Abstract
The Ets2 transcription factor is essential for the development of the mouse placenta and for generating signals for embryonic mesoderm and axis formation. Using a conditional targeted Ets2 allele, we show that Ets2 is essential for trophoblast stem (TS) cells self renewal. Inactivation of Ets2 results in TS cell slower growth, increased expression of a subset of differentiation associated genes and decreased expression of several genes implicated in TS self renewal. Among the direct TS targets of Ets2 is Cdx2, a key master regulator of TS cell state. Thus Ets2 contributes to the regulation of multiple genes important for maintaining the undifferentiated state of TS cells and as candidate signals for embryonic development.
Introduction
The placenta is critical for the intrauterine survival of mammalian embryos, and defective placentation is often the cause of early- to mid-gestation lethality in targeted mouse mutations (Copp, 1995; Rossant and Cross, 2001). Placental failure causes abnormalities in the embryo that can be mistaken for primary embryonic defects (Wu et al., 2003). The trophoblast lineage arises in the preimplantation embryo as a consequence of the mutual inhibition of the homeobox transcription factor, Cdx2 and the embryonic stem cell transcription factor, Oct3/4(Niwa et al., 2005). After implantation, the polar trophectoderm continues to proliferate and gives rise to the diploid extraembryonic ectoderm (ExE) and ectoplacental cone (EPC) of the early conceptus. The outermost cells of EPC differentiate into secondary giant cells, which lie at the periphery of the placenta, forming an interface with maternal decidua. The remaining diploid cells of EPC form a compact layer of spongiotrophoblast (SpT) between the labyrinth and the giant cells. These three layers comprise the mature placenta in mice by E10 and persist throughout the rest of the pregnancy.
The Ets family of transcription factors share the Ets winged helix-turn-helix DNA-binding domain (Wasylyk et al., 1993). Over 30 Ets family members have been identified in organisms ranging from Drosophila to human. These factors have been implicated in a variety of biological processes, including cellular proliferation, differentiation, embryonic development, transformation, immune response and apoptosis (Oikawa and Yamada, 2003). Many members of the family are co-expressed in single cell types (Galang et al., 2004). A subset of Ets proteins, including Drosophila Pointed-P2 and vertebrate Ets1 and Ets2, contain a N-terminal pointed domain that is involved in mediating Ras signaling via a conserved MAPK phosphorylation site (Brunner et al., 1994; Wasylyk et al., 1997; Yang et al., 1996).
Ets2 is expressed in many cell types in developing mouse embryos (Maroulakou et al., 1994) as well as in most adult tissues. Inactivation of Ets2 in mice by deletion of the Ets domain (Ets2db1) caused embryonic lethality before E8.5 (Yamamoto et al., 1998). Analysis of the mutant Ets2db1/db1conceptuses revealed deficiencies in trophoblastic tissues, including a smaller ectoplacental cone and absence of chorion formation. Further analysis has revealed that many Ets2db1/db1 conceptuses fail to form ExE (Georgiades and Rossant, 2006). The ExE has recently been demonstrated to be the origin of signals to the epiblast to initiate mesoderm formation and epiblast axis formation (Georgiades and Rossant, 2006). This signal may be mediated by TGF-β family members that require processing by either of two proteases, Furin and Pace4 (Beck et al., 2002). Two different members of the TGF-β family, Nodal and Activin, have been implicated in maintaining the undifferentiated state of the TS cell (Guzman-Ayala et al., 2004).
Ets2db1/db1 embryos were rescued by aggregation with tetraploid wild-type embryos that provide complementary wild-type function for extraembryonic tissues. The resulting mice were largely normal with the exception of a “waved” hair phenotype resembling that of TGF-α knockout mice (Luetteke et al., 1993; Mann et al., 1993) and the waved-2 mice harboring a mutation in EGF receptor (Luetteke et al., 1994). A point mutation in Ets2 substituting alanine for the conserved MAPK phosphorylation site Thr-72 generated a hypomorphic allele designated Ets2A72 (Man et al., 2003). While mice homozygous for Ets2A72 were viable, the basal activity of Ets2A72 was haploinsufficient for embryonic development as Ets2A72/db1 embryos died around E11. In accordance with the trophoblastic tissue phenotype of Ets2db1/db1 mutants, the Ets2A72/db1 placenta displayed severely underdeveloped labyrinth and SpT. Thus, Ets2 is essential for the early and later development of mouse placental tissues and Ets2 activity is significantly modulated by phosphorylation of Thr-72.
Trophoblast stem (TS) cells can be isolated by the culture of blastocysts or early ExE (Tanaka et al., 1998) and provide an opportunity to investigate self renewal, commitment to differentiation and trans-acting factor production. In the presence of FGF4 and embryonic fibroblast-conditioned medium (EFCM), TS cells proliferate continuously and express characteristic TS marker genes. Upon withdrawal of FGF4 and EFCM, TS cells differentiate into multiple trophoblast subtypes but predominantly trophoblast giant cells (TGCs). While FGF4 signaling is well established to be essential for TS cell maintenance (Rossant and Cross, 2001; Simmons and Cross, 2005), it is less clear what are the key downstream effectors. Most notable among the candidates are two transcription factors, caudal-type homeobox 2 (Cdx2) and Eomesodermin homologue (Eomes). Both show early strong expression in the trophectoderm/ExE, and loss of either gene in embryos causes peri-implantation death (Chawengsaksophak et al., 1997; Russ et al., 2000). In addition, member(s) of the TGF-β superfamily have been implicated as the active component(s) in EFCM contributing to maintenance of TS cell proliferation. While TGF-β and/or activin can replace EFCM in TS cell cultures (Erlebacher et al., 2004), genetic evidence suggest that Nodal may be the embryo-derived growth factor cooperating with FGF4 signaling to promote TS cell maintenance (Guzman-Ayala et al., 2004).
Here we describe the generation of an Ets2 conditional allele and show that Cre-mediated Ets2 inactivation causes a defect in the self renewal of TS cells. Inactivation of Ets2 results in changes in gene expression normally associated with differentiation caused by growth factor withdrawal. Among the genes sensitive to Ets2 inactivation are Cdx2, Pace4, Eomes and Errβ. Ets2 is essential to maintain expression of multiple genes necessary for TS cell self renewal.
Materials and methods
Generation of the Ets2flox mice
BE1, an Ets2 genomic DNA plasmid containing exon 6-10, was used to construct the targeting vector. pM30/pK11 (a gift of Dr. Gail R. Martin), a plasmid containing a Pgk-neo expression cassette flanked by frt sites and a loxP site inserted 3′ of the second frt site (Meyers et al., 1998), was used to insert the frt-neo-frt-loxP fragment into the PshAI site of intron 8. A second loxP site was cloned into the KpnI site of the 3′ UTR by using oligos containing the loxP sequence and KpnI compatible ends. Subsequently an XhoI/SalI fragment containing a herpes simplex virus thymidine kinase cassette was inserted upstream of exon 6 into the unique EcoRI site that had been converted to an XhoI site.
R1 ES cells were electroporated with the linearized targeting vector, and 132 G418- and Gancyclovir-resistant clones were isolated by the Burnham Institute Mouse Genetics service. 37 homologous recombinants were identified by Southern blot analysis using a probe located outside of the targeting vector, an approximately 300 bp EcoRI/BamHI fragment in the 3′ region. Since the recombination event can occur after either of the two loxP sites, all recombinant clones were further screened by PCR using primers flanking the second loxP site. 27 recombinant clones were confirmed to have the second loxP site. After karyotyping, two clones were chosen for injection into C57BL/6J blastocysts, and germ-line transmission of the targeted allele Ets2flox-neo was obtained from one clone, giving rise to the Ets2flox-neo mouse line.
Embryos derived from matings of F1 heterozygous males with superovulated wild-type females were used for microinjection with pCAGGS-Flpe (a gift from Dr. Stewart), an optimized thermostable Flp expression vector (Buchholz et al., 1998). Flp-mediated deletion of the Pgk-neo cassette occurred in five out of eight mice born containing the targeted allele, resulting in the final conditional allele Ets2flox. One of the Ets2flox mice without integration of the Flpe into the genome was used for establishing the Ets2flox line.
Transgenic mice
The R26R Cre reporter mice in 129S3/SvImJ background and the MORE Cre line in C57BL/6 background were obtained from Jackson Laboratory. Genotyping of transgenic mice was performed by standard PCR analysis of tail lysates. The gene-specific primers used were for Cre: CTGGCATTTCTGGGGATTGC and ACGGAAATCCATCGCTCGAC; for Ets2: primer a (GCCACAGCAAACCTCTTTCT), b (GACCCACTTGCTCCAAAGAC) and c (GCTTCCCAGAGACTCTTCCC). Primer b and c produced a 176-bp and a 217-bp fragment from wild-type and Ets2flox-neo/Ets2flox alleles, respectively. Primer a and c produced a 280-bp fragment from the Ets2db2 allele. All animal experiments conformed to NIH guidelines and were approved by the Burnham Animal Research Committee.
Adenovirus propagation and titration
Low passage 293 cells (Microbix Biosystems) were maintained as recommended by the company. Both AdGFP (Qbiogene) and AdCreGFP (a gift from Dr. Serguei V. Kozlov, to be published elsewhere) are replication-deficient, serotype 5 adenoviruses. Viruses were propagated in 293 cells and titrated using a Tissue Culture Infectious Dose 50 (TCID50) method following the manufacturer's instructions. In addition, MEFs derived from the R26R Cre reporter mice (Soriano, 1999) were used to titrate the AdCreGFP virus. MEFs were incubated with 5-fold serially diluted virus and Cre-mediated recombination efficiency was determined by lacZ staining 3 days post infection (dpi). Based on titration result from the TCID50 method, a MOI of 10 is sufficient to achieve more than 95% recombination in MEFs at 3 dpi, whereas a MOI of 20-30 is required for 70-90% infection efficiency in TS cells.
Isolation, culture and analysis of TS cells
TS cells were isolated from blastocysts and grown as previously described (Tanaka et al., 1998). Briefly, Growth medium (GM) was RPMI 1640 supplemented with 20% FBS, 1 mM sodium pyruvate, 100 μM b-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 25 ng/ml human recombinant FGF4 (Sigma), 1 μg/ml heparin, and 70% EFCM. TS cell differentiation was induced by withdrawal of FGF4 and EFCM after trypsinization and replating at appropriate density.
For TS colony formation assay, optimally growing TS cells were trypsinized and replated at 500-2000 cells per 6-cm dish onto feeder cells, and fed with fresh GM every 2-3 days for 1-2 weeks. Colonies were counted following lacZ and Nuclear Fast Red staining as described below.
For Cdx2 rescue experiments, one million Ets2flox/flox;R26R TS cells were transfected with 5 ug of pCX-GR*cdx2iresHygEGFP plasmid, a Cdx2 expressing vector from Dr. Tomilin (Tolkunova et al., 2006), by the Amaxa Nucleofector method. Clones resistant to 200 ug/ml hygromycin B were isolated for analysis. Two clones were tested for rescue by infection with AdCreGFP followed by subculture for up to 20 days without hygromycin. The fraction of cells positive for β-galactosidase was determined at varying times after infection. Cdx2 expression was confirmed by immunocytochemistry.
Semi-quantitative PCR analysis of Ets2flox recombination
Genomic DNA isolated from an Ets2flox/flox mouse tissue and AdCreGFP-infected Ets2flox/flox MEFs was used as 100% Ets2flox and Etsdb2 standard, respectively. The purified DNA samples at the same concentration (30 ng/μl) were mixed to generate a DNA standard set with specified Ets2flox: Ets2db2 ratios. The Ets2-specific primers a, b and c were used for the analysis, and the integrated density value (IDV) of products amplified from Ets2flox and Ets2db2 alleles was measured using the Alpha Imager 2200 software. The ratios of the Ets2flox IDV versus Ets2db2 IDV were plotted as a function of the percentages of Ets2flox content, generating a linear standard curve with a R2 value of 0.96-0.99 (Fig. 2 F). The accuracy of the analysis was confirmed by repeated PCR using different aliquots of the primer mix and the DNA standard set, various amplification cycle numbers (30-40), and different dilutions of the same samples (3-300 ng/μl).
Figure 2. Rescue of Ets2db2 lethality by MORE mice.
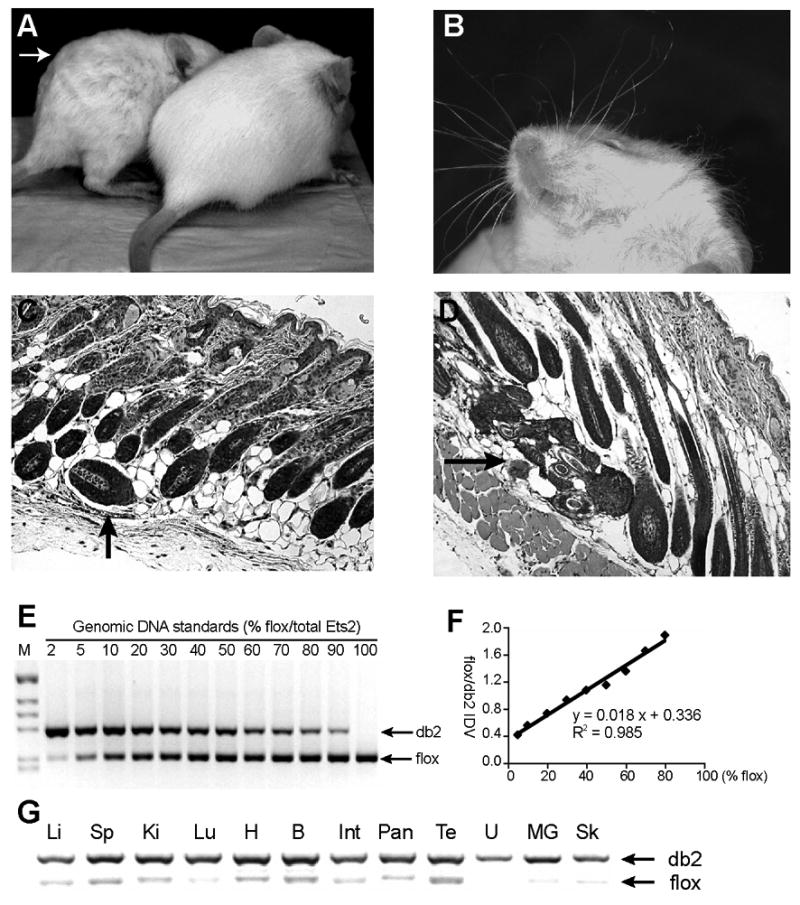
(A-D) Recapitulation of the “waved” hair phenotype in MORE;Ets2db2/db2 mice. (A) Waved hair (arrow) in comparison to a MORE;Ets2db2/wt littermate. (B) Curly whiskers. (C and D) Hematoxylin and eosin-stained (H&E) sections of the skin show misaligned hair follicles and foci of several dysplastic follicles (arrows), respectively. (E) A representative gel picture of the Ets2 PCR result with genomic DNA standards containing denoted allele ratios. Primers a, b and c (depicted in Fig. 1 A) were used to amplify a 217-bp product from Ets2flox and a 280-bp product from Etsdb2. (F) A standard curve was drawn accordingly plotting the ratio of the Ets2flox band density to Ets2db2 band density as a function of Ets2flox content. Also shown are the linear regression equation and R2 value. (G) PCR genotyping of the somatic tissues from the rescued MORE;Ets2db2/db2 adults: Li, liver; Sp, spleen; Ki, kidney; Lu, lung; H, heart; B, brain; Int, intestine; Pan, pancreas; Te, testis; U, uterus; MG, mammary gland; Sk, skin. The percentages of Ets2db2, calculated from the standard curve, were more than 95% for all the samples except for the testis (87%). The genomic DNA was extracted from the tissues of one male and one female mouse. Similar results were obtained with samples from additional mice.
Histology and immunoblotting
For lacZ staining, cells were fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in PBS for 5 minutes, washed once in PBS and twice in lacZ wash buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 in PBS) for 5 minutes. Staining was carried out in 1 mg/ml X-gal, 10 mM potassium ferrocyanide, and 10 mM potassium ferricyanide in lacZ wash buffer at 37°C overnight. Nuclear Fast Red counterstain was used to visualize lacZ-negative cells.
For genotyping of embryos or lacZ-fixed TS colonies, the extraembryonic portions on paraffin sections or the individual colonies were scraped off under dissection microscope and incubated overnight in lysis buffer (100 ug/ml proteinase K, 50 mM KCl, 10mM Tris-HCl, 2.5mM MgCl2, 0.1 mg/ml gelatin, 0.45% NP-40, and 0.45% Tween20, pH 8.3) prior to PCR.
For BrdU incorporation assay, BrdU was added to the medium at 20 μg/ml for 1 hour, washed twice in PBS and fixed in 70% ethanol. After rehydration, DNA was denatured in 2N HCl for 30 min, and blocked in PBS containing 1% goat serum. G3G4, a mouse anti-BrdU antibody (Developmental Studies Hybridoma Bank), was applied at 1:100 and detected with Alexa568 goat anti-mouse antibody (Molecular Probes).
Frozen colon tissues and TS cultures were harvested in cell lysis buffer (Cell Signaling) supplemented with protease inhibitor cocktail (Sigma). Protein concentrations were determined with the Bio-Rad DC protein assay. Endogenous Ets2, Cdx2 and tubulin were detected by immunoblotting using the rabbit polyclonal anti-Ets2 (Santa Cruz, SC-351), monoclonal anti-Cdx2 (BioGenex, MU392-UC) and anti-α-tubulin (Sigma, T6199) antibodies.
mRNA expression analysis
RNA isolation from TS cells, cDNA preparation and real-time PCR were performed as described previously (Galang et al., 2004). Peptidylprolyl isomerase A/cyclophilin A was used as an internal reference for normalization. The primers listed in Supplementary Table S4 were used for cDNA amplification. Primers for Ets factors have been described (Galang et al., 2004).
Gene expression array analyses
We analyzed RNA with Illumina MouseRef-8 and MouseRef-6 Expression BeadChips using the manufacturer's BeadArray Reader and collected primary data using the supplied Scanner software. Data analysis was done in three stages. First, expression intensities were calculated for each gene probed on the array for all hybridizations (26 in total) using Illumina's Beadstudio#1 software. Second, intensity values were quality controlled and normalized: quality control was carried out by using the Illumina Beadstudio detection P-value set to <0.1 as a cutoff. This removed genes which were effectively absent from the array (that is, were not detected). After this step, the initial gene set was reduced to 14888. All the arrays were then normalized using the normalize.quantiles routine from the Affymetrics package in Bioconductor. This procedure accounted for any variation in hybridization intensity between the individual arrays. An assessment of several different normalization techniques using the Bioconductor maCorrPlot routine suggested that normalize.quantiles was the most appropriate for the data.
Finally, these normalized data were imported into GeneSpring and analyzed for differentially expressed genes. The groups of biological replicates were described to the software, and significantly differentially expressed genes were determined on the basis of t-tests and fold difference changes in expression level.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (Hollenhorst et al 2004). Briefly, TS cells in GM at ∼90% confluence were crosslinked, lysed and sonicated followed by incubation with 2 ug of rabbit IgG, anti-Ets2 (Santa Cruz, SC-351) or anti-acetylated histone 3 (Upstate, 06-599) antibody at 4°C overnight. Immunoprecipitated fractions were phenol/chloroform extracted, precipitated and resuspended in 100 ul water. Quantitative PCR of ChIP samples was performed using QuantiTect SYBR Green PCR Kit (Qiagen) on a Mx3000P system (Stratagene). Sequences of primers used to amplify the indicated regions of the mouse Cdx2 promoter are available upon request.
Luciferase reporter genes and assays
The flanking regions of the mouse Cdx2 gene (nt -3031 to +73) was amplified by PCR using Expand High Fidelity PCR kit (Roche) from BglII/NotI digested pcS170 genomic clone containing the Cdx2 locus (Tamai et al., 1999) and cloned into MluI/XhoI sites of pGL3B (Promega). Fragments of Cdx2 promoter were cloned by PCR Supermix High Fidelity (Invitrogen) amplification of pcS170. Primers CGTCTCGAGTTACTAATAGAGTCTTGTAACGT and AGATCTTTCCTTCTTTCCTCCCACC spanned nt -56 to +73 of the Cdx2 promoter and generated XhoI and BglII restriction sites at the 5′ and 3′ ends, respectively, of the amplified product. Overlap PCR using two primer pairs (ACGCGTGGGACAATTAGCCAGCGG and CGAGTGTTTACAAGACTCTATTTAGGACTATCTCCCTCCC, GGGAGGGAGATAGTCCTAAATAGAGTCTTGTAAACACTCG and AGATCTTTCCTTCTTTCCTCCCACC) generated a fusion product of nt -2233 to -1991 and nt -51 to +73 of the Cdx2 promoter. PCR fragments were cloned into the pCR2.1TOPO vector and sequenced. The minimal Cdx2 promoter fragment was cloned into XhoI/BglII sites of pGL3B to generate Cdx2_56. The Cdx2_56DE1 fusion product was cloned into SacI/XhoI sites of pGL3B. The E18 Ets2 reporter gene has been described (Galang et al., 1994).
100,000 Caco-2 cells were plated to each well of 24-well plates in DMEM with 20% fetal calf serum. The next day 0.5 ug of reporter vector, 250 ng of expression vector and 50 ng of pRL-null vector were transfected using Lipofectamine 2000 (Invitrogen). Media was changed to DMEM containing 0.5% serum one day after transfection and cells were incubated overnight before lysis. Luciferase was detected using the Dual Luciferase Assay System kit (Promega,) on a Veritas luminometer (Turner Biosystems).
Results
Generation of an Ets2 conditional allele
To facilitate study of Ets2 function in different cell types, we generated by gene targeting a conditional Ets2 allele that could be inactivated by Cre-mediated recombination (Fig. 1A). Two loxP sites were introduced into the targeting vector to flank exon 9 and 10 that code for a major part of the conserved and essential DNA binding domain. Additionally, a Pgk-neo cassette flanked by Flip recombinase recognitions sites (frt) was inserted into intron 8, which together with a thymidine kinase gene inserted upstream of exon 6, provided both positive and negative selection for homologous recombination. Twenty-seven targeted ES cell clones were identified from a total of 132 clones screened by Southern blot analysis with a 3′ flanking probe (Fig. 1B) and PCR screening for the presence of both loxP sites. After karyotyping, two clones were selected for injection into blastocysts and one transmitted the targeted allele, designated Ets2flox-neo, through the germ line. The Ets2flox-neo mice were phenotypically wild type and transmitted the targeted allele at the expected Mendelian frequency.
Figure 1. Generation of an Ets2 conditional allele.
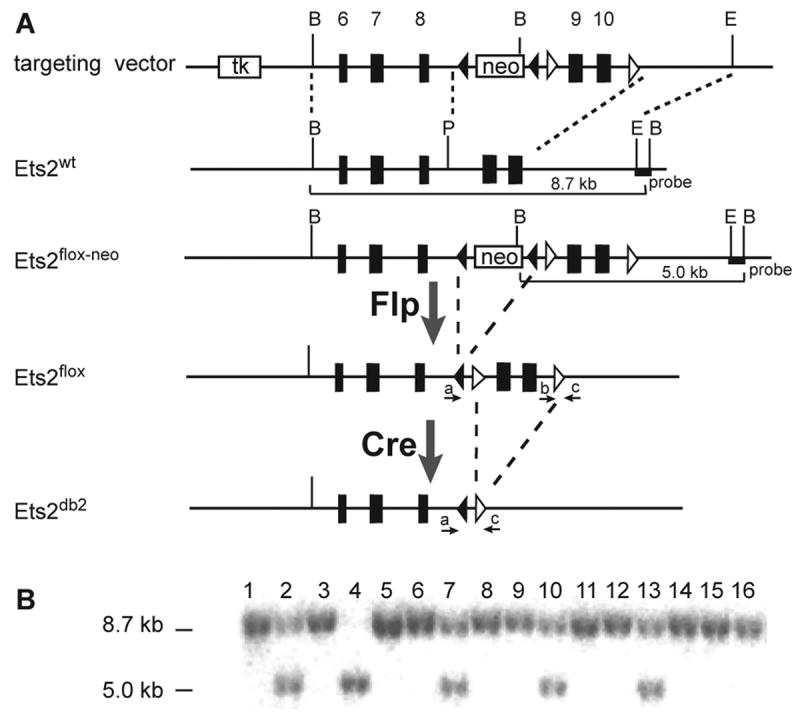
(A) Schematic of the targeting construct, the wild-type Ets2 allele, the targeted Ets2flox-neo allele, the conditional Ets2flox and the inactivated Ets2db2 allele following Flp- and Cre-mediated recombination, respectively. The frt and loxP sites are indicated by filled and open arrowheads, respectively. The 3.8-kb 5′ arm of homology and the 2.6-kb 3′ arm of homology, indicated by the dashed lines, direct the homologous recombination. The diagnostic BamHI restriction fragments are depicted by solid lines. The 3′ probe used for Southern blot analysis and the primers (a, b and c) used for PCR genotyping are indicated by the filled box and arrows, respectively. B, BamHI; E, EcoRI; P, PshAI. (B) Southern blot analysis of BamHI-digested genomic DNA from ES cells hybridized with the 3′ probe. A representative blot of 16 ES cell clones is shown here.
To remove the exogenous Pgk-neo sequence, an optimized thermostable Flip expression vector, Flpe (Buchholz et al., 1998), was injected into embryos carrying the Ets2flox-neo allele. Flpe-mediated recombination occurred in five out of eight mice born containing the targeted allele, resulting in the final conditional allele Ets2flox. Ets2flox homozygotes behaved as wild type with normal development and life span.
Ets2 inactivation and rescue of embryonic lethality by MORE mouse
An epiblast-specific Cre line, Mox2-Cre (MORE), was combined with Ets2flox/+ to recombine the Ets2flox allele in embryonic and germ cells (Tallquist and Soriano, 2000). The resulting recombined allele, designated Ets2db2, caused recessive embryonic lethality around E8 when bred to homozygosity (Table 1). Ets2db2/db2 embryos at E7.5 were very similar to the previously described Ets2db1/db1 mutants, including much smaller conceptuses and unusual cone-shaped yolk sacs (data not shown). We further tested the viability of embryos containing a combination of the Ets2A72 hypomorphic knock-in allele and Ets2db2. As expected, Ets2A72/db2 embryos died later at mid-gestation like previously described Ets2A72/db1 embryos while their Ets2A72/flox littermates developed normally (Table 1).
Table 1.
Embryonic lethality of Ets2db2/db2 and Ets2A72/db2
| Ets2db2/wt x Ets2db2/wt mating | |||||
|---|---|---|---|---|---|
| Age | Ets2wt/db2 | Ets2wt/wt | Ets2db2/db2 | Resorption | Total |
| Adult | 43 | 10 | 0 | 53 | |
| E8.5 | 9 | 5 | 4a | 7 | 25 |
| E7.5 | 10 | 4 | 6a | 0 | 20 |
| E6.5 | 9 | 3 | 4 | 0 | 16 |
|
| |||||
| Ets2A72/A72 x Ets2db2/wt mating | |||||
|
| |||||
| Age | Ets2A72/wt | Ets2A72/db2 | Resorption | Total | |
|
| |||||
| Adult | 24 | 0 | 24 | ||
| E11.5 | 8 | 4a | 8 | 20 | |
| E10.5 | 10 | 7a | 2 | 19 | |
| E9.5 | 17 | 14 | 0 | 31 | |
| E8.5 | 11 | 12 | 0 | 23 | |
apparently retarded or dying embryos
The combination of Ets2flox and the Cre recombinase of MORE mice can be used as an alternative to tetraploid embryo aggregation because of the restriction of Cre expression to only the embryonic tissues. To ensure as complete an embryonic inactivation of Ets2 as possible, we combined the Ets2flox/+ and MORE transgene to generate Ets2db2/+MORE progeny. The Ets2db2/+; MORE mice were mated with Ets2flox/flox to generate Ets2db2/flox; MORE progeny. The Ets2flox allele in the extraembryonic tissues was sufficient to support the development of the largely Ets2db2/db2embryos (Table 2). The near complete inactivation of Ets2 in the embryonic tissues of these mice was confirmed by a PCR analysis that determined the Ets2flox:Ets2db2 ratio in genomic DNA (Fig. 2E-G). Interestingly, these mice recapitulated the “waved” hair phenotype of Ets2db1/db1 mice rescued by tetraploid embryo aggregation. The young animals were distinguished by wavy hair and curly whiskers (Fig. 2A, B), which became less apparent over time. Skin sections revealed misaligned and dysplastic hair follicles (Fig. 2C, D), which is consistent with the observation in Ets2db1/db1 mice. Both sexes of these MORE; Ets2db2/db2 mice were fertile, and without obvious abnormality.
Table 2.
Rescue of Ets2db2/db2 lethality by MORE
| Genotype of mice from MORE;Ets2flox/wt x Ets2flox/flox matings | ||||||
|---|---|---|---|---|---|---|
| MORE
Ets2 |
-
wt/flox |
-
flox/floxb |
-
db2/flox |
+
wt/db2 |
+
db2/db2 |
Total |
| No. | 15 | 3 | 13 | 14 | 15 | 60 |
| Expecta | 20 | 0 | 20 | 20 | 0 | |
Expected is based on 100% of Ets2flox allele would be recombined in mice containing MORE and that Ets2db2/db2 would not be viable (Table 1) resulting in 33% of each remaining genotype.
Detection of Ets2flox/flox without MORE indicates that recombination frequency in the germ line of the MORE:Ets2flox/wt parent is about 13/16 (81%).
Isolation of TS cell lines
The failure to form ExE in Ets2 deficient embryos may be due to a deficiency of TS cells that arise in the same location under the influence of FGF4 produced by the epiblast. An initial attempt to isolate TS cells from Ets2db1/+ heterozygote intercross-derived blastocysts resulted in 19 TS lines but no Ets2db1/db1 TS cells. This result suggested that Ets2 might be required for TS cell maintenance as previously found for Eomes, Cdx2 and Elf5 (Donnison et al., 2005; Russ et al., 2000; Strumpf et al., 2005). TS cell lines carrying wild-type or the conditional Ets2 allele were generated. Some lines also contained the R26R Cre reporter that expressed bacterial β-galactosidase from the broadly active ROSA26 locus when Cre deleted an intervening Neo gene (Soriano, 1999).
Ets2 inactivation restricts TS cell self renewal
To evaluate the effect of loss of Ets2 on TS cells, we infected Ets2flox/flox TS cells with an adenovirus expressing Cre and GFP (AdCreGFP) to achieve optimal efficiency of Ets2 inactivation. A GFP-expressing adenovirus (AdGFP) was used as control to infect the TS cells. Inactivation of Ets2 modestly reduced the cell proliferation rate of the total population (Fig. 3A). However, PCR analysis of Ets2 alleles in the post-infection cells revealed a dramatic selection against the recombined Ets2db2 allele (Fig. 3B). As adenovirus-mediated Cre activity induces rapid recombination of multiple floxed sequences in individual primary mouse cells (Prost et al., 2001), we assumed that the post-infection culture comprised essentially completely recombined Ets2db2/db2 cells and uninfected Ets2flox/flox cells, but no partially recombined Ets2db2/flox cells. Subsequent clonal analysis supported this assumption (see below). Therefore, the percentages of Ets2db2 and Ets2flox alleles represented the percentages of Ets2db2/db2 and Ets2flox/flox cells, respectively. The deduced growth rate of the individual subpopulation indicated a doubling time of Ets2db2/db2 cells close to 62 hours (Fig. 3C). This was in contrast to the co-existing Ets2flox/flox subpopulation as well as the AdGFP- or mock-infected TS cells with doubling times of approximately 30 hours. Similar results were obtained from three independent Ets2flox/flox TS clones using different multiplicities of virus infection (MOI) and cells from various passages (data not shown).
Figure 3. Selection against Ets2db2 allele in AdCreGFP-infected Ets2flox/flox TS cells.
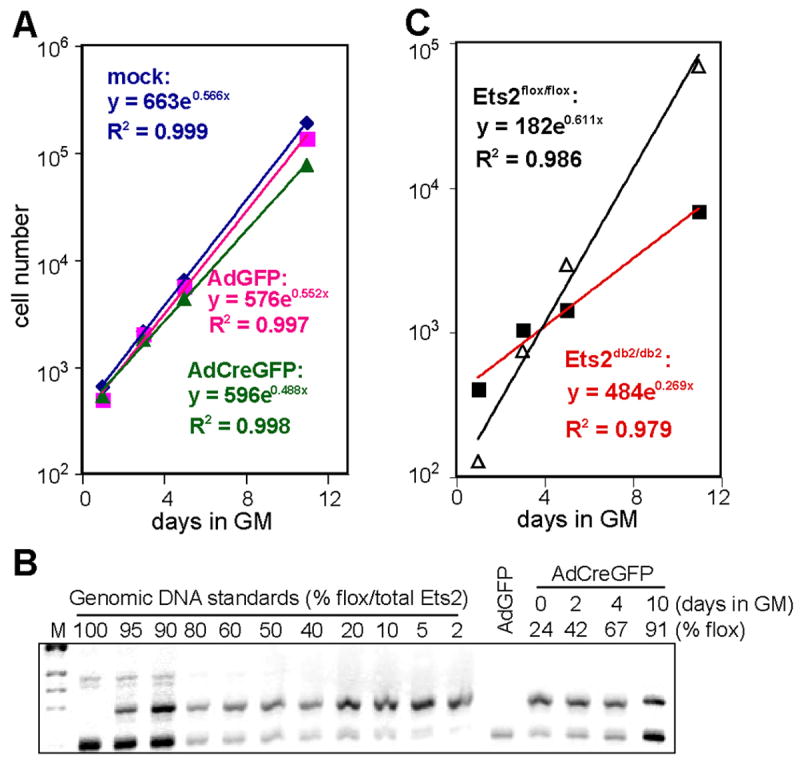
(A) Growth curve of Ets2flox/flox TS cells that were mock infected, infected with AdGFP, or infected with AdCreGFP. Cells were plated at 3 dpi, and averages of cell numbers in duplicate wells were plotted as a function of culture time in GM. Also shown are exponential trend line equations. (B) Agarose gel analysis of Ets2 PCR products in Ets2flox/flox TS cells post AdCreGFP infection. The percent Ets2flox allele was calculated by comparison to the mixed standards. (C) Deduced growth curves of Ets2flox/flox and Ets2db2/db2 subpopulations post AdCreGFP infection. Ets2flox/flox and Ets2db2/db2 cells at each time point calculated based on the PCR result: No. of Ets2flox/flox cells = total cell No. x %flox; No. of Ets2db2/db2 cells = total cell No. x %db2. The doubling time for each group, calculated according to the trend line equations, were 29.4 h (mock infected), 30.2 h (AdGFP-infected), 34.1 h (total AdCreGFP-infected), 27.2 h (Ets2flox/flox subpopulation post AdCreGFP infection), and 61.8 h (Ets2db2/db2 subpopulation post AdCreGFP infection). Similar results were obtained from 5 separate experiments with three independent clones.
The selection against the Ets2db2 alleles was specific to the homozygous TS cells. When heterozygous TS cells were infected with AdCreGFP, the percentage of Ets2db2 allele remained similar over ten days of culture (Fig. 4A). Consistently, β-galactosidase activity staining of TS cells carrying the R26R reporter and Ets2flox/flox confirmed the growth disadvantage of the recombined cells, whereas the Ets2 wild-type cells maintained a high percentage of positively stained cells over 3 weeks of culture (Fig. 4B).
Figure 4. Ets2flox/flox TS cell specific selection.
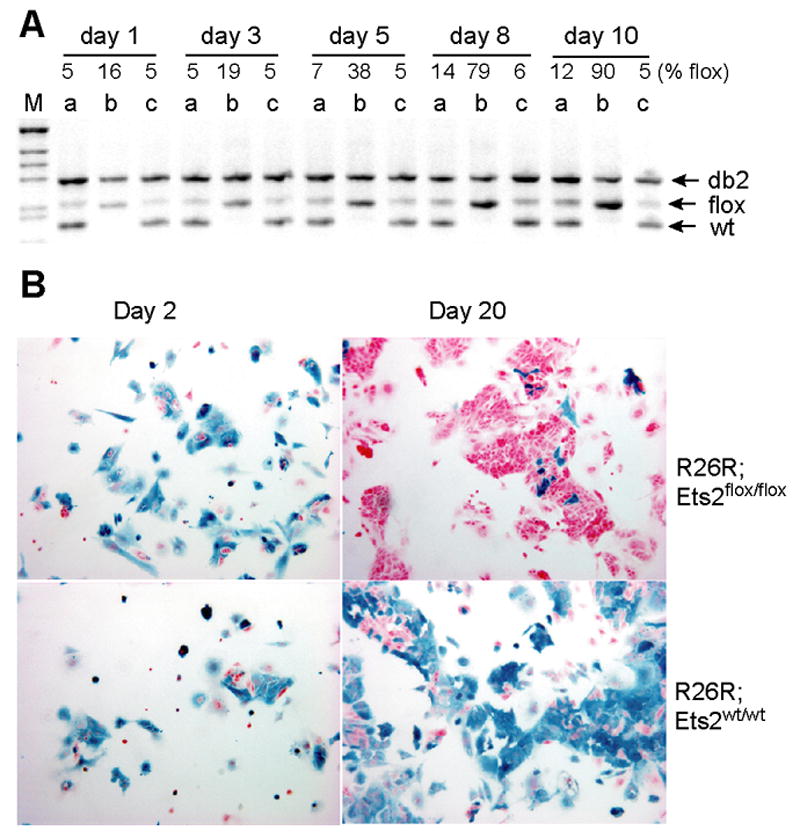
(A) Semi-quantitative PCR analysis of Ets2 alleles in three TS clones: R26R;Ets2flox/wt (a), R26R;Ets2flox/flox (b), and Ets2flox/wt (c) after AdCreGFP infection at the same MOI. The indicated percentages of Ets2flox/(Ets2db2 + Ets2flox) in each clone at indicated time points were calculated from a standard curve. Note the increase of the Ets2flox band in the homozygous TS clone b. (B) TS cells of indicated genotypes were infected with AdCreGFP and replated at 3 dpi (day 0). Cells were cultured in GM for indicated period prior to staining for β-galactosidase (blue). Upper panels show the virtual disappearance of β-galactosidase positive (blue) cells after 20 days of culture in the Ets2flox/flox TS cells. Unrecombined cells were visualized by nuclear fast red counterstain.
The selection against Ets2db2/db2 TS cells was specific to the stem cell culture conditions because only a slight decrease of Ets2db2 allele was detected when the post-infection cells were induced to differentiate by withdrawal of FGF4 and EFCM for the same period of time (Fig. 5A). Furthermore, Ets2flox/flox mouse embryo fibroblasts (MEFs) infected with AdCreGFP did not show significant selection against Ets2db2 allele (data not shown), as might be expected from the relatively normal size of rescued Ets2 deficient animals.
Figure 5. Ets2 dependent TS cell self-renewal.
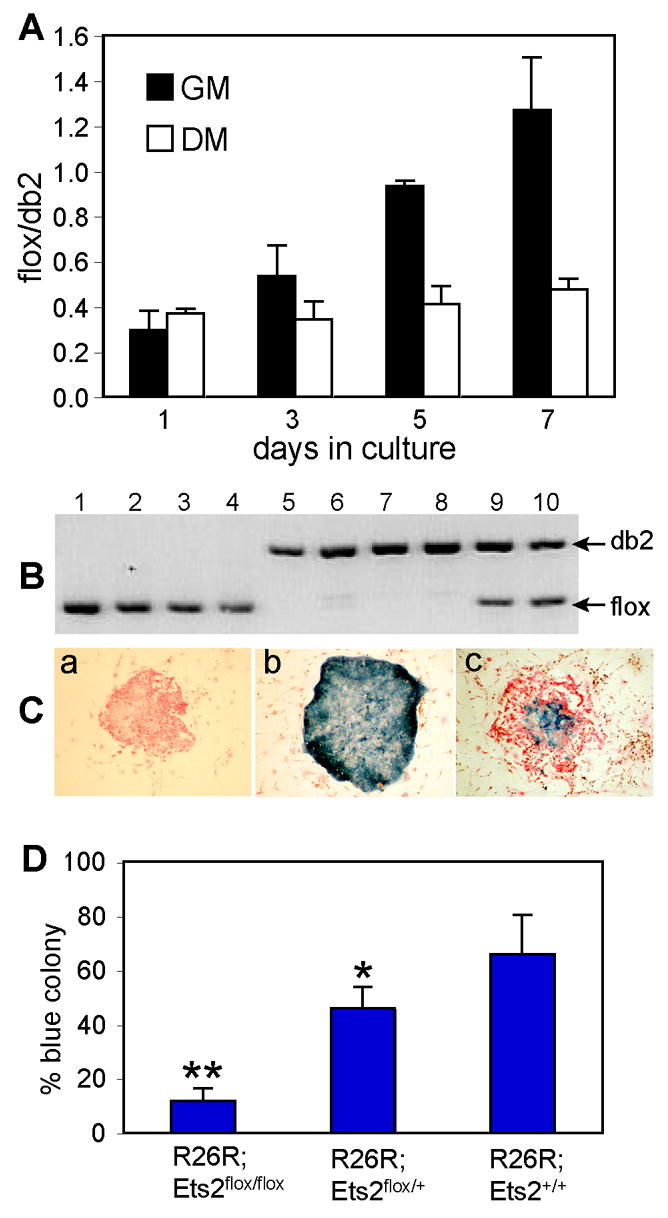
(A) Ets2flox/flox TS cells were cultured either in GM or DM following AdCreGFP infection. Purified DNA samples from indicated time points were analyzed by real-time PCR using primers specific for Ets2flox and Ets2db2. Values represent means ± SEM of three separate experiments. (B and C) Coordinate recombination of Ets2flox and R26R Cre reporter gene. Colonies from AdCreGFP-infected Ets2flox/flox TS cells were stained for β-galactosidase activity and counterstained with nuclear fast red. A representative negative (red), positive (blue) and mixed colony was shown in panel a, b and c, respectively (C). The stained colonies were recovered and analyzed by PCR. The results for 4 red colonies (lanes1-4), 4 blue colonies (lanes 5-8) and 2 mixed colonies (lanes 9-10) are shown (B). Mixed colonies were not scored for the analysis shown in panel D. (D) Colony formation by the recombined cells, indicated by the percentage of blue colonies, was dramatically reduced in the homozygous TS cells. More than 200 colonies were counted from multiple dishes for each genotype. Values represent means + SD of three independent experiments. Statistically significant differences are indicated: * = P < 0.05; ** = P < 0.01, by t test.
When plated at low density (1000 cells per 6-cm dish), only the stem cells of TS cultures form distinct epithelial colonies consisting of several hundred cuboidal cells after 10 days of culture. We used this colony formation assay to evaluate the importance of Ets2 on TS cell self renewal. The AdCreGFP-infected R26R-bearing TS cells were cultured at clonal density and then stained for β-galactosidase activity. PCR analysis of the recombination status of Ets2flox alleles of individual colonies confirmed coordinate Ets2flox and R26R recombination (Fig. 5B, C). Formation of β-galactosidase positive colonies from R26R;Ets2flox/flox TS cells was dramatically reduced compared to R26R;Ets2flox/+ or R26R;Ets2+/+ TS cells (Fig. 5D). R26R;Ets2db2/+ cells also showed a modest but significant decrease in colony formation compared to R26R;Ets2+/+ cells. These results reinforced the specific defect of Ets2 deficient TS cells while Ets2 deficient differentiated trophoblast cells, fibroblasts or rescued whole embryos were not affected similarly.
Ets2 transcriptional activity is stimulated by MAPK-mediated phosphorylation of the threonine-72 residue (Yang et al., 1996). Mutation of Thr-72 to Ala-72 results in a hypomorphic Ets2 allele that is not capable of supporting placental development when combined with the Ets2db1 allele. While Ets2A72/A72 mice are viable, the isolation of TS cell lines from Ets2A72/+ and Ets2A72/A72 intercross generated only 6 Ets2A72/A72 cultures out of 41 blastocysts (29% of expected). In contrast, 17 Ets2A72/+ TS cultures were obtained (83% of expected). In addition, the established Ets2A72/A72 cultures grew slower than Ets2A72/+ TS cells, had increased spontaneous differentiation, and formed fewer colonies when plated at clonal density (data not shown). These findings were consistent with the phosphorylation of Ets2 as a contributing but not absolute requirement for maintenance of TS cell proliferation.
Gene expression changes associated with Ets2 inactivation
The growth disadvantage of the Ets2 deficient TS cells was not due to increased apoptosis, as TUNEL and Annexin-V binding assays revealed rare, but similar numbers of positive cells in AdGFP- or AdCreGFP-infected Ets2flox/flox TS cells (data not shown). BrdU uptake and staining was performed to identify TS cells in S phase of the cell cycle. Under optimal growth conditions, the TS cells were highly proliferative (Supplementary Fig. S1A). The AdCreGFP-infected Ets2fllox/flox cells showed only a modest decrease of labeling index compared to the control AdGFP-infected cells (Supplementary Fig. S1B, C).
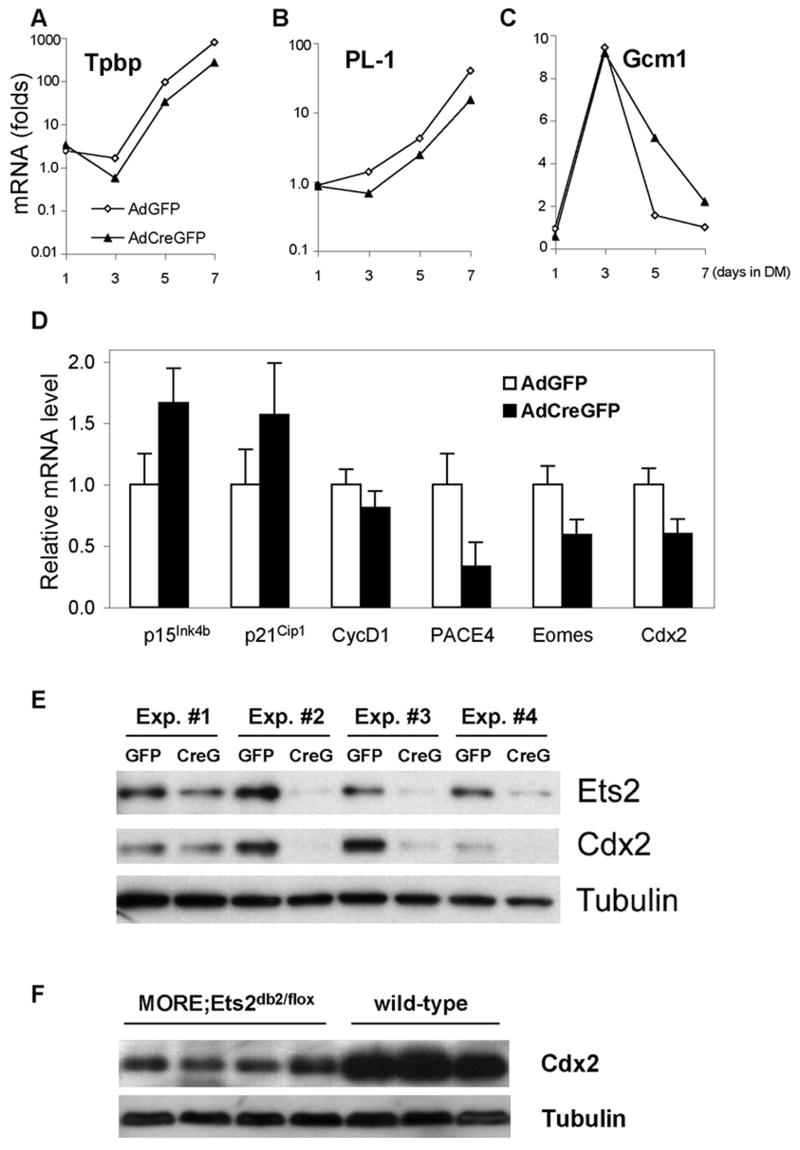
To evaluate gene expression changes due to the inactivation of Ets2, we compared RNA expression profiles of two independently isolated Ets2flox/flox TS cells in normal growth medium (GM), after induction of differentiation by growth factor deficient medium (DM), in GM after infection with AdCreGFP and in GM after infection with control AdGFP viruses. The data are deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE7238. Unsupervised cluster analysis of individual samples revealed that inactivation of Ets2 under normal growth conditions caused only moderate changes in gene expression with the gene expression profile more closely related to control cultures, either not infected or infected with control virus (Fig. 6A). This is consistent with a relatively small number of genes in TS cells that are highly dependent on Ets2. In contrast, RNAs from differentiated TS cells (DM) clustered together and were distinguished from mouse embryo fibroblast RNAs (MEF) and undifferentiated TS cells (GM). To identify individual candidate Ets2 target genes, grouped samples of Ets2flox/flox TS cells, infected with AdCreGFP or AdGFP viruses were compared (Fig. 6B). At the same threshold of statistical significance, the number of genes that changed significantly when infected with Cre expressing virus relative to control virus was much smaller than those changed in differentiated TS cells (Figure 6C). However, 89 of 100 changes in gene expression resulting from Ets2 inactivation were in genes that also changed upon growth factor withdrawal induced differentiation (Fig. 6C, Supplementary Table S1-3 and additional data not shown). At a P≤ 0.05, 113 RNAs were down regulated, and 131 RNAs up regulated by the action of AdCreGFP virus (Table S1 and S2). In contrast over 3000 RNAs were different between TS cells and differentiated TS cells. Thus Ets2 inactivation caused changes in gene expression associated with differentiation, but these genes represented only a subset of differentiation dependent gene expression changes. The magnitudes of the changes in gene expression due to Ets2 inactivation were much less than those upon growth factor withdrawal (Fig. 7A, B and Supplementary Fig. S3). As shown by Figure 6A, virus infection (both AdCreGFP and AdGFP) did not change gene expression profiles greatly. Among the Ets2 sensitive TS genes are: Cdx2, a master regulator of trophoblast specification and self renewal; Esrrb, the gene encoding a nuclear receptor Errβ that has been previously implicated in both TS and ES cell self renewal (Ivanova et al., 2006); Socs2, an inhibitor of intracellular signaling; and Pace4, an extraembryonic protease that activates Nodal to promote TS cell maintenance (Guzman-Ayala et al., 2004). In contrast to Pace4, Furin, another protease implicated in Nodal processing (Guzman-Ayala et al., 2004), is unaffected by Ets2 inactivation (Fig. 7C). Examination of the time course of gene expression reveals that three days after removal of the AdCreGFP virus, changes in these Ets2 sensitive TS markers gradually reverted, as expected from the selective disadvantage of Ets2db2/db2 TS cells (Fig. 3). RNAs for two Ets transcription factors (Etv4/Pea3 and Elf5) are down regulated upon TS cell differentiation (Supplementary Fig. S2). Ets2 inactivation resulted in decreased Etv4 RNA but had no effect on Elf5 RNA (Fig. 7A). Additionally, expression of various trophoblast subtype markers such as SpT-specific trophoblast specific protein (Tpbp), TGC-specific placental lactogen-1 (PL-1) and SynT-specific glial cells missing-1 (Gcm1) (Anson-Cartwright et al., 2000; Faria et al., 1991; Lescisin et al., 1988), were not induced by Ets2 inactivation. Thus Ets2 inactivation clearly does not simply induce precocious differentiation of the TS cells.
Figure 6. Ets2 dependent gene expression in Ets2flox/flox TS cells.
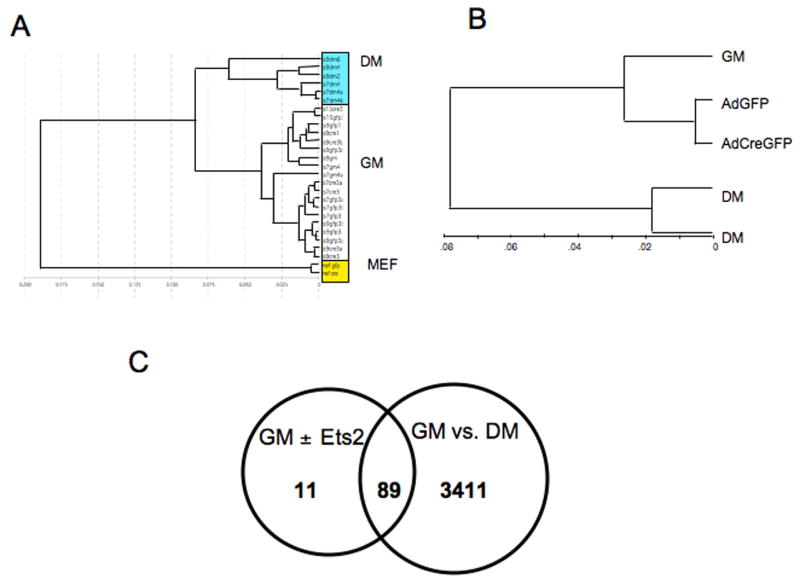
RNAs from two or three independently isolated Ets2flox/flox TS cell lines under four conditions: normal growth medium (GM); in differentiation medium (DM); in GM after infection with AdCreGFP or in GM after infection with AdGFP virus were analyzed by cDNA hybridization to Illumina mouse oligonucleotide arrays. Signal output was analyzed by BeadStudio software. (A) Unsupervised cluster analysis of individual samples shows that replicate individual samples grown in the same medium were similar. Note that RNAs from DM and mouse embryo fibroblasts (MEF) were distinct from cells grown in GM regardless of infection with AdCreGFP or AdGFP virus. (B) Cluster analysis of grouped samples. Results of multiple samples of the same growth and virus conditions were compared for overall similarity by the unsupervised clustering analysis. RNAs from TS cells infected with AdCreGFP or AdGFP were more similar to uninfected TS cells (GM) than cells grown for four days in DM. DM samples were divided into two groups because of the use of Sentrix mouse-6 (24k gene) and mouse-8 (16k gene) chips. (C) Venn diagram representation of overlap of genes most changed during differentiation (GM vs DM) and after infection with AdCreGFP virus (GM ±Ets2). Normalized data of the four groups was analyzed with GeneSpring software. Lists were generated using the same P value threshold for differentially expressed gene sets. 89 of the top 100 genes sensitive to infection with AdCreGFP also changed significantly when TS cells were shifted from GM to DM for four days.
Figure 7. Ets2 dependent gene expression.
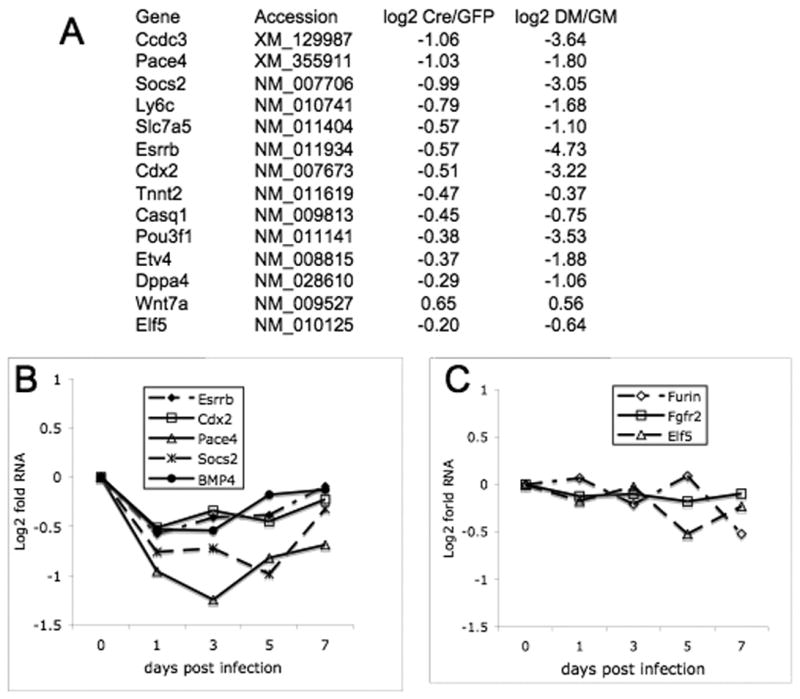
(A) Differences in expression of nine selected genes decreased three days after AdCreGFP infection (Table S1) and known regulators of TS cell or ES cell self renewal are compared to changes in expression of the same genes after 4 days in DM. Ratios of cells infected with Cre virus and control GFP virus are shown as log base 2. All values represent grouped and normalized replicates. (B) Time dependence of selected gene expression. Individual points represent fold difference between Ets2flox/flox TS cells grown for the indicated time after infection with AdCreGFP relative to control cells infected with AdGFP. These values were evaluated in Bead Studio software, normalized to companion samples infected with AdGFP. (C) Expression of Fgfr2, Furin and Elf5 RNAs is not sensitive to AdCreGFP infection.
Ets2-deficient TS cells differentiate normally
To investigate whether Ets2 deficiency affects the differentiation potential of TS cells, we analyzed the expression of trophoblast subtype markers after withdrawal of FGF4 and EFCM from AdCreGFP-infected Ets2flox/flox TS cultures. During induced differentiation, Tpbp and PL-1 were upregulated in Ets2 deficient cultures albeit at lower levels than Ets2 competent, differentiating cultures (Fig. 8A, B). Additional markers of the SpT and TGC lineages, such as proliferin-related protein (PRP), p57Kip2 and placental lactogen-2 (Carney et al., 1993; Hattori et al., 2000), showed similar modest restriction (data not shown), while the level of Gcm1 induction was not distinguishable in Ets2 deficient cultures (Fig. 8C). Additionally, cell morphology changes typical of TGC differentiation was found in Ets2 deficient TS cultures upon growth factor withdrawal. Taken together, these results indicate that Ets2 is not essential for TS cell differentiation but may contribute to the optimal induction of SpT and TGC differentiation.
Figure 8. Molecular changes in Ets2 deficient TS cultures and colon tissues.
(A-C) Adeno-infected Ets2flox/flox TS cells were induced to differentiate after dissociation and replating at equal density at 3 dpi. RNA was isolated every 2 days, and expression of specific mRNAs was quantified by real-time PCR. The results are expressed as fold induction relative to the levels in regular GM culture. Note the logarithmic scale in panel A and B. Similar results were obtained in at least three independent experiments. (D) mRNAs were isolated from three independent Ets2flox/flox TS clones infected with indicated adenoviruses and cultured in GM for 3-4 days. Expression of specific transcripts was quantified with reverse transcription and real-time PCR. Values represent means + SEM normalized relative to the AdGFP-infected controls. The differences are statistically significant by t test (one-tail paired, P<0.05). (E) Endogenous Cdx2, Ets2 and tubulin proteins in cell lysates from TS cultures infected with indicated adenoviruses were detected by SDS-PAGE and immunoblotting. (F) Cdx2 and tubulin protein levels in colon tissues from four “waved” MORE;Ets2db2/flox mice and 3 wild-type mice.
Ets2 regulation of Cdx2
To confirm some of the candidate target genes identified by array analysis and to query previously identified Ets2 target genes, we analyzed the expression of selected potential targets by real-time PCR. RNA levels of several Ets2 targets in other cell types, MMP3, MMP9, Bcl-xL and Cdc2l2 (Hollenhorst et al., 2004; Sevilla et al., 2001; Yamamoto et al., 1998), were not altered in Ets2 deficient TS cells (data not shown), indicating cell type specific regulation of these genes. We found increased expression of the Cdk inhibitors p15Ink4b and p21Cip1 as well as a modest but significant decrease of cyclin D1 mRNA in AdCreGFP-infected Ets2flox/flox TS cultures (Fig. 8D), consistent with the growth defect of such cultures. Similar changes of these genes at protein level were observed (data not shown). Pace4, Eomes and Cdx2 RNA significantly decreased in Ets2 deficient cultures. Protein analysis revealed a remarkable correlation between Ets2 deficiency and Cdx2 down regulation (Fig. 8E). In adult mice, Cdx2 is specifically expressed in the intestinal epithelium and acts as a tumor suppressor (Chawengsaksophak et al., 1997). To determine if Ets2 also regulates Cdx2 expression in colon, we analyzed Cdx2 level in colon tissues from wild-type and MORE;Ets2db2/flox adults. A similar decrease in Cdx2 protein was found in the “waved” mice, suggesting a shared Ets2-dependent regulatory mechanism of Cdx2 expression in TS cells and colon epithelial cells.
Cdx2 transcription is directly regulated by Ets2
Cdx2 is the earliest transcription factor to specify trophoblast cell fate during embryo development, and Cdx2 null mutants fail to implant due to defective trophectoderm formation (Strumpf et al., 2005). Comparison of the Cdx2 gene from human, mouse, rat and dog revealed a high degree of conservation in the proximal promoter (+1 to -400 nt) and in a distal regions (-2300 to -1800) (Fig. 9B), consistent with potential regulatory roles of these regions. Of particular interest, 6 potential Ets binding sites were located with the use of Transcription Element Search System (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess) and the Transfac database of binding. Furthermore, one of these sites, located within the 187 bp of the distal element most conserved between human and mouse sequences, matched 10 nts of the previously determined extended Ets2 binding site {Umezawa, 1997 #5173}.
Figure 9. Ets2 regulation of Cdx2 transcription.
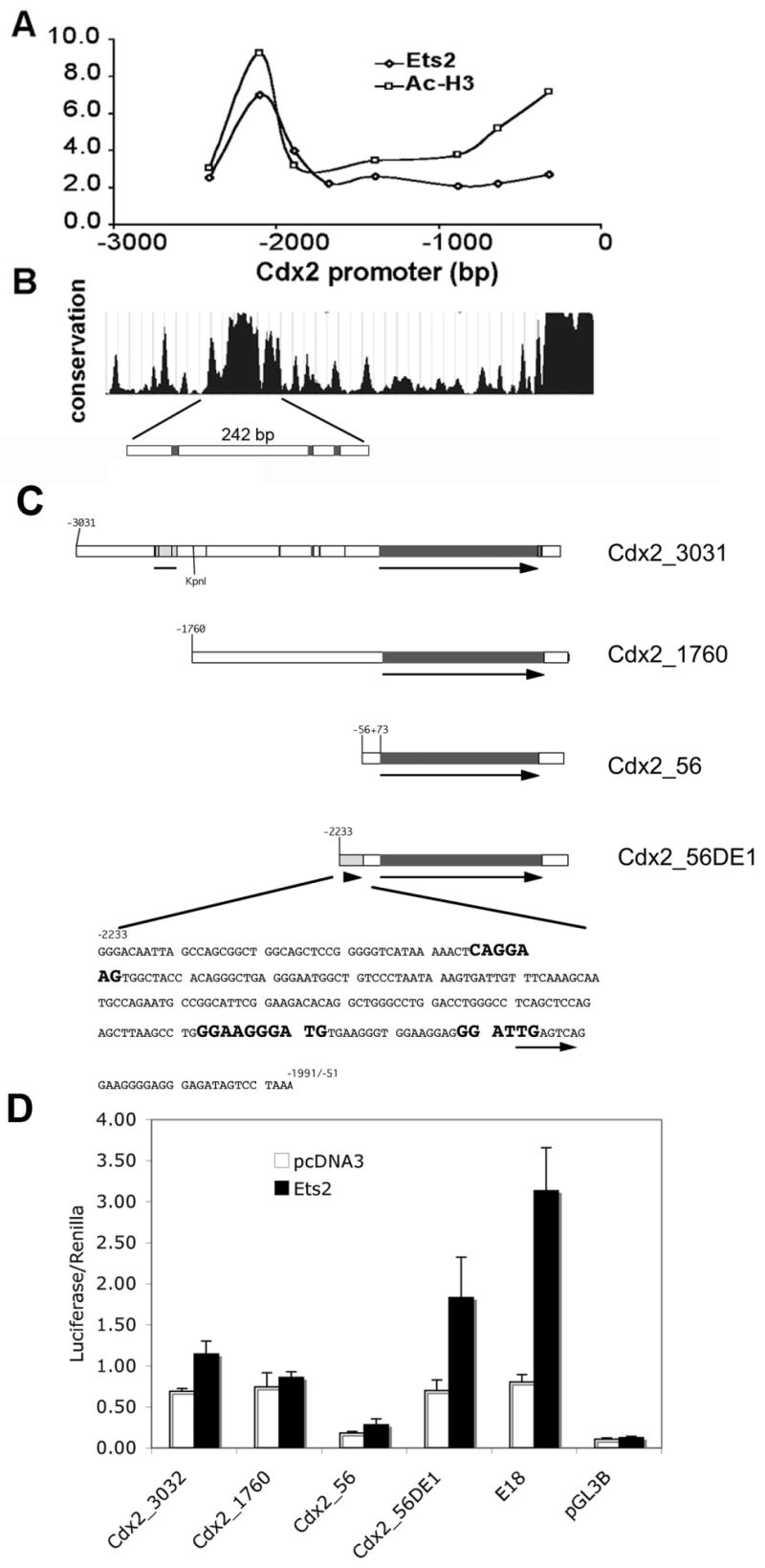
(A) Quantitative PCR analysis of ChIP for Ets2 and acetylated histone H3 (Ac-H3) enrichment relative to non-specific rabbit IgG binding over the 3 kb upstream region of mouse Cdx2 gene. Ets2 is bound to a distal region but not the proximal promoter. Ac-H3 is enriched over both regions. (B) Sequence conservation of the Cdx2 upstream region among mouse, rat, human and dog (http://genome.ucsc.edu). (C) Schematic of luciferase reporter constructs (filled line with arrow): Cdx2_3031, nt -3031 to +73 region of the mouse Cdx2 gene. The hatched region and line indicate the location of the 242 bp conserved region in B and as shown for construct Cdx2_56DE1 below. Vertical lines within the flanking region indicate potential Ets binding sites. Cdx2_1760 represents the 1.7 kb Cdx2 promoter lacking the distal region. Cdx2_56 indicates the minimal Cdx2 promoter from -56 to +73. Cdx2_56DE1 represents the minimal promoter with the additional distel element of 242 bp. The sequence of the DE1 is shown with three potential Ets binding elements highlighted. An arrow indicates an overlapping potential AP1 site. (D) Activation of reporters by co-expressed Ets2. Caco-2 cells were transfected with indicated reporter gene and an Ets2 expression vector or a control plasmid (pcDNA3) and the Renilla luciferase transfection control plasmid. The E18, synthetic Ets responsive reporter (Galang et al., 1994) and pGL3B, a control reporter plasmid without a promoter represent positive and negative controls. Values represent means ± SD of three independent experiments. Values represent the ratio of firefly luciferase activity to the control Renilla luciferase activity.
To determine if Ets2 binds to the Cdx2 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) analysis in TS cells using an Ets2 antibody and PCR primers spanning regions of Cdx2 promoter from nt -2500 to -250 (Fig. 9A). Enrichment for Ets2 was detected in the -2000 distal region but not the proximal promoter region. In comparison, acetylated histone H3 was enriched over both regions.
To test for direct Ets2 regulation of the Cdx2 promoter, portions of the 3 kb upstream region of Cdx2 gene were fused with a luciferase reporter construct (Fig. 9C) and tested in Caco-2 cells, a human intestinal epithelial cell line, previously shown to express Cdx2. Co-expression of Ets2 resulted in activation of the Cdx2 reporter gene containing the conserved flanking region (Fig. 9C, Cdx2_3031). However, deletion of the putative Ets2 regulatory region (Fig. 9C, Cdx2_1760) abolished Ets2 activation of the reporter. Inspection of the conserved distal element of the Cdx2 gene revealed a core region of approximately 300 bp that contained at least three potential binding sites for Ets transcription factors as predicted by the Transcription Element Search System (TESS), (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME). Fusing a 288 bp region of the distal element to the Cdx2 minimal promoter resulted in Ets2 responsiveness to the minimal Cdx2 promoter (Figure 9D). The conserved region is both necessary and sufficient for Ets2-dependent activation of Cdx2 transcription. Variable results due in part to inefficient expression, prevented critical evaluation of the same plasmid constructs in TS cells by transient transfection.
If Ets2 regulation of Cdx2 were solely responsible for the decreased self renewal of Ets2 deficient TS cells, forced expression of Cdx2 might rescue the cellular phenotype. However, several independently isolated Cdx2-overexpressing Ets2flox/flox, R26R TS clones remained at a selective growth disadvantage after Cre inactivation of Ets2 (data not shown). Thus it appears a combination of Ets2 regulated genes may be necessary for TS cell self renewal and that supplementation of Ets2 deficient TS cells with Cdx2 is not sufficient to ensure self renewal.
Discussion
Trophoblast cells initiate the first developmental lineage in the mammalian conceptus and are thus responsible for embryo implantation and placental development, the defining characteristic of eutherians. There is an ever-increasing list of growth factors and their receptors, signaling molecules, transcription factors, and proteases that have been shown to regulate various stages of trophoblast development. We demonstrate here that TS cell self renewal requires Ets2 activity. Inactivation of Ets2 caused placental insufficiency including a smaller ectoplacental cone and an absence of ExE, the area containing TS cells proliferating under the influence of FGF4 produced by embryonic ectoderm (Kunath et al., 2004; Tanaka et al., 1998). We showed evidence of deficient isolation of Ets2db1/db1 TS cells and the selective disadvantage of Ets2db2/db2 TS cells under both high and low density growth conditions, indicating a requirement of Ets2 activity for continuous TS proliferation. TS cells express at least 18 different members of the Ets family of transcription factors (unpublished data). However, deficiency of a single member of the family, Ets2, caused defective self renewal of TS cells, which is not complemented by any other Ets member. In contrast, Ets2 deficient ES cells have no apparent proliferation defect and are able to compose all organ systems (Henkel et al., 1996; Yamamoto et al., 1998). In addition, the generation of nearly normal Ets2 deficient adult mice by complementing extraembryonic cell defects indicates that Ets2 is not essential for the proliferation of other somatic cell types. Thus Ets2 activity is uniquely required for maintenance of TS cell proliferation.
FGF4 signaling is essential for TS cell maintenance, and Erk2 is an important downstream effector during the process. Ets2 is activated downstream of growth factor receptor signaling by phosphorylation of Thr-72 (Yang et al., 1996) (Dankort et al., 2001; Man et al., 2003) and is essential for FGF induction of specific genes in fibroblasts (Yamamoto et al., 1998). Several lines of evidence suggest that Ets2 may act downstream of FGF4-Erk2 signaling to mediate TS cell proliferation and self renewal. In vivo, FGF receptor-dependent Erk phosphorylation occurs specifically within the ExE where the TS cells reside (Corson et al., 2003), and disruption of either Erk2 or Ets2 led to a failure of EPC and ExE formation (Saba-El-Leil et al., 2003), both direct derivatives of the TS cells. In vitro, depriving TS cells of Erk activity (Rossant, 2001) or Ets2 function, as shown here, had similar adverse effects on proliferation and self renewal. Furthermore, the MAPK phosphorylation mutant Ets2A72 is haploinsufficient to support placental development, and isolation of Ets2A72/A72 TS clones showed decreased efficiency. Therefore, Ets2 likely contributes to mediating Erk2 activation by FGF4 in TS cells. However, basal Ets2 activity, independent of activation through phosphorylation of Thr-72, is sufficient for placental development and TS cell isolation albeit with decreased efficiency. Thus Ets2 also acts on additional targets that are not affected by FGF4-Erk2 signaling.
The absence of apoptosis and the gene expression changes associated with differentiation in Ets2 deficient TS cells are consistent with a switch from self renewal to beginning differentiation. However, markers of terminal differentiation, such as PL-1, PL-2 and PRP, were not induced in such TS cells. Consistently, upon growth factor withdrawal, Ets2 deficient TS cells displayed sub-optimal induction of these markers. As Ets2 level increases during induced differentiation (Supplementary Fig. S3D), it is possible that in addition to its essential role in maintaining TS cell proliferation, Ets2 also regulates the transcription of different targets during trophoblast differentiation.
Among the list of potential Ets2 targets in TS cells, indicated by the microarray and quantitative PCR analyses, are several genes previously implicated in the self renewal of TS cells including Cdx2, Eomes and Errβ. Cdx2 is a master regulator of trophoblast commitment and self renewal (Niwa et al., 2005; Strumpf et al., 2005). The decreased Cdx2 RNA and protein in both Ets2 deficient TS cells and colon tissues indicates a requirement for Ets2 for optimal expression of Cdx2. Cdx2 also acts as a tumor suppressor in intestine. It is noteworthy that a second Ets2 candidate target gene, Socs2, also regulates intestinal development (Michaylira et al., 2006).
The physical association of Ets2 with a highly conserved Cdx2 regulatory region and Ets2 activation of reporter constructs containing a portion of the region support a direct regulatory role of Ets2 on Cdx2 transcription. However, inactivation of Ets2 still permits residual Cdx2 RNA expression in both TS cells and colon. Furthermore, during TS differentiation, Cdx2 expression is rapidly turned off, while Ets2 expression rises modestly. Thus Ets2 contributes to the expression of Cdx2 but is not singularly responsible for its expression. The self stimulation of the Cdx2 promoter by Cdx2 protein (Xu et al., 1999) may contribute to a threshold effect of reducing Ets2 stimulation of the Cdx2 gene. Potential binding sites for Ets2, approximately 2-kb upstream of the Cdx2 transcriptional start site represent a region of conserved DNA sequence that is occupied by Oct3/4 and Sox2 in human ES cells (Boyer et al., 2005). The binding of ES cell transcription factors such as Oct4 and Sox2 to conserved DNA regions of key regulators of developmental lineages has been suggested to maintain potential for activation during subsequent differentiation. While the distal regulatory region of Cdx2 can mediate Ets2 activation in transient transfection analysis, the detailed mechanisms by which this region may be used in maintaining, inhibiting or facilitating expression in different cell types remains to be determined.
The forced expression of Cdx2 did not rescue the selective disadvantage of Ets2 deficient TS cells, indicating that Cdx2 down regulation is not solely responsible for the defective proliferation of such TS cells. Other candidate Ets2 target genes that are important for trophoblast development, such as Eomes, Errβ and Pace4, may contribute to Ets2 dependent TS cell self renewal. The decrease of Pace4 mRNA was among the greatest upon Ets2 inactivation. Pace4 is one of two proteases in ExE that activates Nodal, a TGF-β family member that inhibits the differentiation of TS cells (Guzman-Ayala et al., 2004). However, the function of Pace4 is shared with another protease, Furin. As Furin mRNA is not affected in TS cells by Ets2 inactivation, decreased Pace4 expression is unlikely to exclusively account for the self renewal defect. It remains to be determined whether combination of two or more of these candidate targets can rescue the Ets2 dependent defective maintenance of TS cells.
Supplementary Material
(A and B) BrdU immunostaining (red) of TS cells infected with AdGFP and AdCreGFP, respectively. Nuclei were visualized with Hoechst 33258. (C) BrdU labeling index of Adeno-infected Ets2flox/flox TS cells. Values represent means + SD of 3 independent experiments using different TS clones.
RNA isolated from TS cultures exposed to DM for the indicated time were analyzed by array hybridization. Background and gene signals were processed by the cubic spline method of the BeadStudio software and then normalized to cells grown in GM for the same length of time.
Acknowledgments
We thank Dr. Ling Wang and Ms. Robbin Newlin for expert technical assistance, Jacqueline Lesperance and Abraham Gomez for animal husbandry care, Dr. A. F. Stewart for providing the Flpe construct, Dr. S. V. Kozlov for the gift of AdCreGFP, Dr. Y. Tamai for providing the mouse Cdx2 genomic clone, Dr. Tomilin for providing the Cdx2 expressing vector, Dr. C. A. Hauser for stimulating discussions and providing real-time PCR primers, Dr. A. Terskikh for helpful comments.
This work was supported by National Cancer Institute grants CA74547 and CA98778 (R.G.O.). F. Wen was supported in part by a predoctoral fellowship award (DAMD17-02-1-0316) from Department of Defense Breast Cancer Research Program.
Abbreviations
- Cdx2
caudal-type homeobox 2
- dpi
days post infection
- DM
differentiation medium
- E
embryonic day
- EFCM
inactivated embryonic fibroblast-conditioned medium
- Eomes
eomesodermin homologue
- EPC
ectoplacental cone
- Erk
extracellular signal-regulated kinase
- ExE
extraembryonic ectoderm
- Gcm1
glial cells missing-1
- GM
growth medium
- MEF
mouse embryonic fibroblast
- MOI
multiplicity of infection
- MORE
Mox2-Cre
- PL-1
placental lactogen 1
- PRP
proliferin-related protein
- SpT
spongiotrophoblast
- SynT
syncytiotrophoblast
- TGCs
trophoblast giant cells
- Tpbp
trophoblast specific protein
- TS cell
trophoblast stem cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet. 2000;25:311–4. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol. 2002;4:981–5. doi: 10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–9. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–62. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Carney EW, Prideaux V, Lye SJ, Rossant J. Progressive expression of trophoblast-specific genes during formation of mouse trophoblast giant cells in vitro. Mol Reprod Dev. 1993;34:357–68. doi: 10.1002/mrd.1080340403. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–7. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- Copp AJ. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–37. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Dankort D, Jeyabalan N, Jones N, Dumont DJ, Muller WJ. Multiple ErbB-2/Neu Phosphorylation Sites Mediate Transformation through Distinct Effector Proteins. J Biol Chem. 2001;276:38921–8. doi: 10.1074/jbc.M106239200. [DOI] [PubMed] [Google Scholar]
- Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275:158–69. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Faria TN, Ogren L, Talamantes F, Linzer DI, Soares MJ. Localization of placental lactogen-I in trophoblast giant cells of the mouse placenta. Biol Reprod. 1991;44:327–31. doi: 10.1095/biolreprod44.2.327. [DOI] [PubMed] [Google Scholar]
- Galang CK, Der CJ, Hauser CA. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-ets-2 binding sites. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–92. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Rossant J. Ets2 is necessary in trophoblast for normal embryonic anteroposterior axis development. Development. 2006;133:1059–68. doi: 10.1242/dev.02277. [DOI] [PubMed] [Google Scholar]
- Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci U S A. 2004;101:15656–60. doi: 10.1073/pnas.0405429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Davies TC, Anson-Cartwright L, Cross JC. Periodic expression of the cyclin-dependent kinase inhibitor p57(Kip2) in trophoblast giant cells defines a G2-like gap phase of the endocycle. Mol Biol Cell. 2000;11:1037–45. doi: 10.1091/mbc.11.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel GW, McKercher SR, Yamamoto H, Anderson KL, Oshima RG, Maki RA. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood. 1996;88:2917–26. [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–8. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kunath T, Strumpf D, Rossant J. Early trophoblast determination and stem cell maintenance in the mouse--a review. Placenta. 2004;25 A:S32–8. doi: 10.1016/j.placenta.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 1988;2:1639–46. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGFα deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Man AK, Young LJ, Tynan JA, Lesperance J, Egeblad M, Werb Z, Hauser CA, Muller WJ, Cardiff RD, Oshima RG. Ets2-dependent stromal regulation of mouse mammary tumors. Mol Cell Biol. 2003;23:8614–25. doi: 10.1128/MCB.23.23.8614-8625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann GB, Fowler KJ, Gabriel A, Nice EC, Williams RL, Dunn AR. Mice with a null mutation of the TGFα gene have abnormal skin architecture, wavy hair, and curly whiskers and often develop corneal inflammation. Cell. 1993;73:249–261. doi: 10.1016/0092-8674(93)90227-h. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–65. [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–41. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Michaylira CZ, Ramocki NM, Simmons JG, Tanner CK, McNaughton KK, Woosley JT, Greenhalgh CJ, Lund PK. Haplotype insufficiency for suppressor of cytokine signaling-2 enhances intestinal growth and promotes polyp formation in growth hormone-transgenic mice. Endocrinology. 2006;147:1632–41. doi: 10.1210/en.2005-1241. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Prost S, Sheahan S, Rannie D, Harrison DJ. Adenovirus-mediated Cre deletion of floxed sequences in primary mouse cells is an efficient alternative for studies of gene deletion. Nucleic Acids Res. 2001;29:E80. doi: 10.1093/nar/29.16.e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. Stem cells from the Mammalian blastocyst. Stem Cells. 2001;19:477–82. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–48. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, Wilson V, Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Saba-El-Leil MK, Vella FD, Vernay B, Voisin L, Chen L, Labrecque N, Ang SL, Meloche S. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 2003;4:964–8. doi: 10.1038/sj.embor.embor939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla L, Zaldumbide A, Carlotti F, Dayem MA, Pognonec P, Boulukos KE. Bcl-XL expression correlates with primary macrophage differentiation, activation of functional competence, and survival and results from synergistic transcriptional activation by Ets2 and PU.1. J Biol Chem. 2001;276:17800–7. doi: 10.1074/jbc.M008270200. [DOI] [PubMed] [Google Scholar]
- Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol. 2005;284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–5. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, Tomilin A. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells. 2006;24:139–44. doi: 10.1634/stemcells.2005-0240. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hahn SL, Giovane A. The ets family of transcription factors. European Journal of Biochemistry. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Bradford AP, Gutierrez-Hartmann A, Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–7. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Xu F, Li H, Jin T. Cell type-specific autoregulation of the Caudal-related homeobox gene Cdx-2/3. J Biol Chem. 1999;274:34310–6. doi: 10.1074/jbc.274.48.34310. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes and Development. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activity of c-Ets1 and c-Ets2. Molecular and Cellular Biology. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A and B) BrdU immunostaining (red) of TS cells infected with AdGFP and AdCreGFP, respectively. Nuclei were visualized with Hoechst 33258. (C) BrdU labeling index of Adeno-infected Ets2flox/flox TS cells. Values represent means + SD of 3 independent experiments using different TS clones.
RNA isolated from TS cultures exposed to DM for the indicated time were analyzed by array hybridization. Background and gene signals were processed by the cubic spline method of the BeadStudio software and then normalized to cells grown in GM for the same length of time.