Abstract
The lysine demethylase JMJD2A has the unique property of binding trimethylated peptides from two different histone sequences (H3K4me3 and H4K20me3) through its tudor domains. Here we show using X-ray crystallography and calorimetry that H3K4me3 and H4K20me3, which are recognized with similar affinities by JMJD2A, adopt radically different binding modes, to the extent that we were able to design single point mutations in JMJD2A that inhibited the recognition of H3K4me3 but not H4K20me3 and vice versa.
The methylation of histones at lysine residues participates in the regulation of fundamental biological processes, such as gene transcription and cellular response to DNA damage, by attracting effector proteins that bind chromatin only after a target histone has been methylated at a specific lysine1. Therefore, there is considerable interest in understanding how specialized domains of these regulatory proteins recognize histone amino acid sequences in a methylation site–specific and sometimes methylation state–specific manner. The previously determined three-dimensional structures of chromo, double-chromo, tandem tudor, hybrid tudor and PHD domains in complex with methylated histone peptides have all revealed a binding cage2, made of two to four aromatic amino acid side chains, that accounts for the specific interaction with a methyllysine at the positively charged methylated ammonium group through cation-π interactions3–9. In one instance, it was shown that the selectivity for a particular methylation state—a dimethyllysine over a trimethyllysine—is mediated by the carboxylate side chain of an aspartate that forms a hydrogen bond with the dimethylammonium hydrogen7. Besides the methylation state, interactions of the effector protein with several histone amino acids adjacent to the methyllysine confer high specificity for a given methylated histone site3–9.
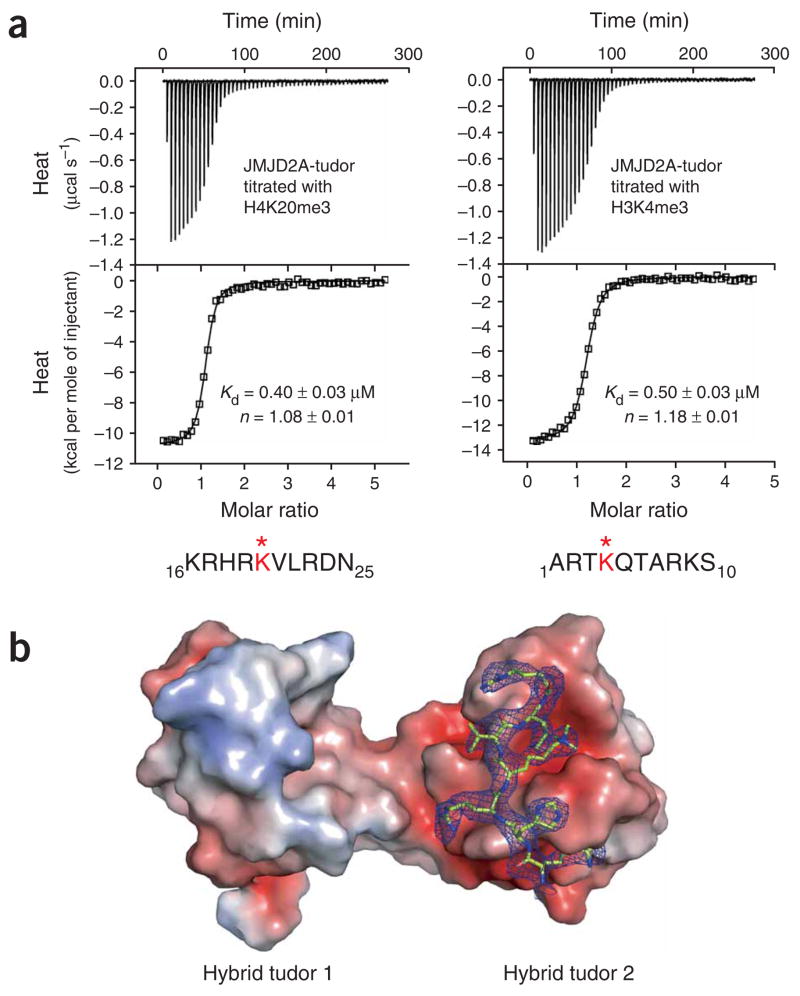
The tandem hybrid tudor domains of the human histone demethylase JMJD2A (JMJD2A-tudor) can recognize H3K4me3 (residues 1–10) and H4K20me3 (residues 16–25), two histone peptides that do not share any amino acid sequence similarity except the trimethylated lysine10. H3K4me3 and H4K20me3 bound with about the same Kd to JMJD2A-tudor, as shown by isothermal titration calorimetry (ITC) (Fig. 1a and Table 1). To better understand the histone binding mechanism of JMJD2A and probe possible differences between the recognition of histones H3K4me3 (ref. 6) and H4K20me3 by JMJD2A-tudor, we determined the crystal structure of human JMJD2A-tudor in complex with H4K20me3 at a resolution of 2.8 Å (Figs. 1b and 2; see Supplementary Methods and Supplementary Table 1 online for details). In the crystal asymmetric unit, there are two copies of the JMJD2A-tudor–H4K20me3 complex. In one complex, seven residues (residues 17–23) of the peptide were unambiguously modeled in the electron density map (Fig. 1b and Supplementary Fig. 1 online), whereas in the other complex five residues (residues 17–21) of the peptide were readily modeled. The two H4K20me3 peptides bound the same surface of each JMJD2A-tudor protein, both in the same orientation. However, whereas in one complex the trimethyllysine was caged by a canonical arrangement of the aromatic side chains of Phe932, Trp967 and Tyr973 of JMJD2A-tudor (Fig. 2b), in the other complex the trimethyllysine was pulled out of the aromatic pocket due to crystal packing interactions; this peptide was located at the interface between the two JMJD2A-tudor molecules in the crystal asymmetric unit (not shown). We also determined the three-dimensional structure of JMJD2A-tudor in the absence of peptide at a resolution of 1.8 Å (Supplementary Fig. 2a and Supplementary Table 1 online). Notably, the two molecules of JMJD2A-tudor in the crystal asymmetric unit form a head-to-tail dimer, with the guanido group of Arg913 of one molecule occupying the aromatic cage of the other molecule (Supplementary Fig. 2b). We identified exactly the same dimeric arrangement for two of the four proteins in the asymmetric unit of free JMJD2A-tudor crystallized in a different crystalline lattice6. This might correspond to a native arrangement of JMJD2A-tudor in the context of the full-length protein unbound to a methylated ligand. Native JMJD2A is a multimer, although it is not known whether the tudor domains participate in the oligomerization of the protein11. Comparison of the structure of JMJD2A-tudor in the free state with that in complex with H4K20me3 did not reveal any substantial difference in backbone and side chain conformations (data not shown).
Figure 1.
Interaction of JMJD2A hybrid tudor domains with trimethylated histones H3 and H4 peptides. (a) ITC of JMJD2A-tudor with H4K20me3 (left) and with H3K4me3 (right). The peptide amino acid sequences are indicated with the methylated (*) lysine in red. Raw titration data and integrated heat measurements are shown in the upper and lower plots, respectively. The Kd and stoichiometry numbers (n) obtained by fitting a standard one-interaction-site model are reported with the associated s.d. determined by nonlinear least-squares analysis. (b) Molecular surface and electrostatic potential representation of JMJD2A-tudor in complex with the H4K20me3 peptide. The electrostatic potential is shown in red for negatively charged and blue for positively charged surfaces. The peptide is in stick representation with the 2Fo– Fc electron density map displayed at the 1.0σ contour level.
Table 1.
Effect of mutations on JMJD2A binding to H3K4me3 and H4K20me3 peptides
|
Kd (μM)
|
||
|---|---|---|
| Hybrid tudor domains of JMJD2A | H3K4me3 peptide | H4K20me3 peptide |
| Wild type | 0.50 ± 0.03 | 0.40 ± 0.03 |
| D939R | 3.39 ± 0.14 | 85.87 ± 4.24 |
| D945R | 99.01 ± 5.84 | 0.77 ± 0.04 |
| N940R | 23.15 ± 1.45 | 1.00 ± 0.06 |
| Y942A | 1.52 ± 0.04 | 0.50 ± 0.03 |
| Y942R | 1.87 ± 0.08 | 0.76 ± 0.10 |
| T968A | 0.94 ± 0.05 | 0.21 ± 0.07 |
| T968R | 4.67 ± 0.10 | 0.70 ± 0.05 |
|
| ||
| H4K20me3 H18G peptide | H4K20me3 R19G peptide | |
|
| ||
| Wild type | 2.10 ± 0.11 | 21.93 ± 0.91 |
The Kd values were derived using ITC by fitting a one-site binding model and are reported with the associated s.d. determined by nonlinear least-squares analysis.
Figure 2.
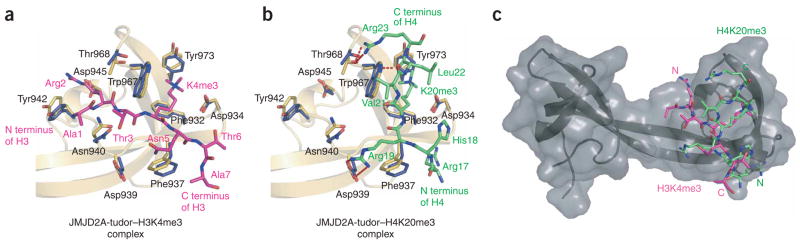
Two different binding modes of JMJD2A hybrid tudor domains with H3K4me3 and H4K20me3 peptides. (a, b) Close-up views of the JMJD2A-tudor interaction sites with H3K4me3 (a, PDB 2GFA6) and H4K20me3 (b). Amino acids of JMJD2A-tudor involved in binding H3K4me3 and H4K20me3 are shown. (c) Overall view of JMJD2A-tudor in complex with superimposed H3K4me3 (pink) and H4K20me3 (green), illustrating the opposite orientations of these peptides.
Analysis of the JMJD2A-tudor–H4K20me3 structure in light of the recently published structure of JMJD2A-tudor bound to H3K4me3 (ref. 6) unexpectedly revealed two very different binding modes (Fig. 2a–c). Although the two histone peptides were bound to the second hybrid tudor domain with the methyllysine caged by the same three aromatic residues, they adopted opposite relative orientations and contacted different surfaces of the hybrid tudor domain. Comparison of the two structures immediately suggested that mutations could be designed to impair the interaction of JMJD2A-tudor with H3K4me3 but not with H4K20me3, and vice versa.
Different aspartate side chains of JMJD2A established coulombic and hydrogen bond interactions with an arginine of each peptide. JMJD2A Asp945 interacted with Arg2 of the H3K4me3 peptide but did not contact the H4K20me3 peptide. JMJD2A Asp939, on the other hand, was salt-bridged to Arg19 of H4K20me3 but was not involved in any interaction with H3K4me3. As shown in Table 1, a D945R mutation increased the Kd of JMJD2A for H3K4me3 by ~200-fold—in agreement with previously published data6—but did not affect the interaction with H4K20me3. A D939R mutant had the opposite effect of increasing the Kd of JMJD2A for H4K20me3 by ~200-fold but did not markedly change the affinity for H3K4me3. Like Asp945, Asn940 of JMJD2A interacted with H3K4me3 but not with H4K20me3. Asn940 formed hydrogen bonds with the backbone amide and hydroxyl groups of H3K4me3 Thr3. Consistent with these observations, mutating Asn940 into an arginine did not perturb JMJD2A binding to H4K20me3 but decreased its affinity for H3K4me3 by ~46-fold.
Two other amino acids of JMJD2A, Tyr942 and Thr968, involved in weaker intermolecular hydrogen bonds, selectively contacted one peptide and not the other; we also mutated these. Through its hydroxyl group, Tyr942 in the wild-type structure formed a hydrogen bond with the terminal amino group of the H3K4me3 peptide but was far from any residue of H4K20me3. Replacing Tyr942 by an alanine or an arginine had no marked effect on the Kd of JMJD2A for H3K4me3 or H4K20me3. The hydroxyl group of Thr968 in the wild-type structure formed a hydrogen bond with the guanido group of H4K20me3 Arg23 but did not contact H3K4me3. Changing Thr968 to an alanine or arginine did not appreciably alter the interaction of JMJD2A with either H3K4me3 or H4K20me3. Thus, Tyr942 and Thr968 are not essential for the tight binding of JMJD2A to H3K4me3 and H4K20me3.
We also introduced mutations in the H4K20me3 peptide at the His18 and Arg19 positions. Replacing His18 by a glycine increased the Kd by fivefold. This small change in Kd is in agreement with the structure, in which only the carbonyl group of His18 contacted JMJD2A, through a hydrogen bond with the amide hydrogen atom of Phe937. If H4K20me3 were oriented similarly to H3K4me3 in the complex with JMJD2A-tudor, His18 would occupy the position of the key Arg2 residue of H3K4me3 (ref. 6), and mutation of His18 into a glycine would have been likely to lead to a larger increase in Kd. Consistent with the structure of the JMJD2A-tudor–H4-K20me3 complex, changing Arg19 to a glycine decreased the affinity of H4K20me3 for JMJD2A by ~55-fold. As mentioned above, Arg19 of H4K20me3 forms a salt bridge with JMJD2A Asp939.
These mutations, summarized in Table 1, validate in solution the different histone binding modes of JMJD2A for H3K4me3 and H4K20me3. In the future, they may also become useful for deciphering the relative roles of H3K4me3 and H4K20me3 in vivo.
The precise function of JMJD2A is unknown, but it was shown to demethylate histone H3 dimethylated or trimethylated at lysine 9 and lysine 36, by means of an N-terminal catalytic domain11–17. As the hybrid tudor domains at the C terminus of JMJD2A are necessary for the demethylation activity of JMJD2A in vivo, it is plausible that they could recruit JMJD2A to chromatin through their interaction with methylated H3K4, H4K20 or both. The full-length JMJD2A protein is a multimer, most likely a dimer11, and could thus combinatorially interact with methylated H3K4 and H4K20.
In summary, this study has uncovered an unsuspected complexity in the recognition of methylated histones by an effector protein. The hybrid tudor domains of the histone demethylase JMJD2A can recognize equally well two unrelated histone peptides, H3K4me3 and H4K20me3, by means of two very different binding mechanisms. There is virtually no change in the conformation of JMJD2A-tudor in the free or H3K3me3- or H4K20me3-bound states. Although the only residue common to H3K4me3 and H4K20me3, the trimethyllysine, occupies the same binding cage in JMJD2A-tudor, the specificity of interaction is driven by opposite orientations of the peptides that contact different surfaces of JMJD2A. These binding modes ensure dual specificity while preventing the two peptides from simultaneously interacting with the same JMJD2A monomer. The biological significance of this observation remains to be established. Tudor domains as well as structurally related chromodomains and MBT domains appear in a wide variety of proteins1, several of which can bind more than one methylated histone target10 and can probably bind methylated proteins other than histones. It will be interesting to see whether some of these effector domains recognize more than one methylated amino acid sequence by means of different binding mechanisms, similarly to JMJD2A.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We acknowledge the use of beamlines 19BM and 19ID at Argonne National Laboratory’s Advance Photon Source (APS). We thank Y. Kim and C. Chang at APS for their assistance with X-ray data collection. Use of Argonne National Laboratory Structural Biology Center beamlines at APS is supported by the US Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported by grants from the Human Frontier Science Program and from the US National Institutes of Health (CA109449) to G.M.
Footnotes
Accession codes. Protein Data Bank: The atomic coordinates and structure factors have been deposited with the accession codes 2QQR for JMJD2A-tudor in the free state and 2QQS for H4K20me3-bound JMJD2A-tudor.
References
- 1.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Selenko P, et al. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs SA, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen PR, et al. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan JF, et al. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 7.Botuyan MV, et al. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, et al. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pena PV, et al. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, et al. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin S, Janknecht R. Biochem Biophys Res Commun. 2007;353:973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 12.Whetstine JR, et al. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, et al. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, et al. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Ng SS, et al. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 16.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z, et al. Proc Natl Acad Sci USA. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.