition along their genomes [17]. It is known that betaherpesvirus genomes are composed of core genes located in the middle of their genomes and shared by different subfamilies of Herpesviridae, plus betaherpesvirus-specific genes located at the extremities of the genomes [45]. As shown in Fig. 5, many of the beta-family genes were identified as cA genes making genomic islands.
Figure 5.
Gene organization and the locations of the cA genes for herpesviruses. The phylogenetic reconstruction (on the left) is based on the DNA polymerase catalytic subunit sequences. Red and pink dots (on the right) correspond to the cA genes and the remaining genes, respectively. Green lines show orthologous gene relationships defined by BLAST reciprocal best hit.
Correlation between cA genes and functional properties
Functional constraint on specific genomic regions can be a cause of atypical nucleotide compositions of genes. We selected five genomic regions to examine if there were correlations between cA genes and certain of their functional attributes. For the first three cases (gamma- and betaherpseviruses), the correlation between the nucleotide compositional biases and gene functions has already been described, and could serve as a positive control for our method. For the last two cases (Emiliania huxleyi virus-86 and mimivirus), no such correlation has been previously reported.
Human herpesvirus 4 (Epstein-Barr virus) of the gammaherpesvirus subfamily has two different life cycle modes: latent and productive. Nine genes are known to be expressed during latency [46]. Those genes are hardly expressed during the productive mode. They are highly A+T rich compared to the remaining genes. Karlin et al. hypothesized that the unique codon usage of those genes helps to minimize the competition with host genes for various host resources during latency [19]. We found that eight of the nine latency genes were indeed detected as cA genes (89%). In contrast, the cA gene proportion was significantly lower for the remaining genes (29/81, 36%, p = 0.003).
The immediate-early transcription region (≈10-kb) of the murid herpesvirus 1 (murine cytomegalovirus, betaherpesvirus subfamily) genome is known to be CpG suppressed [47]. Experimental data suggest that the methylation of the cytosines in CpG dinucleotides in the enhancer/promoter of these regions has a regulatory role in gene expression [48]. The CpG suppressed region of the murid herpesvirus 1 genome encodes 10 genes. We found that the cA gene proportion in this region (6/10 genes, 60%) was twice as important as in the remaining regions (46/146, 32%) of the genome, though their difference might be due to chance (p = 0.085).
The murid herpesvirus 1 genome possesses three (19-kb, 10-kb and 17-kb) regions that are A+T rich relative to other parts of the genome [47]. Those regions are enriched in genes encoding membrane glycoprotein (e.g. m02 and m145 families). Some of those proteins were suggested as responsible for the evasion from natural killer cell-mediated immune surveillance through their interaction with inhibitory natural killer cell receptors [49,50]. Of 37 genes within the A+T rich regions, 67% (24 genes) were detected as cA genes, which is significantly higher than the cA gene proportion for the remaining genes (23.5%, 28/119 genes, p < 10-5). It is uncertain if the increased A+T levels of these regions have functional roles. However, it should be noted that these A+T rich regions contain five of the seven genes exhibiting significant sequence polymorphisms between strains of murid herpesvirus 1 [51].
Emiliania huxleyi virus-86 (EhV-86) of the Phycodnaviridae family exhibits two distinct transcription phases during its lytic infection to the host alga, E. huxleyi [52]. The primary phase is characterized by the transcriptions of a group of genes by 1 hour post-infection. The secondary phase involves the transcriptions of other genes between 2 and 4 hours post-infection. Respectively thirty-eight and 253 genes in our data set (ORF length ≥ 100 codons) correspond to the primary and the secondary transcription phases. We found a significantly greater fraction of cA genes in the primary phase (8/38, 21%) than in the secondary phase genes (19/253, 7.5%; p = 0.014). Genes expressed during the primary phase map in the 104-kb central genomic region (bases 200,000 to 304,000), which shows a similar G+C content as the rest of the genome [53]. It has been suggested that promoter-like elements (family A repeats) uniquely found in this region control this early expression pattern of EhV-86 [7]. The functions of the early expression are unknown as most of the transcribed genes lack detectable homologs in the databases. Allen et al. postulated that an ancestral EhV acquired the 104-kb region by horizontal transfer [53].
Mimivirus has unusually well conserved promoter-like AAAATTGA motifs in the upstream regions of half of its predicted genes in the genome [39]. Based on the putative associated gene functions, Suhre et al. predicted that these promoter-like elements regulate gene expression during the early stages of the viral infection, whilst most of the genes contributing to the virus particle are devoid of this motif [54]. Of 402 genes with the promoter-like motifs in our data set, 43 (10.6%) were detected as cA genes. Of 508 genes lacking the motifs, 40 (7.9%) were detected as cA genes. The difference is not statistically significant (p = 0.16).
In summary, we found a significant correlation between cA genes and previously described functional categories of genes for three of the five cases. It should be noted that in the case of EhV, no relationship between expression timing and gene nucleotide composition was previously reported. This suggests the possible use of nucleotide composition analysis to predict expression patterns of viral genes. These results indicate that anomalous nucleotide compositions of some of the cA genes can be due to functional constraints, although the distinction between functional constrains and horizontal transfer events is generally difficult given the known bias in functions of horizontally transferred genes in prokaryotes [24,26].
Many eukaryotic LDVs exhibit cA genes with putative functions associated with host defense systems
We classified the cA genes according to their putative functions based on their annotations and further similarity searches against sequence databases (Additional file 8, Fig. 6). The vast majority of the cA genes have no predicted functions. A notable feature among the remaining cA genes concerns eukaryotic LDVs. We observed that many eukaryotic LDVs possess cA genes putatively associated with the control of host defense systems, such as innate/adaptive immune systems or apoptosis pathways (Additional file 9). Poxviruses, betaherpesviruses and gammaherpesviruses are particularly rich in cA genes of this category (p ≤ 10-5). They present cA genes having host homologs such as cytokine (interleukin), chemokine and MHC class I genes [55]. Remarkably, in nearly all the families of eukaryotic viruses (except for phycodnaviruses), we found species exhibiting cA genes encoding proteins putatively involved in apoptosis pathways (Additional file 9). This suggests a possible central role of apoptotic pathways in eukaryotic LDV-host interactions. Those cA genes may endow these large DNA viruses a capacity to prolong the integrity of the host cells and thereby "buy time" to pursue their replication cycles. Alternatively, viruses may induce host apoptosis to facilitate host cell exit as previously suggested for EhV-86 [7]. The biased nucleotide compositions of cA genes associated with host defense systems may be due to their relatively recent origins (i.e. horizontal gene transfer) and/or particular functional constraints on their nucleotide compositions.
Figure 6.
Functional categories of the cA genes detected in the 67 LDV genomes.
We found that the protein product from a cA gene in mimivirus (MIMI_L211 previously annotated as unknown function) exhibits significant sequence similarities to the C-terminal half of the etoposide-induced 24 (EI24) proteins (pfam07264, E-value < 10-7). EI24 (also known as PIG8) is directly induced by p53, a critical tumor suppressor coordinating DNA repair, cell-cycle arrest and apoptosis in response to cellular stresses. It has been suggested that EI24 acts as a pro-apoptotic factor and prevents tumor spreading in mammals [56]. The N-terminal part of the EI24, which is missing in MIMI_L211, binds to Bcl-2, while the function of the C-terminus of EI24 is unknown. This is the first identification of an EI24-like domain in a viral genome. The presence of such a domain in mimivirus is puzzling since, to the best of our knowledge, no apoptotic phenomenon has been described for its unicellular host Acanthamoeba polyphaga. It is notable that another amoeba Dictyostelium discoideum, which carries out apoptotic processes, possesses a hypothetical ORF (Q54PW9_DICDI) matching the EI24 domain (E-value < 10-19).
Phylogenetic evidences for horizontal gene transfers
Horizontally acquired genes may or may not exhibit anomalous nucleotide compositions depending on their origin. Phylogenetic reconstruction is a more powerful approach to examine horizontal gene transfers. However, the approach could be used only for a limited part of our data set, as most of the cA genes have no detectable homologs in the databases. Wherever possible, we systematically conducted phylogenetic reconstructions for the cA genes. Out of a total of 1633 phylogenetic reconstruction, only eight candidate horizontal gene transfer events were supported by the reconstructed trees (using conservative criteria, see Materials and Methods). All these trees revealed striking phylogenetic incongruities (Additional file 10): glutathione peroxidase of molluscum contagiosum virus (Additional file 10) (A), interleukin 10 of Cercopithecine herpesvirus 15 (B), hypothetical protein (R1R) of monkeypox virus (C), G protein-coupled receptor of fowlpox virus (D), photosystem II D2 protein (E) and site-specific DNA methylase (F) of bacteriophage S-PM2, thymidylate synthase of cyanophage P-SSM2 (G) and cytosine methyl-transferase of cyanophage P-SSM4 (H).
Discussion
We demonstrated that there is no significant correlation between the G+C content of LDV genomes and that of their modern hosts, and that the genomic signatures of LDVs are also significantly different from those of the hosts. Recently, Mrázek and Karlin reported a similar result for large DNA viruses based on a smaller data set than the one used in this study [27]. This simple observation does not favor the "gene pickpockets" depiction of viruses which supposedly frequently acquire genetic material from their hosts [57,58]. Various factors can potentially account for the existence of species-specific global nucleotide compositions [18,22]. Most importantly, LDVs carry their own genes for the major components of replication machinery. Several authors have suggested that viruses are ancient and that their evolutionary trajectory could be largely dissociated from those of their present hosts [2-4]. Accordingly, specific genomic signatures of LDV genomes may originate from intrinsic properties of their replication machinery. As an alternative, we speculate that viruses may take advantage of the compositional differences to re-orient host machineries towards viral DNA/RNA molecules.
We found no significant correlation between the cA gene proportion and the size of the LDV genomes. This observation suggests that extremely large sizes of the genomes of some LDVs such as mimivirus are not due to recent accretion of foreign genes. By extrapolation, the capacity to capture foreign genes is unlikely to be the major factor that determines the tremendous variation in genome size for DNA viruses [6].
We showed that the degree of intra-genome heterogeneity was highly variable across LDVs (i.e. the proportion of cA genes ranging from 1% to 53%). In prokaryotes, anomalous gene nucleotide composition is usually attributed to horizontal gene acquisition events. It has been argued that traces of horizontal gene transfer events as old as 100 million years can be detected through the identification of anomalies in nucleotide composition for bacterial genes [44]. The composition-based method used in this study identified horizontally transferred gene candidates for prokaryotes, which constitute 0% to 25% of their gene complements. Surprisingly the same method identified only a comparable level of cA gene proportions for half of the analyzed LDVs. Furthermore, several LDVs exhibited a cA proportion as low as that of small parasitic intracellular bacteria, rarely exchanging genes with other bacteria. Thus, the nucleotide compositions of LDVs could indicate no general differences between LDVs and prokaryotes in the frequency of horizontal gene acquisitions.
Most of the identified cA genes were of unknown functions and did not exhibit cellular homologs in the databases. This also argues against massive HGTs from hosts to LDVs. One may speculate that cellular homologs could not be detected due to a faster viral sequence evolution. However, a recent study showed that evolutionary rates are similar between LDV genes with database homologs and those lacking detectable homologs (i.e. ORFans) [8].
Furthermore, functional and structural constraints can also cause intra-genome compositional heterogeneity. In this study, we explored several mechanisms that could potentially cause compositional anomaly in LDV genomes. These mechanisms include specific gene functions and replication associated strand asymmetry. We validated a significant enrichment of cA genes in specific functional categories of genes for some of the LDVs including EhV-86. We evidenced the presence of a nucleotide composition bias in the EhV-86 genes expressed in the primary phase that was previously overlooked. In contrast, we showed that replication associated compositional asymmetry is not a major cause of cA gene compositional bias for most of the LDVs.
It should be noted, that highly expressed genes exhibiting biased codon usages are usually pre-excluded in quantifying the number of horizontally transferred genes in prokaryotic genomes [26]. Due to the lack of such a general knowledge on viral codon usage bias with functional consequences, we did not apply such a filter in our computational identification of cA genes. In this regard, possible functional constraints on the nucleotide compositions will tend to contribute to an overestimation of recent horizontal gene transfers in viruses relative to prokaryotes.
The lack of database homologs or phylogenetic evidences does not exclude the possible HGT-origins of cA genes. The hugely diverse world of viruses is probably the most underrepresented in the current database [59]. Thus, we speculate that a significant part of these cA genes might have been transferred from other viruses that are not yet sequenced. Such inter-virus gene exchange can be achieved by illegitimate or homologous recombination [60-62] occasionally leading to the fusion of two viral genomes [63], or with the aid of mobile elements [64] upon co-infection of a cell by multiple virus species. Li et al. described a clear case of HGT from Xestia c-nigrum granulovirus to Mamestra configurata nucleopolyhedrovirus B [65], though this HGT event was not detected by our study due to the similar nucleotide compositions of these two genomes.
Goldenfeld and Woese pointed out the important role of viruses in the gene flux among microbial communities [66]. They conceptualized the viral gene pool as a large dynamic repository of genetic information accessible to microorganisms. Inter-virus gene transfer may be central to the dynamics of the viral gene pool. Given the known diversity of genetic material in the virus world and their under-representation in the current sequence databases [67], this hypothesis appears as plausible as the functional/structural scenario in explaining the existence of compositionally anomalous genes in large DNA viruses.
Conclusion
The genomes of large DNA viruses exhibit nucleotide compositions largely differing from their host genomes. Our results based on nucleotide composition analyses, database searches and phylogenetic tree reconstructions suggest that horizontal gene transfer to these viruses from their current hosts is infrequent and does not account for the large variation in their genome size. However, the viral genomes still show a variable proportion of compositionally anomalous genes. Such compositional biases potentially arise from particular biological functions at the nucleic acid level and/or inter-virus gene transfers.
Methods
Viral genome data
Viral genome data were downloaded from the viral section of the NCBI Reference Sequence (RefSeq) database [68]. We have selected 67 non-redundant double-strand DNA viral genomes larger than 150 kb from the dataset (Table 1).
Host sequence data
We prepared 32 sets of protein coding sequence (CDS) data for viral hosts (Table S6). For viral hosts species for which complete genome sequences are available, we downloaded sequence data from KEGG [69]. For other viral hosts, we retrieved CDS data from GenBank [70]. We used only CDS longer than or equal to 300 bp. For some of the CDS data sets built from GenBank, we included sequences from organisms that are closely related to the viral host species (Additional file 11). We removed the sequence redundancy in each CDS data set from GenBank by keeping only one representative from a cluster of homologs. Clustering of homologous protein sequences was performed using BLASTCLUST [71]; we used 40% for the minimal sequence identity (-S option) and 75% for the minimal coverage by alignment (-L option) for at least one sequence (-b option). Of the 32 host sequence data sets, 26 sets were composed of more than 20 sequences, and used in this study. These represent the host sequence data for 61 LDV genomes.
Viral ORF and intergenic sequence data
Based on the RefSeq annotation, we prepared a dataset for CDSs and non-coding sequences (NCDSs) for viral genomes. We retained only CDSs longer than or equal to 300 bp, and discarded shorter ones.
Genomic signature distance
For the comparison of statistical nucleotide sequence properties of CDSs among viruses and their hosts, we used the method proposed by Sandberg et al. [30]. This method defines a genomic signature as a set of frequencies of all overlapping oligonucleotides of specific length k (k = 4 in this study) in the forward strand of the whole nucleotide sequences available for a given genome. The distance between two genomic signatures (X and Y) is calculated as an Euclidian distance DE, using the following formula:
where n = 256 for k = 4.
Identification of compositionally anomalous genes
To detect cA genes in LDV genomes, we used the computer program developed by Nakamura et al. [26]. We used a Markov order of 3 with a 96 bp window sliding with a step size of 12 bp along the genome. A Monte-Carlo simulation was used to compute the statistical significance of the nucleotide composition bias in a gene as in Nakamura et al.. The threshold for the statistical was set to 1% significance (unilateral statistical test). In addition to the dataset derived from RefSeq, a control dataset was generated from the viral genomic sequences by conserving only open reading frames longer than or equal to 300 bp that exhibit significant sequence similarity hits to database sequences from cellular organisms or distantly related virus, using BLASTP [71] against SwissProt/TrEMBL [72] and the viral RefSeq databases. The proportions of the cA genes for the control dataset varied from 0% to 54% across different viruses, and correlated with the cA gene proportions for the dataset derived from RefSeq (R2 = 0.73, p < 0.001, not shown). In the manuscript we show only the results obtained from the RefSeq dataset.
Statistical test for cA genes co-localization
We conducted two distinct statistical tests for physical proximity of cA genes in a viral genome (i.e. clustering of genes along the chromosome) using a Monte-Carlo simulation. First, we counted the number of pairs (n) of cA genes that were consecutively encoded in the genome. We shuffled the order of genes in the genome 1000 times and obtained the distribution of n in the randomized data. The distribution was used to determine p-value for n from the real data. Second, we recorded the size (number of genes, N) of each cA gene cluster. From the Monte-Carlo simulation, we obtained the p-value for the observation of at least one cA gene cluster with the size equal to or greater than N genes.
Phylogenetic tree reconstruction
All the cA genes were searched against the SwissProt/TrEMBL database using BLASTP (E-value < 10-5) to identify homologous sequences. For each gene, we retrieved 50 best hits at the maximum from the database, and aligned them to the candidate sequence using MUSCLE [73]. We improved these initial alignments by removing highly divergent or fragmented sequences with visual inspection of the alignments. The alignments were used to generate neighbor-joining (NJ) trees using CLUSTALW [74] with Kimura's correction for distance. Some of the NJ trees were selected for further analyses by the neighbor-joining method with Jones-Taylor-Thornton (JTT) substitution model [75] using MEGA [76] and the maximum likelihood method by PHYML [77]. The results of the BLAST searches were also served to assign putative functions to the cA genes where the annotations in the RefSeq database were inadequate for our analysis.
Phylogenetic evidences for HGT
Horizontal gene transfer (HGT) is usually invoked, when a phylogenetic tree based on gene sequences is incongruent with a species tree. In this study, we assumed deep phylogenetic origins for large DNA virus lineages (i.e. before the divergence of major phyla of cellular organisms), which have been indicated by the analyses conserved genes (e.g. DNA polymerase) [5,78]. To list up candidates of HGT, we first identified cases where the grouping of viral sequences, V, and their cellular homologs, I, is supported with a high bootstrap value (i.e. ≥ 80% for (V, I)]). As the reconstructed tree is unrooted, V could represent an outgroup of I and other cellular homologs. In this case, HGT between viruses, V, and cellular organisms, I, is not required to account for the gene tree topology. To eliminate this possibility, we demanded one of the following two additional criteria for potential HGT events.
Additional criteria 1
- From independent evidences, one can chose a group of sequences, O1, representing an outgroup of I.
- From independent evidences, one can chose a group of sequences, O2, representing an outgroup of I and O1. [Consequently, the tree topology will become (O2, (O1, (V, I))), when rooted by O2.]
Additional criteria 2
- There are only a few species and/or genera in I.
- Other sequences outside the (V, I) group are from a wide variety of genera.
- The branches from the root of (V, I) to V and I are relatively shorter than the branches from the root of (V, I) to other peripheral nodes.
Authors' contributions
AM performed most of the analyses. HO designed most of the experiments and performed part of the analyses. JMC designed part of the experiments. All authors contributed to the writing of the manuscript.
Supplementary Material
Genomic G+C content of different groups of LDVs.
Genomic G+C content of the 67 LDVs.
Comparison of the cA gene proportions with the total number of genes encoded in each LDV genomes.
Comparison of the cA gene proportions with genomic G+C content.
Comparison of the nucleotide composition between cA genes and host genes. We used genes from 61 LDV genomes with sufficient amount of host CDS data. For every viral gene, genomic signature distances (i.e. Euclidian distances based on di-nucleotide frequencies) from viral genome (horizontal axis) and from host genome (vertical axis) were computed. (A): non-cA genes. (B) and (C): cA genes. In (C), the vertical axis corresponds to the distance between the cA genes and a randomly chosen host genome.
Sub-strand classification, according to their nucleotide-skew, for 15 LDV genomes.
cA gene clusters of significant size in 11 LDV genomes.
List of the cA genes in the 67 LDV genomes.
Summary of viral genes involved in the control of host defense system.
Phylogenetic trees supporting the HGT events for the cA genes detected by a composition-based approach. (A) Glutathione peroxidase, (B) Interleukin 10, (C) R1R hypothetical protein, (D) G protein-coupled receptor, (E) Photosystem II D2 protein, (F) Site-specific DNA methylase, (G) Thymidylate synthase and (H) Cytosine methyltransferase. Each node is labeled with SWISS-PROT/TrEMBL entry, the taxonomic group (A for Archaea, B for Bacteria, E for Eukaryota and V for Virus) and the specie name. The viral genes detected as cA genes by the method of the Nakamura et al. are highlighted in red. "I" corresponds to a clade branching with the viral cA genes; "O1" corresponds to the outgroup of I, and "O2" corresponds to the outgroup of I and O1. Paralogs, orthologs and homologs indicate the relationships between the outgroups (O1 or O2) and the internal groups (I or (I, O1)). (A-E) satisfy the criterion 1, and (F-H) satisfy the criterion 2 (see Methods). Tree reconstruction was carried out by the maximum likelihood method. Bootstrap values are indicated as the branch levels.
Nucleotide sequence data for the hosts (or its close relatives) of the 67 LDVs.
Acknowledgments
Acknowledgements
We thank Prof. Y. Nakamura for providing the computer program for the identification of cA genes. We also thank Dr. Pascal Hingamp for his detailed reading of the manuscript. We thank three anonymous reviewers for their comments. AM is partially supported by the EuroPathoGenomics European network of excellence. This work was partially supported by Marseille-Nice Genopole and the French National Network (RNG)
Contributor Information
Adam Monier, Email: Adam.Monier@igs.cnrs-mrs.fr.
Jean-Michel Claverie, Email: Jean-Michel.Claverie@igs.cnrs-mrs.fr.
Hiroyuki Ogata, Email: Hiroyuki.Ogata@igs.cnrs-mrs.fr.
References
- Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman DJ, Fraser CM, Taubenberger JK, Salzberg SL. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc Natl Acad Sci U S A. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of "giant" virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Ogata H, Claverie JM. Unique genes in giant viruses: Regular substitution pattern and anomalously short size. Genome Res. 2007;17:1353–1361. doi: 10.1101/gr.6358607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Viral Genome Resources http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/viruses.html
- Donelli G. Isolamento di un batteriofago di eccezionali dimensioni attivo su B.megatherium. Atti Accad Naz Lincei--Rend Clas Sci Fis Mat Nat. 1968. pp. 95–97.
- Suhre K. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J Virol. 2005;79:14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, Martins N, Poirie M, Periquet G, Drezen JM. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science. 2004;306:286–289. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The human genome: organization and evolutionary history. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell BE, Campbell AM, Karlin S. Similarities and dissimilarities of phage genomes. Proc Natl Acad Sci U S A. 1996;93:5854–5859. doi: 10.1073/pnas.93.12.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L, Kledal TN, Karlin S, Mocarski ES. Predicting coding potential from genome sequence: application to betaherpesviruses infecting rats and mice. J Virol. 2005;79:7570–7596. doi: 10.1128/JVI.79.12.7570-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol. 1998;1:598–610. doi: 10.1016/S1369-5274(98)80095-7. [DOI] [PubMed] [Google Scholar]
- Karlin S, Blaisdell BE, Schachtel GA. Contrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J Virol. 1990;64:4264–4273. doi: 10.1128/jvi.64.9.4264-4273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao KN, Gu W, Fang NX, Saunders NA, Frazer IH. Gene codon composition determines differentiation-dependent expression of a viral capsid gene in keratinocytes in vitro and in vivo. Mol Cell Biol. 2005;25:8643–8655. doi: 10.1128/MCB.25.19.8643-8655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence G. Gene transfer, speciation, and the evolution of bacterial genomes. Curr Opin Microbiol. 1999;5:519–523. doi: 10.1016/S1369-5274(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Karlin S. Distinctive features of large complex virus genomes and proteomes. Proc Natl Acad Sci U S A. 2007;104:5127–5132. doi: 10.1073/pnas.0700429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Mocarski ES, Schachtel GA. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J Virol. 1994;68:1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Cardon LR. Computational DNA sequence analysis. Annu Rev Microbiol. 1994;48:619–654. doi: 10.1146/annurev.mi.48.100194.003155. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Branden CI, Ernberg I, Coster J. Quantifying the species-specificity in genomic signatures, synonymous codon choice, amino acid usage and G+C content. Gene. 2003;311:35–42. doi: 10.1016/S0378-1119(03)00581-X. [DOI] [PubMed] [Google Scholar]
- Kliman RM, Bernal CA. Unusual usage of AGG and TTG codons in humans and their viruses. Gene. 2005;352:92–99. doi: 10.1016/j.gene.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Glazko G, Mushegian A. Protein repertoire of double-stranded DNA bacteriophages. Virus Res. 2006;117:68–80. doi: 10.1016/j.virusres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/S0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/PL00006428. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A, Viari A. Universal replication biases in bacteria. Mol Microbiol. 1999;32:11–16. doi: 10.1046/j.1365-2958.1999.01334.x. [DOI] [PubMed] [Google Scholar]
- Rocha E. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 2002;10:393–395. doi: 10.1016/S0966-842X(02)02420-4. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Ongoing evolution of strand composition in bacterial genomes. Mol Biol Evol. 2001;18:1789–1799. doi: 10.1093/oxfordjournals.molbev.a003966. [DOI] [PubMed] [Google Scholar]
- Suhre K, Audic S, Claverie JM. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci U S A. 2005;102:14689–14693. doi: 10.1073/pnas.0506465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J, Karlin S. Strand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci U S A. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. Strand-specific compositional asymmetries in double-stranded DNA viruses. Virus Res. 1999;60:1–19. doi: 10.1016/S0168-1702(98)00139-7. [DOI] [PubMed] [Google Scholar]
- Freeman JM, Plasterer TN, Smith TF, Mohr SC. Patterns of genome organization in bacteria. Science. 1998;279:1827a. doi: 10.1126/science.279.5358.1827a. [DOI] [Google Scholar]
- Hsiao WW, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FS. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1:e62. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- Young LS, Dawson CW, Eliopoulos AG. The expression and function of Epstein-Barr virus encoded latent genes. Mol Pathol. 2000;53:238–247. doi: 10.1136/mp.53.5.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosch S, Stein J, Staak K, Liebenthal C, Volk HD, Kruger DH. Inactivation of the very strong HCMV immediate early promoter by DNA CpG methylation in vitro. Biol Chem Hoppe Seyler. 1996;377:195–201. doi: 10.1515/bchm3.1996.377.3.195. [DOI] [PubMed] [Google Scholar]
- Oliveira SA, Park SH, Lee P, Bendelac A, Shenk TE. Murine cytomegalovirus m02 gene family protects against natural killer cell-mediated immune surveillance. J Virol. 2002;76:885–894. doi: 10.1128/JVI.76.2.885-894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- Smith LM, Shellam GR, Redwood AJ. Genes of murine cytomegalovirus exist as a number of distinct genotypes. Virology. 2006;352:450–465. doi: 10.1016/j.virol.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Allen MJ, Forster T, Schroeder DC, Hall M, Roy D, Ghazal P, Wilson WH. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol. 2006;80:7699–7705. doi: 10.1128/JVI.00491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Schroeder DC, Holden MT, Wilson WH. Evolutionary history of the Coccolithoviridae. Mol Biol Evol. 2006;23:86–92. doi: 10.1093/molbev/msj010. [DOI] [PubMed] [Google Scholar]
- Renesto P, Abergel C, Decloquement P, Moinier D, Azza S, Ogata H, Fourquet P, Gorvel JP, Claverie JM. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Latour S. The SLAM family of immune-cell receptors. Curr Opin Immunol. 2003;15:277–285. doi: 10.1016/S0952-7915(03)00041-4. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ayer RE, Davis SL, Ames SJ, Florence B, Torchinsky C, Liou JS, Shen L, Spanjaard RA. Apoptosis factor EI24/PIG8 is a novel endoplasmic reticulum-localized Bcl-2-binding protein which is associated with suppression of breast cancer invasiveness. Cancer Res. 2005;65:2125–2129. doi: 10.1158/0008-5472.CAN-04-3377. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Virology: Gulliver among the Lilliputians. Curr Biol. 2005;15:R167–9. doi: 10.1016/j.cub.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. Comment on "The 1.2-megabase genome sequence of Mimivirus". Science. 2005;308:1114; author reply 1114. doi: 10.1126/science.1110820. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F. Recombination in alphaherpesviruses. Rev Med Virol. 2005;15:89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- Tyler SD, Severini A. The complete genome sequence of herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J Virol. 2006;80:1214–1221. doi: 10.1128/JVI.80.3.1214-1221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The organization of the herpes simplex virus genomes. Annu Rev Genet. 1979;13:25–57. doi: 10.1146/annurev.ge.13.120179.000325. [DOI] [PubMed] [Google Scholar]
- Karlin S, Blaisdell BE. A model for the development of the tandem repeat units in the EBV ori-P region and a discussion of their possible function. J Mol Evol. 1987;25:215–229. doi: 10.1007/BF02100015. [DOI] [PubMed] [Google Scholar]
- Li L, Donly C, Li Q, Willis LG, Keddie BA, Erlandson MA, Theilmann DA. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology. 2002;297:226–244. doi: 10.1006/viro.2002.1411. [DOI] [PubMed] [Google Scholar]
- Goldenfeld N, Woese C. Biology's next revolution. Nature. 2007;445:369. doi: 10.1038/445369a. [DOI] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. The Marine Viromes of Four Oceanic Regions. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–4. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–7. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A mutation data matrix for transmembrane proteins. FEBS Lett. 1994;339:269–275. doi: 10.1016/0014-5793(94)80429-X. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Villarreal LP, DeFilippis VR. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74:7079–7084. doi: 10.1128/JVI.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic G+C content of different groups of LDVs.
Genomic G+C content of the 67 LDVs.
Comparison of the cA gene proportions with the total number of genes encoded in each LDV genomes.
Comparison of the cA gene proportions with genomic G+C content.
Comparison of the nucleotide composition between cA genes and host genes. We used genes from 61 LDV genomes with sufficient amount of host CDS data. For every viral gene, genomic signature distances (i.e. Euclidian distances based on di-nucleotide frequencies) from viral genome (horizontal axis) and from host genome (vertical axis) were computed. (A): non-cA genes. (B) and (C): cA genes. In (C), the vertical axis corresponds to the distance between the cA genes and a randomly chosen host genome.
Sub-strand classification, according to their nucleotide-skew, for 15 LDV genomes.
cA gene clusters of significant size in 11 LDV genomes.
List of the cA genes in the 67 LDV genomes.
Summary of viral genes involved in the control of host defense system.
Phylogenetic trees supporting the HGT events for the cA genes detected by a composition-based approach. (A) Glutathione peroxidase, (B) Interleukin 10, (C) R1R hypothetical protein, (D) G protein-coupled receptor, (E) Photosystem II D2 protein, (F) Site-specific DNA methylase, (G) Thymidylate synthase and (H) Cytosine methyltransferase. Each node is labeled with SWISS-PROT/TrEMBL entry, the taxonomic group (A for Archaea, B for Bacteria, E for Eukaryota and V for Virus) and the specie name. The viral genes detected as cA genes by the method of the Nakamura et al. are highlighted in red. "I" corresponds to a clade branching with the viral cA genes; "O1" corresponds to the outgroup of I, and "O2" corresponds to the outgroup of I and O1. Paralogs, orthologs and homologs indicate the relationships between the outgroups (O1 or O2) and the internal groups (I or (I, O1)). (A-E) satisfy the criterion 1, and (F-H) satisfy the criterion 2 (see Methods). Tree reconstruction was carried out by the maximum likelihood method. Bootstrap values are indicated as the branch levels.
Nucleotide sequence data for the hosts (or its close relatives) of the 67 LDVs.