Abstract
As the principal iron-regulatory hormone, hepcidin plays an important role in systemic iron homeostasis. The regulation of hepcidin expression by iron loading appears to be unexpectedly complex and has attracted much interest. The GPI-linked membrane protein hemojuvelin (GPI-hemojuvelin) is an essential upstream regulator of hepcidin expression. A soluble form of hemojuvelin (s-hemojuvelin) exists in blood and acts as antagonist of GPI-hemojuvelin to downregulate hepcidin expression. The release of s-hemojuvelin is negatively regulated by both transferrin-bound iron (holo-Tf) and non-transferrin bound iron (FAC), indicating s-hemojuvelin could be one of the mediators of hepcidin regulation by iron. In this report, we investigate the proteinase involved in the release of s-hemojuvelin and show that s-hemojuvelin is released by a proprotein convertase through the cleavage at a conserved polybasic RNRR site.
Keywords: hepcidin, iron, furin convertase inhibitor, GPI anchor, repulsive guidance molecule C
Introduction
The juvenile hemochromatosis gene hemojuvelin (HJV, RGMc) belongs to the repulsive guidance molecule (RGM) family. All three members of the RGM family, RGMa, RGMb and hemojuvelin (RGMc) are GPI-linked proteins, and have been shown to be co-receptors for BMP2 and 4 [2;3;34;36]. RGMa and RGMb are predominantly expressed in the central nervous system in a mostly non-overlapping pattern, and are proposed to function in axon guidance and neural tube closure [25;29;30]. In contrast, hemojuvelin is predominantly expressed in the liver and skeletal muscles [31]. Hemojuvelin has been shown to be essential for the regulation of hepcidin production and iron homeostasis [15;22;28;31].
Hepcidin is secreted from the liver into blood circulation and acts as the principal iron regulatory hormone. Dysregulation of hepcidin expression has been shown as the underlying cause of many common iron-related disorders [10]. Homozygous or compound heterozygous mutations of hemojuvelin cause juvenile hemochromatosis, the most severe form of hereditary hemochromatosis. Patients with juvenile hemochromatosis due to hemojuvelin mutations have very low hepcidin levels and are phenotypically indistinguishable from patients with homozygous mutations which disrupt the hepcidin gene itself [31]. Like their human counterparts, hemojuvelin knockout mice also had very low hepatic hepcidin expression and severe iron overload [15;28]. Using antisense RNA to suppress hemojuvelin synthesis, we demonstrated in the hepatoma cell line Hep3B that hemojuvelin is indeed required for hepcidin mRNA expression [22]. Hemojuvelin likely influences hepcidin synthesis through the BMP signaling pathway, since it was recently shown that hemojuvelin binds BMP2/4 and functions as a BMP coreceptor [2], and that the SMAD4 transcription factor common to the TGFβ/BMP receptor pathway is required for hepcidin expression in the liver [46].
Hemojuvelin protein exists both as a cell-associated GPI form (GPI-hemojuvelin) and a soluble form (s-hemojuvelin) that can be detected in cell culture supernatants as well as in human [22] and rat [47] blood. The amount of s-hemojuvelin in the cell culture supernatant was reduced by treatment with inorganic non-transferrin-bound-iron or holotransferrin [22]. Based on the observation that s-hemojuvelin suppressed hepcidin production in primary hepatocytes in a dose-dependent manner [22], we proposed a model in which s-hemojuvelin and GPI-hemojuvelin reciprocally regulate hepcidin expression in response to iron-load. GPI-hemojuvelin generates a positive signal for hepcidin production. In contrast, s-hemojuvelin competes with GPI-hemojuvelin for BMP2/4 ligands, which in turn modulates the BMP2/4 signaling and hepcidin production. To understand how hemojuvelin regulates hepcidin in response to iron signals, we characterized the enzyme that generates s-hemojuvelin and examined how iron regulates the cleavage process.
It is not yet certain whether s-hemojuvelin originates from cellular secretion, from the shedding of GPI-hemojuvelin on the membrane, or from a combination of the two mechanisms. Silvestri et al. showed that hemojuvelin mutants defective in plasma membrane presentation (with significant ER retention) can still release s-hemojuvelin. They argue that s-hemojuvelin does not originate from membrane bound GPI-hemojuvelin, but is independently secreted by the cell [39]. However, definitive evidence supporting the secretion model is lacking. A recent study using cell surface protein biotinylation provided evidence that s-hemojuvelin can originate from cell surface GPI-hemojuvelin [18]. Cell surface GPI-hemojuvelin is composed of two distinct forms.
The major form is autocatalytically cleaved at a labile Asp-Pro bond (within the partial von Willebrand factor domain) in the late secretory pathway, resulting in a two-chain GPI-hemojuvelin structure linked by disulfide-bonds [18;22;39;48]. A second, minor population of the cell surface GPI-hemojuvelin is an uncleaved one-chain form rapidly released as s-hemojuvelin [18].
The release of soluble extracellular domain fragments (ectodomains) from membrane proteins through regulated proteolytic cleavage is a common process referred to as ectodomain shedding [11;12]. A variety of integral membrane proteins (transmembrane proteins and lipid-anchored proteins) including cell adhesion molecules, leukocyte antigens, receptors/ligands and viral membrane proteins are targets for this proteolytic release [14]. Ectodomain shedding can downregulate receptors and adhesion molecules on the cell surface, which prevents further receptor-ligand interactions [11;14]. Proteinases involved in ectodomain shedding (ectodomain sheddases) have been shown to comprise a heterogeneous population that function near (Golgi) or at the cell surface (plasma membrane) [11;26].
Two subfamilies of the metzincin metalloproteinases (ADAMs and MMPs) have been shown to be involved in ectodomain shedding. ADAMs (a disintegrin and metalloproteinase) are a family of transmembrane proteins characterized by a metalloproteinase catalytic domain and a disintegrin-like adhesive domain. MMPs (matrix metalloproteinase) are a family of matrix-degrading proteinases existing in either transmembrane or secreted forms. In common with other members of the metzincin metalloproteinase superfamily, proteolytic activities of both ADAMs and MMPs are zinc-dependent, and are readily inhibited by chelating agents such as 1,10-phenanthroline (PNT) and EDTA [9;11], as well as hydroxamate-based inhibitors including MMP inhibitor 2 and TAPI-2 (tumor necrosis factor α protease inhibitor-2) [1;1]. ADAMs and MMPs are typically synthesized with a prodomain that helps maintain their inactive state, and is located N-terminally to the metalloproteinase catalytic domain. Activation of the enzymatic activity requires prodomain removal through autocatalysis, or limited endoproteolysis by other metalloproteinases or proprotein convertases [9;26].
Proprotein convertases (PCs) are a family of calcium-dependent serine proteinases. They are also implicated in the ectodomain shedding of some substrates such as type XIII collagen [43]. The involvement of proprotein convertases in ectodomain shedding could be either through direct cleavage of the substrate, or indirect activation of ADAMs and MMPs. Seven members of mammalian subtilisin (bacterial)/Kex2p(yeast)-like proprotein convertases have been identified, namely furin, PC2, PC1/PC3, PC4, PACE4, PC5/PC6 and PC7/LPC. Furin was the first to be identified and has been the subject of extensive studies [27]. Proprotein convertases play an essential role in the processing of a variety of protein precursors including hormones, proteases, growth factors, receptors, bacterial toxins and viral glycoproteins. Distinct from ADAMs and MMPs which lack a consensus cleavage motif [16], proprotein convertase family members share a consensus cleavage site at the carboxyl side of polybasic sequence -Arg-X-Lys/Arg-Arg↓-(X: any amino acid, ↓: cleavage site). Although variations have been reported, a minimal cleavage sequence Arg-X-X-Arg is required. Arg in positions P1 and P4 are essential, while Arg/Lys in P2 enhances processing efficiency [42]. One of the most widely used inhibitors of the proprotein convertase family members is the peptidyl inhibitor decanoyl–Arg–Val–Lys–Arg–CH2Cl (furin convertase inhibitor, FCI), which blocks all proprotein convertase activity with a low nanomolar Ki.
In the current study, we assessed the properties of the proteinase involved in the release of s-hemojuvelin. Using HEK293 cell lines expressing either wild-type human hemojuvelin protein or a human hemojuvelin-alkaline phosphatase chimera protein, we identified the cleavage site for the release of human s-hemojuvelin as the C-terminus of the polybasic segment RNRR (aa332-335), a conserved cleavage site for the proprotein convertases.
Materials
Reagents and mice
Tumor necrosis factor α protease inhibitor-2 (TAPI-2), matrix metalloproteinase inhibitor 1 (MMPI-1) and inhibitor 2 (MMPI-2) were from Calbiochem (San Diego, CA). Furin convertase inhibitor (FCI; decanoyl-Arg-Val-Lys-Arg-CH2Cl) was from Alexis Biochemicals (San Diego, CA) and 1, 10-phenanthroline (PNT) was from Sigma-Aldrich (St. Louis, MO).
C57/Bl6J mice were from Charles River Laboratories (Wilmington, MA). All mice were maintained on NIH 31 rodent diet (iron content 336 mg/kg; Harlan Teklad, Indianapolis, Indiana, USA).
Vector construction and stable cell lines
The plasmid pHJV was generated by cloning full length human hemojuvelin cDNA into the pcDNA3.1(+) plasmid vector (Invitrogen, Carlsbad, CA) [22]. The plasmid pHJV-AP was generated by cloning human hemojuvelin cDNA (truncated by 81 nt at the 3′ end) and fusing it to pAPtag-5 (GenHunter, Nashville, TN) at the 5′-end of AP gene coding region, to express a human hemojuvelin ectodomain (aa1-400)-alkaline phosphatase chimera protein with a C-terminal 6xHis tag. Hemojuvelin-alkaline phosphatase chimera was purified using Ni-NTA resin (Invitrogen, Carlsbad, CA). The plasmids pRRL-mHJV-FL, pRRL-mHJV-ΔFur and pRRL-mHJV-ΔGPI were generated by cloning mouse hemojuvelin cDNA (FL: full-length cDNA, expressing full-length mouse hemojuvelin; ΔFur: truncated by 279 nt at the 3′ end with an added stop codon, expressing mouse hemojuvelin aa1-328; ΔGPI: truncated by 84 nt at the 3′ end with an added stop codon, expressing mouse hemojuvelin aa1-393) into a lentiviral transfer vector pRRL-sin-hCMV-MCS(R)-pre-cPPT. All plasmids were transfected into HEK293 cells using Lipofectamine™ 2000 Transfection Reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s protocol for 24 hours prior to further treatment. HEK293 stable cell lines were generated for pHJV, pHJV-AP and pAPtag-5 (pHJV, pHJV-AP and pAP HEK293 stable cultures) by co-transfecting pBabe-Puro vector (GenHunter, Nashville, TN) and selected by puromycin resistance.
Recombinant mouse soluble hemojuvelin production and purification
Lentiviral particles were packaged with pRRL-mHJV-ΔFur plasmid by the UCLA Vector Core Facility. A permanent cell line expressing mouse soluble hemojuvelin (mHJV-ΔFur HEK293 cell line) was generated by transducing HEK293 cell line with the packaged lentivirus overnight. This cell line was maintained in an equal volume mixture of Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA) and Pro-293a-CDM (Cambrex), and supplemented with 5% fetal bovine serum (FBS). Conditioned culture medium was purified by cation exchange chromatography (CM Prep, Biorad, Richmond, CA), followed by high performance liquid chromatography on a C4 reverse phase column (Vydac, 214TP54) eluted with an acetonitrile gradient.
Western blot analysis and antibody
Cellular protein was extracted with 150 mM NaCl, 10 mM EDTA, 10 mM Tris (pH 7.4), 1% Triton X-100 (NETT) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO) to generate a whole cell lysate. Conditioned cell culture medium was prepared for analysis by incubating cells in serum-free medium for 24 hours and 100 μl Conditioned cell culture medium was vacuum-dried before SDS-PAGE. Protein samples were separated on 4–20% SDS-Tris-Glycine PAGE (iGels, Gradipore, Hawthorne, NY) with or without dithiothreitol (DTT), and transferred to Immobilon-P membrane (Millipore Corp., Bedford, Massachusetts, USA). Two different anti-hemojuvelin polyclonal antibodies were prepared by immunizing rabbits with: Ab-HJV-N: against peptide antigen, target sequence N-CDYEGRFSRLHGRPPG-C, Ab-HJV-FL: against purified recombinant soluble human hemojuvelin (aa36-403) [22]. Ab-AP is a purified IgG 2a monoclonal antibody against human placental alkaline phosphatase (GenHunter, Nashville, TN). β-Actin was detected by goat anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA). Western blots were visualized by chemiluminescence.
Mouse primary hepatocyte isolation
Mouse primary hepatocytes were isolated using a Krebs-Henseleit solution/collagenase perfusion protocol published by Amaxa Biosystems (Amaxa Inc. Gaithersburg, MD). Viable hepatocyte population was further isolated by a Percoll purification step described in Lee et al [20]. Hepatocytes were plated in collagen-coated plates at 3×105 cells per well in 6-well plates, in William’s E medium supplemented with 5% fetal bovine serum and antibiotics. Cells were allowed to attach for 2 hours before switching to fresh medium and adding all treatments. All cells were treated for 18 to 24 hours before collection in TRIzol (Invitrogen).
RNA isolation and mRNA assay
RNA was prepared using TRIzol (Invitrogen) according to the manufacturer’s instructions. Single-pass cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using iQ SYBR Green Supermix (Bio-Rad). Murine hepcidin 1 (Hepc1) mRNA concentrations were normalized to murine β-actin. The following primers were used in qRT-PCR: murine hepcidin1: forward: 5′-TTGCGATACCAATGCAGAAGA –3′; reverse: 5′-GATGTGGCTCTAGGCTATGTT -3′ [21]; murine β-actin: forward 5′-ACCCACACTGTGCCCATCTA -3′; reverse: 5′-CACGCTCGGTCAGGATCTTC -3′.
Using a mathematical method described by Pfaffl [32], we first normalized mRNA concentrations of the target gene (Hepc1) to a reference stable housekeeping gene (β-actin), and then presented the measurement as a ratio to the control treatment within each experiment. The average relative expression value of the triplicate control treatments were assigned as 1 in each experiment.
Results
Regulation of soluble hemojuvelin release by proteinase inhibitors
To identify the proteinase involved in soluble hemojuvelin (s-hemojuvelin) production, we tested a group of proteinase inhibitors (Table 1) for their effects on s-hemojuvelin release in the pHJV HEK293 stable cell cultures. The various forms of hemojuvelin protein are described in Table 2. At 25 μM concentration, furin convertase inhibitor (FCI), 1, 10-phenanthroline (PNT), and matrix metalloproteinase inhibitor 2 (MMPI-2) markedly decreased s-hemojuvelin concentrations in the conditioned cell culture media (Figure 1A, upper panel). In contrast, tumor necrosis factor α protease inhibitor-2 (TAPI-2) and matrix metalloproteinase inhibitor 1 (MMPI-1) had no effect on s-hemojuvelin release. However, further analyses of the cell-associated GPI-hemojuvelin in whole cell lysate showed that the decrease in s-hemojuvelin release upon 1,10-phenanthroline (PNT) or matrix metalloproteinase inhibitor 2 (MMPI-2) treatment was probably due to the decrease in the cell-associated GPI-hemojuvelin (Figure 1A, middle panel), rather than a specific effect on the release of s-hemojuvelin. While s-hemojuvelin release was suppressed by furin convertase inhibitor (FCI), an alternative soluble form of hemojuvelin, ecto-hemojuvelin, was detected in the conditioned cell culture medium (Figure 1A and 1B, top panels). Ecto-hemojuvelin showed a similar migration pattern on reducing SDS-PAGE as the cell-associated GPI-hemojuvelin in whole cell lysate (Figure 1A and 1B, middle panels). This suggests that an alternative shedding mechanism became dominant when the furin convertase inhibitor-sensitive process was suppressed. Moreover, furin convertase inhibitor (FCI) at a concentration as low as 100 nM not only suppressed s-hemojuvelin release, but also slightly elevated the cell-associated GPI-hemojuvelin level (Figure 1A and 1B, middle panels). Since whole cell lysate contained not only the plasma membrane-bound GPI-hemojuvelin but also hemojuvelin biosynthetic intermediates before transport to the plasma membrane, it is likely that our system underestimates the relative increase in the cell-surface form of GPI-hemojuvelin upon furin convertase inhibitor treatment. These results suggest that the s-hemojuvelin release is mediated by a proteinase sensitive to furin convertase inhibitor.
Table 1.
Effects of proteinase inhibitors on soluble hemojuvelin (s-hemojuvelin) release.
| Proteinase inhibitor | Abbreviation | Concentration | s-hemojuvelin release inhibition | Target proteinases |
|---|---|---|---|---|
| Furin convertase inhibitor | FCI | 100 nM | Yes | Furin-like proprotein convertases |
| Tumor necrosis factor α protease inhibitor-2 | TAPI-2 | 50 μM | No | Metalloproteinases (MMPs and ADAMs) |
| 1,10-Phenanthroline | PNT | 25 μM | Yes* | Metalloproteinases (MMPs and ADAMs) |
| Matrix metalloproteinase inhibitor 1 | MMPI-1 | 50 μM | No | MMPs (MMP-1, MMP-3, MMP-8, and MMP-9) |
| Matrix metalloproteinase inhibitor 2 | MMPI-2 | 25 μM | Yes* | Metalloproteinases (MMPs and ADAMs) |
Also showed decrease in cell-associated GPI-hemojuvelin protein level
Table 2.
Terminology used for different hemojuvelin forms.
| Hemojuvelin forms | Abbreviation | Human hemojuvelin final product aa Sequence | Description |
|---|---|---|---|
| GPI-hemojuvelin | GPI-HJV | aa 36-400 * | Hemojuvelin associated with the plasma membrane through GPI anchor (Figure 1A lane 1, middle panel), including both one-chain structure (~46 kD, upper band), and two-chain structure (~30 kD, not shown, and ~16 kD, lower band) autocatalytically cleaved at a labile 172Asp-Pro bond [22;48]. |
| s-hemojuvelin | SHJV | aa 36-335 | Mature soluble hemojuvelin (Figure 1A, lane 1, upper panel). One-chain structure (~40 kD). |
| ecto-hemojuvelin | ecto-HJV | aa 36-400 | Alternative soluble forms of hemojuvelin only seen when s-hemojuvelin release is suppressed by furin convertase inhibitor (Figure 1A, lane 2, upper panel). They are possibly generated by endogenous lipolytic cleavage. They include both one-chain structure (~46 kD, top band), and two-chain structure (~30 kD, not shown, and ~16 kD, bottom band) autocatalytically cleaved at a labile 172Asp-Pro bond [22;48] |
| hemojuvelin-alkaline phosphatase | HJV-AP | aa 36-400 | Human hemojuvelin ectodomain (aa36-400)-alkaline phosphatase chimera protein (Figure 2B, left lane). It includes both one-chain structure( ~150 kD, Band 1), and two-chain structure (~130 kD, not shown, and ~16 kD, #) autocatalytically cleaved at a labile 172Asp-Pro bond [22;48]. A portion of mature protein is cleaved into s-hemojuvelin (~40 kD, Figure 2B, left lane, *) and an alkaline phosphatase fragment with a small stretch of the hemojuvelin ectodomain C-terminus still attached (~97 kD, HJV-C-term-AP, Figure 2A, B and 2). |
We assume that that hemojuvelin behaves like other GPI-linked proteins and that the transmembrane segment is lost when the GPI anchor is generated, with a cleavage site analogous to other GPI-linked proteins.
Figure 1. Soluble hemojuvelin release is inhibited by furin convertase inhibitor (FCI).
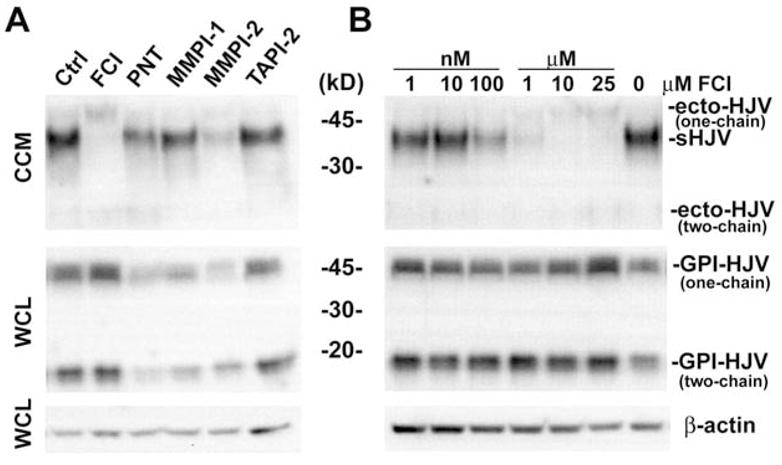
pHJV HEK293 stable cell cultures were treated in serum-free medium for 24 hours with (A) different proteinase inhibitors at 25 μM concentrations or (B) furin convertase inhibitor (FCI) ranging from 1 nM to 25 μM. Conditioned cell culture medium (CCM) and whole cell lysate (WCL) were analyzed on a reducing SDS-PAGE. Western blots were probed with Ab-HJV-N and anti-β-actin antibody. Visible bands correspond to: sHJV: s-hemojuvelin, ecto-HJV: ecto-hemojuvelin and GPI-HJV: GPI-hemojuvelin. “One-chain” refers to the one-chain form and “two-chain” to the ~16 kD small subunit of the two-chain form generated by autocatalytic cleavage, dissociated from the large subunit under reducing conditions, and detected with the N-terminal peptide-specific Ab-HJV-N antibody.
Soluble hemojuvelin is released at a conserved polybasic RNRR cleavage site
Using an alkaline phosphatase specific antibody (Ab-AP) and two different hemojuvelin-specific antibodies (Ab-HJV-N targeting aa 148-163 in human hemojuvelin and Ab-HJV-FL targeting recombinant human hemojuvelin aa 36-403), we analyzed hemojuvelin-alkaline phosphatase protein (schematic structure shown in Figure 2E) purified on the nickel column from the conditioned media of pHJV-AP HEK293 stable culture. The soluble human alkaline phosphatase expressed by pAP HEK293 stable culture was also purified and used as a control.
Figure 2. Soluble hemojuvelin release from the hemojuvelin-alkaline phosphatase chimera protein is sensitive to furin convertase inhibitor (FCI) and iron loading.

(A) Conditioned cell culture media from pHJV-AP and pAP HEK293 stable cell cultures were purified using a nickel-column, and analyzed on a non-reducing SDS-PAGE. Western blot was probed with Ab-HJV-N, Ab-HJV-FL and Ab-AP antibodies. (B) Conditioned cell culture media from pHJV-AP and pHJV HEK293 stable cell cultures were analyzed on a reducing SDS-PAGE. Western blot was probed with Ab-HJV-N antibody. (C) pHJV-AP HEK293 stable cell cultures treated with ferric ammonium citrate (FAC) at 0, 10, 30 and 100 μM. Conditioned cell culture media were analyzed on a reducing SDS-PAGE. Western blot was probed with Ab-HJV-N antibody. (D) pHJV-AP HEK293 stable cell cultures treated with or without 25 μM furin convertase inhibitor (FCI). Conditioned cell culture media were analyzed on a non-reducing SDS-PAGE. Western blot was probed with Ab-HJV-N antibody. (E) Schematic illustration of human hemojuvelin ectodomain (aa 36-400)-alkaline phosphatase chimera protein (HJV-AP). SP: signal peptide (aa 1-35); RGD: RGD motif (aa 98-100); vWF: partial von Willebrand factor domain (aa 167-253). Visible bands correspond to mature full-length hemojuvelin-alkaline phosphatase chimera protein (band 1), HJV-C-term-AP fragment (band 2), human alkaline phosphatase (band 3), s-hemojuvelin produced by pHJV-AP HEK293 cells (sHJV, *) and the N-terminal fragment of hemojuvelin-alkaline phosphatase (N-term-HJV, #).
Hemojuvelin-alkaline phosphatase chimera is comprised of human hemojuvelin ectodomain (aa36-400) tagged with alkaline phosphatase protein. Surprisingly, purified hemojuvelin-alkaline phosphatase migrated as at least two different forms under non-reducing conditions (Figure 2A). The first form was the full-length hemojuvelin-alkaline phosphatase chimera (Figure 2A, Band 1, apparent MW 150 kD, by mass spectrometry 107.1 kD, Figure 2E). The second form was a fragment of the hemojuvelin-alkaline phosphatase chimera cleaved near the C-terminus of the hemojuvelin ectodomain. It was purified as an alkaline phosphatase fragment with a small stretch of the hemojuvelin ectodomain C-terminus still attached (HJV-C-term-AP fragment, Figure 2A, Band 2, apparent MW 97 kD, by mass spectrometry 71.3 kD, Figure 2E). Bands 1 and 2 could be detected with both Ab-HJV-FL and Ab-AP antibody, while Ab-HJV-N antibody only detected band 1 but not band 2, presumably due to the absence of the N-terminus of the human hemojuvelin.
We also compared the conditioned cell culture media from pHJV-AP and pHJV HEK293 stable cell cultures on a reducing SDS-PAGE. Using Ab-HJV-N, three specific protein bands were detected in the media conditioned by the pHJV-AP cell line (Figure 2B, left lane). One protein band (Figure 2B, Band 1) with an apparent molecular weight of 150 kD corresponded to the full-length hemojuvelin-alkaline phosphatase chimera (Figure 2A, Band 1). A smaller protein band (#) with an apparent molecular weight of 16 kD corresponded to the N-terminal fragment of hemojuvelin-alkaline phosphatase chimera protein (N-term-HJV, Figure 2E). This was similar to what was observed for human recombinant soluble hemojuvelin (aa1-403 or aa1-401) [22;48], as a result of an autocatalytic cleavage at the conserved labile 172Asp-Pro bond (Figure 2E). A third protein band (*) with an apparent molecular weight of ~40 kD co-migrated with the single protein band (sHJV, Figure 2B, right lane) detected in the pHJV HEK293 stable cell line.
We further showed that the release of the ~40 kD fragment in pHJV-AP cell line progressively decreased with increasing concentration of ferric ammonium citrate (FAC) (Figure 2C). Moreover, this cleavage event could be inhibited by the furin convertase inhibitor (FCI) (Figure 2D). Both observations resemble the regulation of s-hemojuvelin release in HEK293 cell line overexpressing full-length human hemojuvelin (Figure 1A) [22].
Taken together, we conclude that pHJV-AP cell line is producing a protein identical to s-hemojuvelin, presumably through the same mechanism that releases s-hemojuvelin from membrane associated GPI-hemojuvelin.
To determine the actual cleavage site, we circumvented the technical difficulty of C-terminal protein sequencing. Instead, we used N-terminal protein sequencing to identify the HJV-C-term-AP fragment (Figure 2A, Band 2 and Figure 2E). The N-terminal sequence of the HJV-C-term-AP fragment was GAITIDT, the amino acid sequence immediately following a conserved polybasic cleavage site to the carboxyl side of the sequence RNRR (human hemojuvelin aa 332-335, mouse hemojuvelin aa 325-328, Figure 2E). This result, combined with the observation that the release of human s-hemojuvelin in cell cultures can be blocked by furin convertase inhibitor, strongly supports the proposed role of a furin-like proprotein convertase in the release of s-hemojuvelin.
Additional experiments with vectors expressing various mouse hemojuvelin forms also confirmed RNRR328 as the cleavage site for mouse s-hemojuvelin release. Mouse s-hemojuvelin produced by HEK293 cells transfected with pRRL-mHJV-ΔFur vector (encoding a C-terminally-truncated mouse hemojuvelin, aa 1-328) had an identical migration pattern as the mature mouse s-hemojuvelin released by HEK293 cells transfected with pRRL-mHJV-FL (encoding full-length mouse hemojuvelin) (Figure 3A). These data establish the importance of the RNRR polybasic cleavage site in s-hemojuvelin release.
Figure 3. Western blot analysis of mouse and human soluble hemojuvelin produced by HEK293 cells.

HEK293 cells were transiently transfected with expression vectors carrying cDNAs encoding mouse and human hemojuvelin. Lane 1–4: pRRL: empty vector; pΔFur: pRRL-mHJV-ΔFur, mouse s-hemojuvelin vector truncated at R328 (RNRR polybasic cleavage site); pΔGPI: pRRL-mHJV-ΔGPI, mouse s-hemojuvelin vector truncated at D393 (GPI anchor processing site); pFL: pRRL- mHJV -FL, mouse hemojuvelin full-length vector. Lane 5–6: pHJV: human hemojuvelin full-length vector; pcDNA: pcDNA3.1 (+) empty vector. (A) Soluble mouse and human hemojuvelin forms in conditioned cell culture medium (CCM) and (B) mouse and human hemojuvelin in whole cell lysate (WCL) were analyzed on a reducing SDS-PAGE. Western blots were probed with Ab-HJV-N antibody.
In contrast, mouse s-hemojuvelin produced by HEK293 cells transfected with pRRL-mHJV-ΔGPI vector (encoding a C-terminally-truncated mouse hemojuvelin, aa 1-393, pΔGPI cells) migrates similarly as the forms detected in pΔGPI and pFL cells whole cell lysate (Figure 3B), as well as the ecto-hemojuvelin detected when s-hemojuvelin release was inhibited by furin convertase inhibitor (FCI, Figure 1A and 1B), but different from mature s-hemojuvelin produced by pFL cells.
Mouse soluble hemojuvelin specifically suppresses hepcidin production in mouse primary hepatocyte cultures in a dose-dependent manner
To describe the biological effects of mouse s-hemojuvelin derived from the iron/furin convertase inhibitor sensitive cleavage of membrane associated GPI-hemojuvelin, we produced recombinant mouse s-hemojuvelin with the same C-terminus (ms-hemojuvelin, Gln33-Arg328) using the stably lentivirus-transduced mHJV-ΔFur HEK293 cell line.
In primary mouse hepatocyte cultures treated with purified ms-hemojuvelin, hepcidin mRNA was suppressed in a dose-dependent manner (Figure 4A), similar to what was shown previously in human primary hepatocytes with a longer form (aa 1-403) of recombinant soluble human hemojuvelin [22]. At 10 μg/ml, ms-hemojuvelin caused a >2-fold reduction in hepcidin mRNA (p=0.007, paired t-test) while heat-denatured (boiled) ms-hemojuvelin and the irrelevant protein bovine serum albumin (BSA) did not affect hepcidin mRNA concentrations (Figure 4B).
Figure 4. Mouse soluble hemojuvelin (ms-hemojuvelin) specifically suppresses hepcidin production in primary mouse hepatocyte cultures.
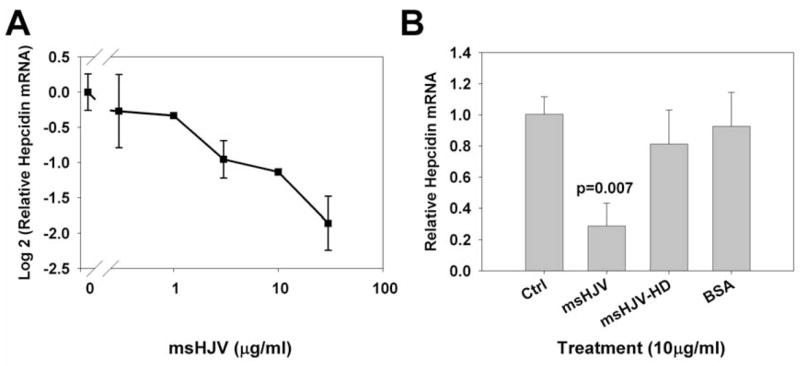
Primary mouse hepatocytes were isolated from wild-type C57/Bl6J mice. (A) Hepatocyte cultures were treated with 3-fold serial dilution of ms-hemojuvelin (msHJV) ranging from 0.3–30 μg/ml. (B) Hepatocyte cultures were treated with 10 μg/ml ms-hemojuvelin (msHJV), heat denatured ms-hemojuvelin (msHJV-HD), or bovine serum albumin (BSA).
Discussion
The release of soluble forms of membrane proteins by ectodomain shedding has long been recognized as an important regulatory process involved in a variety of biological processes. Downregulation of receptor activity through decreased cell surface receptor density and simultaneous generation of soluble antagonist has also been shown in many instances. A well-known example is the LPS receptor CD14 [4;37]. CD14 is a GPI-linked monocyte surface molecule. It functions as a pattern recognition receptor for serum LPS-binding protein/LPS complex that cooperates with toll-like receptor 4 (TLR4) to mediate monocyte activation and TNF-α synthesis. Soluble CD14 detected in human sera has been reported to serve as soluble antagonist for monocyte and macrophage activation [17;37]. Soluble CD14 was shown to be released from cell surface by multiple alternative mechanisms including proteolytic processing dependent on MMP12/MMP9 and other MMPs [38], human leukocyte elastase (HLE) [19] and a leukocyte carboxyl/aspartate protease [7]; lipolytic release from the GPI-linker [37] or direct secretion without the GPI moiety [5].
As demonstrated in the studies of soluble CD14, the identification of the specific ectodomain sheddases can be difficult considering the possible involvement of multiple shedding mechanisms. Similar to CD14, hemojuvelin is also a GPI-linked membrane protein whose soluble form antagonizes the effect of the membrane-associated GPI-form on hepcidin mRNA production[22]. In the current study, we examined the process that releases s-hemojuvelin.
General metalloproteinase inhibitor tumor necrosis factor α protease inhibitor-2 (TAPI-2) and MMP-specific inhibitor matrix metalloproteinase inhibitor 1 (MMPI-1) had no significant effect on the s-hemojuvelin production. These results argue against the major involvement of ADAMs and MMPs in the release of s-hemojuvelin. Although other general inhibitors of metalloproteinases including 1, 10-phenanthroline (PNT) and matrix metalloproteinase inhibitor 2 (MMPI-2) inhibited s-hemojuvelin release, they also reduced the GPI-hemojuvelin level. Analysis by qRT-PCR showed that both 1,10-phenanthroline (PNT) and matrix metalloproteinase inhibitor 2 (MMPI-2) suppressed hemojuvelin mRNA levels in the cell line studied (data not shown), which could also result in the decrease of GPI-hemojuvelin level. It was reported that high dose 1, 10-phenanthroline (PNT, 250–500 μM) had an inhibitory effect on GPI anchor synthesis. However, at 50 μM concentration, this inhibitory effect is small [24]. In summary, there is no convincing evidence to suggest any involvement of ADAMs and MMPs in s-hemojuvelin release.
Only the proprotein convertase (PC) inhibitor decanoyl–Arg–Val–Lys–Arg–CH2Cl (furin convertase inhibitor, FCI) had a consistent and potent inhibitory effect on s-hemojuvelin release in our cellular assay system. The inhibition of s-hemojuvelin release by furin convertase inhibitor takes place at concentrations as low as 100 nM and the inhibitor exerts this activity without suppressing GPI-hemojuvelin concentration. Notably, when s-hemojuvelin release was suppressed completely by furin convertase inhibitor, a second form of soluble hemojuvelin (ecto-hemojuvelin, see Figure 1) became predominant in the cell culture supernatant, but at a much lower concentration. This form is most likely generated by the low level lipolytic activity of the endogenous glycosylphosphatidylinositol phospholipase D (GPI-PLD) [13]. Although fetal bovine serum in cell culture could be a potential source of soluble GPI-PLD, it has been reported that serum GPI-PLD is inactive and unable to cleave GPI-anchors under physiological conditions [23]. Moreover, the culture media used in this study were serum-free. These results showed that lipolytic cleavage could contribute to the release of the minor hemojuvelin soluble forms (ecto-hemojuvelin, one-chain and two-chain) that are distinct from the major one-chain soluble form (s-hemojuvelin) detected in the cell culture supernatant (Figure 1), but resemble the soluble mouse hemojuvelin forms truncated at the GPI anchor processing site (pΔGPI, Figure 3A). The appearance of both one-chain and two-chain ecto-hemojuvelin also agrees with the heterogeneous GPI-hemojuvelin populations model proposed by Kuninger et al [18], showing the lack of preference for lipolytic cleavage.
To further characterize this proteolytic process, we utilized a human hemojuvelin-alkaline phosphatase chimera protein, which could be cleaved into a soluble form of hemojuvelin with an identical migration pattern as s-hemojuvelin on SDS-PAGE. Using this unique tool, we confirmed that the furin convertase inhibitor-sensitive cleavage site on hemojuvelin-alkaline phosphatase was also sensitive to iron concentration. Protein sequencing further confirmed the actual cleavage site as the carboxyl side of the polybasic RNRR consensus sequence conserved among human (aa 332-335), mouse (aa 325-328) and rat (aa 328-331) versions of hemojuvelin. This is distinct from the ADAMs and MMPs mediated cleavage which lacks a consensus cleavage site. Comparisons between the migration patterns of mouse s-hemojuvelin showed no difference in cultures transfected with mouse hemojuvelin vectors coding for the full-length or the RNRR truncated forms (aa1-328). Functionally, the mouse s-hemojuvelin (ms-hemojuvelin) generated by the direct truncation up to the RNRR cleavage site is a specific and potent inhibitor of GPI-hemojuvelin and suppresses hepcidin mRNA expression in primary mouse hepatocyte cultures. All these results support the conclusion that the proteinase that cleaves GPI-hemojuvelin to release s-hemojuvelin is a furin-like proprotein convertase (PC).
Brefeldin A (BFA) is a general inhibitor of protein transport from ER to Golgi apparatus. Silvestri et al. showed s-hemojuvelin release is a BFA-sensitive process. 24-hour BFA treatment of a hemojuvelin-expressing cell line resulted in decreased s-hemojuvelin release but unchanged cell surface one-chain GPI-hemojuvelin. Based on these results, Silvestri et al. suggested that s-hemojuvelin did not originate from cell surface one-chain GPI-hemojuvelin [39]. However, furin-like proprotein convertases are located in trans-Golgi network (TGN) and on the cell surface, and BFA treatment also interferes with furin-like proprotein convertases localization and target processing [6;44]. The involvement of proprotein convertases in s-hemojuvelin processing could account for the BFA sensitivity of s-hemojuvelin release. Thus, BFA sensitivity is not conclusive evidence for the direct-secretion model proposed by Silvestri et al. In contrast, the cell surface protein biotin-labeling results by Kuninger et al. provided a strong argument that s-hemojuvelin can be released from the cell surface [18].
Furin (encoded by the fur gene) is the most extensively studied member of the proprotein convertase family. Furin has been shown to be ubiquitously expressed. Although mainly localized in the trans-Golgi network (TGN), a subpopulation of the furin molecules has been shown to cycle between TGN and the cell surface [27;40]. This cellular localization profile suggests that furin or one of the furin-like proprotein convertases could be a hemojuvelin sheddase.
The further identification of this furin-like proprotein convertase is challenging, because of the common cleavage pattern and overlapping substrate specificity of the proprotein convertase family. Some proprotein convertases showed restricted tissue-specific or developmental specific expression pattern and can be ruled out. They are PC4 with germ cells exclusive expression, and PC1 and PC2 which are specifically expressed in neuroendocrine cells. However, the remaining members of the proprotein convertase family (furin, PACE4, PC5/6, PC7/LPC) are ubiquitously expressed and share overlapping cellular localizations [41].
Knockout mouse models have been generated for most individual proprotein convertase family members for functional studies. However, due to their critical roles in embryonic development, the absence of furin, PC5/6, and PACE4 (but with less penetrance) in mice have been shown to be embryonic lethal [35;41;41], thus would not provide useful information for this particular study. Two proprotein convertase knockout animals, PC7 null mice [45] and a conditional liver-specific furin knockout mouse [33] did not show any developmental abnormality and any phenotype. These were explained as the results of specialized function of PC7 and the functional redundancy of furin. However, the study of PC7 null mice was mostly focused on substrates in the central nervous system, and the conditional liver-specific furin knockout mice were only analyzed 10 days after the furin inactivation in adult liver. Thus, it is possible that any iron metabolism-related phenotypes have not been evaluated or may have been overlooked in these knockout animal models. Further investigation in these proprotein convertase null mice could provide insights on the processing of s-hemojuvelin. Of course, any phenotype observed in these models needs to be interpreted with caution, since proprotein convertases are reported to be involved in the maturation of hepcidin itself (Valore and Ganz, manuscript in preparation) as well as BMP ligands [8] which are crucial for hepcidin expression [2].
Apart from our finding that the s-hemojuvelin is released by a furin-like proprotein convertase, it was recently reported that neogenin, a widely expressed transmembrane binding partner of hemojuvelin [48], is also required for s-hemojuvelin release [47]. However, the mechanism by which iron loading affects s-hemojuvelin production is still unclear. Because proprotein convertases process many substrates, it is unlikely that their activities are directly regulated by iron. Instead, iron probably interacts with other proteins, such as transferrin and its receptors, to regulate the accessibility of the polybasic site on hemojuvelin to cleavage by the proprotein convertase. Although it remains to be determined how neogenin is involved in this proprotein convertase mediated cleavage process, it is possible that neogenin could bind to hemojuvelin [47;48] and thereby enhance the accessibility of its cleavage site to proprotein convertase.
Using an in vivo rat model, Zhang et. al. [47] also confirmed our in vitro observation that iron loading suppresses s-hemojuvelin release [22] and showed an increase in serum s-hemojuvelin during the early phase of dietary iron deficiency in weanling rats. The fusion protein s-hemojuvelin/Fc also significantly suppressed hepatic hepcidin expression when injected into mice (Babitt et. al., in press), confirming the inhibitory effect of s-hemojuvelin on hepcidin expression [22] in vivo. These studies further emphasize the likely role of s-hemojuvelin in regulating hepcidin expression. The identification of the s-hemojuvelin-releasing enzyme and elucidation of how iron regulates this proteolytic cleavage event will advance our understanding of the regulation of iron metabolism, and could lead to the development of more specific therapies for iron-related disorders.
Acknowledgments
The study was supported by NIH grants RO1 DK065029 and R21 DK073226 to T.G., KO1 DK07538 to E.N. and by the Will Rogers Fund. We gratefully thank Dr. Ernest Beutler from the Scripps Research Institute for sharing a hepatocyte purification protocol, and Dr. Renata Stripecke from UCLA Vector Core Facility for lentiviral packaging. We would also like to thank Yen Phung and Chun-Ling Jung for their assistance on this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Arribas J, Coodly L, Vollmer P, Kishimoto TK, RoseJohn S, Massague J. Diverse cell surface protein ectodomains are shed by a system sensitive to metalloprotease inhibitors. Journal of Biological Chemistry. 1996;271:11376–11382. doi: 10.1074/jbc.271.19.11376. [DOI] [PubMed] [Google Scholar]
- 2.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nature Genetics. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 3.Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. Journal of Biological Chemistry. 2005;280:29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- 4.Bazil V, Strominger JL. Shedding As A Mechanism of Down-Modulation of Cd14 on Stimulated Human Monocytes. Journal of Immunology. 1991;147:1567–1574. [PubMed] [Google Scholar]
- 5.Bufler P, Stiegler G, Schuchmann M, Hess S, Kruger C, Stelter F, Eckerskorn C, Schutt C, Engelmann H. Soluble Lipopolysaccharide Receptor (Cd14) Is Released Via 2 Different Mechanisms from Human Monocytes and Cd14 Transfectants. European Journal of Immunology. 1995;25:604–610. doi: 10.1002/eji.1830250244. [DOI] [PubMed] [Google Scholar]
- 6.Cao J, Rehemtulla A, Pavlaki M, Kozarekar P, Chiarelli C. Furin directly cleaves proMMP-2 in the trans-Golgi network resulting in a nonfunctioning proteinase. Journal of Biological Chemistry. 2005;280:10974–10980. doi: 10.1074/jbc.M412370200. [DOI] [PubMed] [Google Scholar]
- 7.Coyne CP, Howell T, Smodlaka H, Willetto C, Fenwick BW, Chenney E. Alterations in membrane-associated CD14 expression and the simultaneous liberation of soluble CD14 fragment in adherent macrophages mediated by a leukocyte carboxyl/aspartate protease. Journal of Endotoxin Research. 2002;8:273–283. doi: 10.1179/096805102125000489. [DOI] [PubMed] [Google Scholar]
- 8.Cui YZ, Jean F, Thomas G, Christian JL. BMP-4 is proteolytically activated by furin and/or PC6 during vertebrate embryonic development. Embo Journal. 1998;17:4735–4743. doi: 10.1093/emboj/17.16.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowlkes JL, Winkler MK. Exploring the interface between metallo-proteinase activity and growth factor and cytokine bioavailability. Cytokine & Growth Factor Reviews. 2002;13:277–287. doi: 10.1016/s1359-6101(02)00005-9. [DOI] [PubMed] [Google Scholar]
- 10.Ganz T, Nemeth E. Regulation of iron acquisition and iron distribution in mammals. Biochimica et Biophysica Acta-Molecular Cell Research. 2006;1763:690–699. doi: 10.1016/j.bbamcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Garton KJ, Gough PJ, Raines EW. Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J Leukoc Biol. 2006;79:1105–1116. doi: 10.1189/jlb.0106038. [DOI] [PubMed] [Google Scholar]
- 12.Hattori M, Osterfield M, Flanagan JG. Regulated Cleavage of a Contact-Mediated Axon Repellent. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- 13.He XT, Hannocks MJ, Hampson I, Brunner G. GPI-specific phospholipase D mRNA expression in tumor cells of different malignancy. Clinical & Experimental Metastasis. 2002;19:291–299. doi: 10.1023/a:1015545407700. [DOI] [PubMed] [Google Scholar]
- 14.Hooper NM, Karran EH, Turner AJ. Membrane protein secretases. Biochem J. 1997;321:265–279. doi: 10.1042/bj3210265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang FW, Babitt JL, Wrighting DM, Samad TA, Xia Y, Sidis Y, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Hemojuvelin acts as a bone morphogenetic protein co-receptor to regulate hepcidin expression. Blood. 2005;106:153A. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 16.Huovila APJ, Turner AJ, Pelto-Huikko M, Karkkainen L, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends in Biochemical Sciences. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Kitchens RL, Thompson PA. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. Journal of Endotoxin Research. 2005;11:225–229. doi: 10.1179/096805105X46565. [DOI] [PubMed] [Google Scholar]
- 18.Kuninger D, Kuns-Hashimoto R, Kuzmickas R, Rotwein P. Complex biosynthesis of the muscle-enriched iron regulator RGMc. J Cell Sci. 2006;119:3273–3283. doi: 10.1242/jcs.03074. [DOI] [PubMed] [Google Scholar]
- 19.Le Barillec K, Si-Tahar M, Balloy V, Chignard M. Proteolysis of monocyte CD14 by human leukocyte elastase inhibits lipopolysaccharide-mediated cell activation. Journal of Clinical Investigation. 1999;103:1039–1046. doi: 10.1172/JCI5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee P, Peng HF, Gelbart T, Beutler E. The IL-6- and lipopolysaccharide-induced transcription of hepcidin in HFE-, transferrin receptor 2-, and beta(2)-microglobulin-deficient hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9263–9265. doi: 10.1073/pnas.0403108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee P, Peng HF, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1906–1910. doi: 10.1073/pnas.0409808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 23.Low MG, Huang KS. Factors Affecting the Ability of Glycosylphosphatidylinositol-Specific Phospholipase-D to Degrade the Membrane Anchors of Cell-Surface Proteins. Biochem J. 1991;279:483–493. doi: 10.1042/bj2790483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann KJ, Sevlever D. 1,10-Phenanthroline Inhibits Glycosylphosphatidylinositol Anchoring by Preventing Phosphoethanolamine Addition to Glycosylphosphatidylinositol Anchor Precursors. Biochemistry. 2001;40:1205–1213. doi: 10.1021/bi0024512. [DOI] [PubMed] [Google Scholar]
- 25.Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, Mueller BK. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- 26.Moss ML, Bartsch JW. Therapeutic benefits from targeting of ADAM family members. Biochemistry. 2004;43:7227–7235. doi: 10.1021/bi049677f. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. Journal of Clinical Investigation. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niederkofler V, Salie R, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. Journal of Neuroscience. 2004;24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldekamp J, Kramer N, Alvarez-Bolado G, Skutella T. Expression pattern of the repulsive guidance molecules RGM A, B and C during mouse development. Gene Expression Patterns. 2004;4:283–288. doi: 10.1016/j.modgep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald MLE, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nature Genetics. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roebroek AJM, Taylor NA, Louagie E, Pauli I, Smeijers L, Snellinx A, Lauwers A, Van de Ven WJM, Hartmann D, Creemers JWM. Limited Redundancy of the Proprotein Convertase Furin in Mouse Liver. Journal of Biological Chemistry. 2004;279:53442–53450. doi: 10.1074/jbc.M407152200. [DOI] [PubMed] [Google Scholar]
- 34.Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ. DRAGON, a bone morphogenetic protein co-receptor. Journal of Biological Chemistry. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- 35.Scamuffa N, Calvo F, Chretien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 36.Schmidtmer J, Engelkamp D. Isolation and expression pattern of three mouse homologues of chick Rgm. Gene Expression Patterns. 2004;4:105–110.26. doi: 10.1016/s1567-133x(03)00144-3. [DOI] [PubMed] [Google Scholar]
- 37.Schutt C. CD14. Int J Biochem Cell Biol. 1999;31:545–549. doi: 10.1016/s1357-2725(98)00153-8. [DOI] [PubMed] [Google Scholar]
- 38.Senft AP, Korfhagen TR, Whitsett JA, Shapiro SD, Levine AM. Surfactant protein-D regulates soluble CD14 through matrix metalloproteinase-12. Journal of Immunology. 2005;174:4953–4959. doi: 10.4049/jimmunol.174.8.4953. [DOI] [PubMed] [Google Scholar]
- 39.Silvestri L, Pagani A, Fazi C, Gerardi G, Levi S, Arosio P, Camaschella C. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007 doi: 10.1182/blood-2006-08-041004. blood-2006. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of Furin to the Trans-Golgi Network and Recycling from the Cell-Surface Involves Ser and Tyr Residues Within the Cytoplasmic Domain. Journal of Biological Chemistry. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- 41.Taylor NA, Van de Ven WJM, Creemers JWM. Curbing activation: proprotein convertases in homeostasis and pathology. Faseb Journal. 2003;17:1215–1227. doi: 10.1096/fj.02-0831rev. [DOI] [PubMed] [Google Scholar]
- 42.Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Väisänen MR, Väisänen T, Pihlajaniemi T. The shed ectodomain of type XIII collagen affects cell behaviour in a matrix-dependent manner. Biochem J. 2004;380:685–693. doi: 10.1042/BJ20031974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vey M, Schafer W, Berghofer S, Klenk HD, Garten W. Maturation of the trans-Golgi network protease furin: compartmentalization of propeptide removal, substrate cleavage, and COOH-terminal truncation. J Cell Biol. 1994;127:1829–1842. doi: 10.1083/jcb.127.6.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villeneuve P, Feliciangeli S, Croissandeau G, Seidah NG, Mbikay M, Kitabgi P, Beaudet A. Altered processing of the neurotensin/neuromedin N precursor in PC2 knock down mice: a biochemical and immunohistochemical study. Journal of Neurochemistry. 2002;82:783–793. doi: 10.1046/j.1471-4159.2002.00988.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang RH, Li CL, Xu XL, Zheng Y, Xiao CY, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepicidin expression. Cell Metabolism. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Zhang AS, Anderson SA, Meyers KR, Hernandez C, Eisenstein RS, Enns CA. Evidence that inhibition of hemojuvelin shedding in response to iron is mediated through neogenin. Journal of Biological Chemistry. 2007;282:12547–12556. doi: 10.1074/jbc.M608788200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang AS, West AP, Wyman AE, Bjorkman PJ, Enns CA. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. Journal of Biological Chemistry. 2005;280:33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]


