Abstract
Humans play little role in the epidemiology of Escherichia coli O157:H7, a commensal bacterium of cattle. Why then does E. coli O157:H7 code for virulence determinants, like the Shiga toxins (Stxs), responsible for the morbidity and mortality of colonized humans? One possibility is that the virulence of these bacteria to humans is coincidental and these virulence factors evolved for and are maintained for other roles they play in the ecology of these bacteria. Here, we test the hypothesis that the carriage of the Stx-encoding prophage of E. coli O157:H7 increases the rate of survival of E. coli in the presence of grazing protozoa, Tetrahymena pyriformis. In the presence but not the absence of Tetrahymena, the carriage of the Stx-encoding prophage considerably augments the fitness of E. coli K-12 as well as clinical isolates of E. coli O157 by increasing the rate of survival of the bacteria in the food vacuoles of these ciliates. Grazing protozoa in the environment or natural host are likely to play a significant role in the ecology and maintenance of the Stx-encoding prophage of E. coli O157:H7 and may well contribute to the evolution of the virulence of these bacteria to colonize humans.
Keywords: Shiga toxin, Escherichia coli O157:H7, prophage, protozoa predation, Tetrahymena pyriformis
1. Introduction
Escherichia coli O157:H7 is responsible for a significant amount of human morbidity and mortality in the developed world (Besser et al. 1999; Rangel et al. 2005) and a recurrent source of economic privation for the food industry (DeWaal et al. 2006). However, E. coli O157:H7 is not a human pathogen in the sense that human-to-human transmission is rare and not sufficient to sustain populations of these bacteria indefinitely. Cattle and other ungulates, in which these enteric bacteria rarely cause disease, are considered to be the natural host of E. coli O157:H7 (Dean-Nystrom et al. 1998a; Hancock et al. 1998). Why then do E. coli O157:H7 and the prophage they carry code for toxins and other virulence factors that are responsible for morbidity and mortality of colonized humans?
A probable answer is that the Shiga toxins (Stx) and other factors responsible for the virulence of these bacteria in humans evolved and are maintained by selection for other roles they play in the ecology of these bacteria. In this interpretation, the virulence of E. coli O157:H7 in colonized humans is coincidental, an inappropriate or over response to these factors. But what other roles do the virulence determinants of E. coli O157:H7 play?
One possibility is that the virulence factors responsible for human disease facilitate E. coli O157's colonization, replication or transmission in their bovine hosts. There is evidence for this being the case. For example, E. coli O157 bear a pathogenicity island containing the eae locus, which promotes colonization in cattle via attaching and effacing lesions (Dean-Nystrom et al. 1998b; Cornick et al. 2002; Sheng et al. 2006). Moreover, bovine intestinal cells have receptors for the Stxs (Menge et al. 2004) expressed by these bacteria and, at least in a mouse model, Stx 2 promotes intestinal colonization (Robinson et al. 2006).
Another possibility is that some of the virulence factors enhance the survival of these bacteria by providing protection against predation by bactivorous protozoa, nematodes or other predators in the soil, water or the gastrointestinal tract of their bovine hosts. Bacteria, including pathogenic coliforms (King et al. 1988), are grazed upon by bactivorous protozoa, and this predation is considered to be important in shaping the structure of bacterial communities in planktonic as well as terrestrial ecosystems (Hahn & Hofle 2001; Jurgens & Matz 2002). If indeed predation by protozoa contributes to bacterial mortality, it would not be surprising at all that these bacteria have evolved ways to resist this predation. There are, in fact, a variety of mechanisms that bacteria use to either evade or survive protozoan predation (Matz & Kjelleberg 2005). There is also evidence for within- and between-species variation in the susceptibility of bacteria to protozoan predation. Some serotypes of Salmonella enterica are more resistant to predation by amoeba than others (Tezcan-Merdol et al. 2004; Wildschutte et al. 2004). Salmonella enterica serovar Thompson survive better than Listeria monocytogenes within the food vacuoles of Tetrahymena pyriformis (Brandl et al. 2005). In a recent review, it has been suggested that E. coli O157:H7 may have a survival advantage in the food vacuoles of these bactivorous ciliates (Brandl 2006).
Here, we present the results of experiments testing the hypothesis that the Stx-encoding prophage of E. coli O157 provides protection against predation by grazing protozoa. We demonstrate that in the presence but not the absence of T. pyriformis, E. coli K-12, as well as O157:H7 and O157:H−, that is lysogenic for this lambda-like phage has an advantage over non-lysogens. Our results suggest that part, but not all, of this protection is associated with Stx 2 (Stx2) and can be attributed to the prophage coding for this toxin, increasing the rate of survival of these bacteria in the food vacuoles of these ciliate predators.
2. Material and methods
(a) Strains
All strains are listed in table 1. Strains include human clinical isolates from stool and laboratory-made lysogens created using the K-12 derivative, C600, lysogenized with Stx2-encoding prophages from 933W (CDC) and 3538/95 (Schmidt et al. 1999); these strains are called C600P and C600PT+, respectively, in this paper. Additionally, C600 was lysogenized with the 3538/95Δtox::cat prophage in which the toxin genes have been deleted and a chloramphenicol resistance cassette inserted in its place (Schmidt et al. 1999), and this strain will be known as C600PT−. The identity of bacterial genotypes was confirmed by biotyping and with PCR markers, and that of phage genotypes was confirmed by restriction digest analysis of purified phage DNA (M. Reynolds 2000, unpublished data).
Table 1.
List of E. coli strains and constructs used in this investigation.
| strain designation | strain | Shiga toxin | motility | markera | source |
|---|---|---|---|---|---|
| C600 | C600 | — | — | Lac- | this study |
| C600P | C600Φ933W | 2 | — | Ara- | this study |
| C600PT+ | C600Φ3538/95 | 2 | — | this study | |
| C600PT− | C600Φ3538/95Δtox::cat | — | — | CamR | this study |
| C600gfp | C600Pgfp | — | — | Ara-/AmpR | this study |
| C600Pgfp | C600Φ933WPgfp | 2 | — | Ara-/AmpR | this study |
| PT+ | O157:H7 3538/95 | 2 | + | Schmidt et al. (1999) | |
| PT− | O157:H7 3538/95Δtox::cat | — | + | CamR | Schmidt et al. (1999) |
| P2T+ | O157:H7 86-24 | 2 | + | Gunzer et al. (1998) | |
| P2T− | O157:H7 TUV86-2 | — | + | NalR | Gunzer et al. (1998) |
| 00-3032 | O157:H7 00-3032 | 1,2 | + | CDC | |
| 98-3126 | O157:H− 98-3126 | 1,2 | — | CDC | |
| 00-3150 | O157:H7 00-3150 | — | + | CDC | |
| 89-3360 | O157:H7 89-3360 | 1 | + | CDC | |
| 85-3476 | O157:H− 85-3476 | 1 | — | CDC |
Used for sampling.
The C600 and C600P strains were transformed with a commercially available green fluorescent protein (gfp)-encoded plasmid (Clontech) and are referred to as C600gfp and C600Pgfp, respectively. Transformation protocols can be found in Sambrook et al. (1989). Cultures were grown in Luria–Bertani (LB) broth with 5% ampicillin until reaching a density of 0.3 at 600 nm, at which point they were induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated for an additional 16 h at 37°C on a rotary shaker.
For all experiments, bacteria were grown in LB broth (with appropriate antibiotics) on a rotary shaker at 37°C. In mixed culture experiments, arabinose, lactose, chloramphenicol and nalidixic acid markers were used to distinguish toxin-positive from toxin-negative bacteria by selective plating on tetrazolium agar with arabinose or lactose, or LB agar with chloramphenicol (2.5%) or nalidixic acid (2.0%). Axenic cultures of T. pyriformis were obtained from Carolina Biological Supply Co. (Burlington, NC). Tetrahymena pyriformis is a freshwater and soil ciliate that commonly inhabits lakes and streams. These organisms will grow in axenic or defined medium and tolerate temperatures up to 22°C. For all experiments, Tetrahymena were grown in proteose peptone/tryptone media (5 g proteose peptone, 5 g tryptone and 0.2 g K2HPO4 in 1 l H2O, pH adjusted to 7.2) in a stationary culture flask to a density of 105 cells ml−1 at 24–27°C.
(b) Mixed culture experiments
Mixed cultures were maintained in the presence and absence of T. pyriformis. The cultures were maintained without agitation at room temperature, 22–27°C. Samples were taken every 24 h for 7 days, and the total cell densities and relative frequencies of prophage (or toxin)-positive and -negative bacteria were estimated by counting the number of colony-forming units (CFUs). Tetrahymena densities were estimated by counting the number of ciliates in 10 μl samples stained with 10 μl of Lugol's iodine using a Neubauer improved haemocytometer and an inverted microscope. Toxin production was measured using a commercially available EHEC ELISA kit (Meridian, Inc.).
(c) Vacuole production and bacterial cell number per vacuole
To determine the number of vacuoles produced per Tetrahymena and measure the number of bacterial cells per vacuole, we used a modified version of the protocol described in Brandl et al. (2005). Briefly, stationary-phase bacteria from cultures of C600gfp and C600Pgfp were washed twice, resuspended in sterile distilled water and serially diluted in sterile distilled water to 108 cells ml−1. One millilitre of prophage-positive or prophage-negative bacteria was placed in sterile Eppendorf tubes, and 5×104 Tetrahymena cells were added to the tubes. Tubes were incubated for 24 h at room temperature without shaking. At 12 and 24 h, samples were taken and bacterial densities were estimated from the CFU data. At 24 h, 10 μl aliquots were placed on a clean Polysine slide and viewed under an epifluorescent microscope. The numbers of green cells per vacuole were counted for 30 vacuoles from duplicate tubes. Bacterial viability within vacuoles was assessed using the Live/Dead BacLight viability system (Molecular Probes, Eugene, OR).
(d) Statistical methods
The t-tests assuming unequal variances were used to test the significance between the initial bacterial ratios and densities and the ratios and densities after day 3. This test was also used to compare the mean numbers of bacterial cells per vacuole.
3. Results
(a) The Stx-encoding prophage of E. coli O157 augments the fitness of E. coli K-12 in the presence of T. pyriformis
An E. coli K-12 derivative, C600 (Ara+) and C600 (Ara−) lysogenized with a Stx2-encoding prophage, C600P, were separately grown overnight in LB broth, washed and resuspended in sterile distilled water to full volume. Equal frequencies of prophage-positive (C600P) and prophage-negative (C600) suspensions totalling 2 ml were added to the 5 ml wells of 12-well macrotitre plates and approximately 500 unstarved (10 μl) T. pyriformis from axenic cultures were added to each experimental well for total densities of approximately 250 ciliates and 109 bacteria ml−1. These plates were maintained at room temperature (approx. 22°C) without agitation. At 24 h intervals, the densities of C600P and C600 were estimated from the CFU data on tetrazolium arabinose agar and that of Tetrahymena microscopically with a haemocytometer.
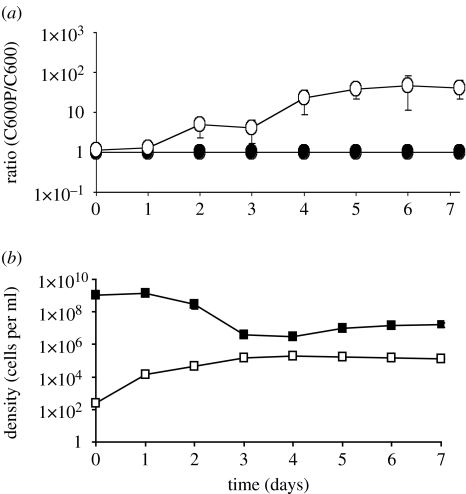
As shown in figure 1a, there is no change in the ratio of C600P to C600 in the Tetrahymena-free controls but a nearly two order of magnitude increase in the ratio of C600P to C600 in the cultures with these grazing ciliates (p<0.002). The C600P/C600 ratio (figure 1a) increases when the density of Tetrahymena increases and levels off when there is no longer a change in the density of Tetrahymena.
Figure 1.
Mixtures of E. coli C600 with and without the Stx2-encoding prophage, C600 and C600P, respectively, in the presence and absence of Tetrahymena. Means ±s.e. for two control (Tetrahymena-free) and six experimental (with Tetrahymena) cultures. (a) Change in ratio of C600P/C600 in the presence and absence of Tetrahymena. Open circles, with Tetrahymena; filled circles, without Tetrahymena. (b) Change in the total densities of bacteria and Tetrahymena in the culture shown in (a). Filled squares, total bacteria; open squares, Tetrahymena.
(b) Predation dynamics
On first consideration, it may seem curious that the Tetrahymena population appears to stop growing while there are still substantial numbers of bacteria, approximately 106 ml−1. This had been observed and explained by Watson and colleagues in their studies with T. pyriformis and washed E. coli from chemostats (Watson et al. 1981). These authors suggest that there is a threshold density of approximately 105 bacteria ml−1 below which Tetrahymena no longer feed on the E. coli. To ascertain whether this threshold also exists under the conditions of our experiments, we diluted washed stationary-phase C600 cultures to 105 cells ml−1 and added 250 Tetrahymena ml−1. The density of E. coli remained at 105 cells ml−1 and there was no growth of the protozoa (data not shown). Moreover, if we consider the initial density of bacteria in these experiments and the maximum density Tetrahymena achieve, approximately 109 and 105 cells ml−1 (see figure 1b), our data are also consistent with that of Watson and colleagues (Watson et al. 1981); it requires consumption of approximately 104 E. coli to produce a single Tetrahymena.
(c) The Stx-encoding prophage augments the fitness of E. coli O157:H7 and E. coli O157:H− in the presence of Tetrahymena
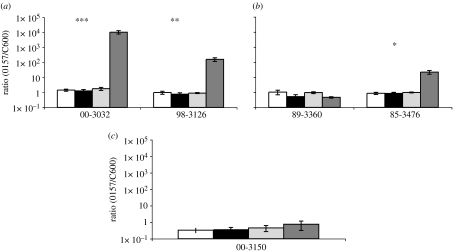
To ascertain whether the advantage of E. coli lysogenic for the Stx-encoding prophage in the presence of Tetrahymena obtains for E. coli O157 lysogens as well as E. coli K-12 lysogens, we performed these pairwise mixed culture experiments with C600 and clinical isolates of E. coli O157:H7 and E. coli O157:H−. In figure 2, we plot the ratio of E. coli O157 and C600 at the start of these experiments and after 3 days in the presence and absence of Tetrahymena.
Figure 2.
Mixtures of clinical isolates of E. coli O157:H7 or E. coli O157:H− and C600 in the presence and absence of Tetrahymena. Ratio of O157 to C600 initially and after 3 days with and without Tetrahymena, means ±s.e. (a) Clinical isolates with Stx1- and Stx2-encoding prophage. (b) Clinical isolates with Stx1-encoding prophage. (c) Clinical isolates with no Stx-encoding prophage. For each experiment, there were two control and six experimental cultures (*p<0.05, **p<0.005 and ***p<0.0005). White bars, initial without Tetrahymena; black bars, after 3 days without Tetrahymena; light grey bars, initial with Tetrahymena; dark grey bars, after 3 days with Tetrahymena.
When E. coli O157 carry prophage that codes for Stx1 and Stx2 (figure 2a), the results are consistent with those of the analogous experiments with C600P and C600. In the presence but not the absence of Tetrahymena, these lysogenic clinical isolates have a fitness advantage over the C600. The results of the experiments with clinical E. coli O157 isolates that carry prophage coding for Stx1 but not Stx2 or no prophage are more ambiguous (figure 2b,c). One clinical isolate of E. coli O157:H−, 85-3476 (stx1), had an advantage over C600 in the presence but not the absence of Tetrahymena. However, two E. coli O157:H7 strains, 89-3360 (stx1) and 00-3150 (stx−), had no apparent advantage in the presence or absence of Tetrahymena.
(d) Role of the stx genes in augmenting the fitness of E. coli in the presence of Tetrahymena
The results of the above-described experiments with clinical strains of E. coli O157 support a role for stx2 in augmenting the fitness of E. coli carrying prophage with this gene in the presence of Tetrahymena. Further evidence for this comes from mixed culture experiments we performed with E. coli carrying prophage in which the stx2 locus was inactivated.
Four E. coli lysogens were used in these experiments: (i) C600PT+ which carry a prophage with a functional stx2, (ii) C600PT− which carry the same prophage as C600PT+, but in which the stx2 locus was replaced with a chloramphenicol resistance cassette (Schmidt et al. 1999), (iii) P2T+, an E. coli O157:H7 strain which carry a prophage with a functional stx2 locus, and (iv) P2T−, the above E. coli O157:H7 which carry the same prophage as P2T+, but in which the stx2 locus is deleted (Gunzer et al. 1998). These Stx+ and Stx− lysogens were mixed with C600 (figure 3). We also performed these pairwise mixed culture experiments with mixtures of otherwise isogenic Stx+ and Stx− lysogens (figure 4).
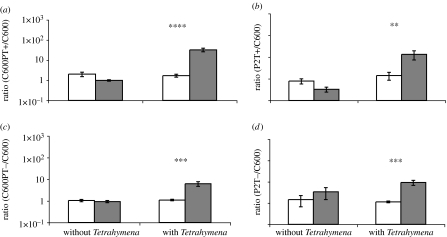
Figure 3.
Mixtures of lysogens and non-lysogens in the presence and absence of Tetrahymena initially and after 3 days, means ±s.e. (a) Ratio of C600 with an Stx2-encoding prophage, C600PT+, and C600. (b) Ratio of E. coli O157:H7 with an Stx2-encoding prophage, P2T+, and C600. (c) Ratio of C600 with the toxin-negative construct of the Stx2-encoding prophage, C600PT−, and C600. (d) Ratio of the toxin-negative E. coli O157:H7 construct of the Stx2-encoding prophage, P2T−, and C600. In all experiments, there were three control and nine experimental cultures (**p<0.005, ***p<0.0005 and ****p<0.00005). White bars, initial; grey bars, after 3 days.
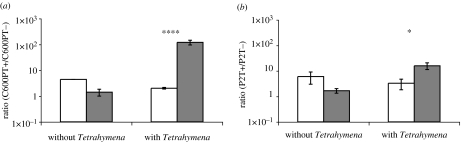
Figure 4.
Mixtures of stx2-positive strains and isogenic stx2-negative constructs in the presence and absence of Tetrahymena initially and after 3 days, means ±s.e. (a) Ratio of C600PT+ and C600PT−. (b) Ratio of P2T+ and P2T−. For each experiment, there were two control and six experimental cultures (*p<0.05 and ****p<0.00005). White bars, initial; grey bars, after 3 days.
For both the C600 and E. coli O157:H7 constructs, bacteria bearing prophage with a functional stx2 gene have an advantage over C600 in the presence but not the absence of Tetrahymena (figure 3a,b). The results of experiments with lysogens bearing prophage with inactivated stx2 loci suggest that even without a functional Stx2, this prophage increases the fitness of E. coli in the presence but not the absence of Tetrahymena (figure 3c,d). The magnitude of the advantage appears to be greater when the stx2 locus is functional. Additional support for this interpretation comes from experiments where the stx-positive and stx-negative lysogens are mixed with each other. In the presence but not the absence of Tetrahymena, the stx2-positive bacteria have a fitness advantage over the stx2-negative bacteria (figure 4).
(e) The Stx-encoding prophage increases the survival of E. coli in the food vacuoles of Tetrahymena
How do Tetrahymena select for prophage-bearing E. coli? One possibility is that the carriage of the prophage increases the rate of survival of E. coli ingested by Tetrahymena (Brandl et al. 2005) relative to bacteria not carrying this element. During grazing, Tetrahymena fill organelles known as food vacuoles with bacteria which are lysed and digested over a period of time. Bacteria-filled food vacuoles are also released into the environment (King et al. 1988; Schlimme et al. 1995). In accordance with this hypothesis, bacteria with higher rates of survival in food vacuoles would be more likely to be recovered as CFUs by plating either vacuoles or bacteria released from vacuoles, than bacteria with lower rates of survival.
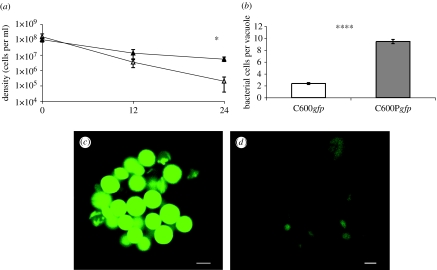
To test this hypothesis, we labelled C600 and C600P with gfp (pEGFP, Clontech), C600gfp and C600Pgfp, respectively. To washed, overnight, single clone cultures of C600gfp and C600Pgfp, we added approximately 5×104 Tetrahymena and incubated the suspensions at room temperature for 24 h. At 12 and 24 h, we estimated densities of bacteria by plating (CFU) and counting the number of bacteria per food vacuole using an epifluorescent microscope. To test the viability of the bacteria in the food vacuoles, we used the Live/Dead BacLight viability system (Molecular Probes, Eugene, OR).
The results of our plating experiments suggest that the colonies on the plates arise from single cells rather than vacuoles containing multiple bacteria. The colonies on the tetrazolium arabinose agar did not contain Ara− and Ara+ sectors and when restreaked on this indicator agar, the colonies produced were monomorphic Ara− or Ara+. As estimated from either colony count data or number of fluorescing bacteria per vesicle, there are more surviving prophage-bearing E. coli in the presence of Tetrahymena than prophage-free bacteria (figure 5a,b). The survival of the bacteria within the food vacuoles can be seen from confocal micrographs of Tetrahymena in figure 5c,d. In figure 5c, where C600 carries the prophage, there are an abundance of viable cells within the food vesicles. This is clearly not the case when C600 does not carry the prophage (figure 5d). In the absence of Tetrahymena, there is no difference in the rate of survival of the bacteria with the gfp with and without the prophage (data not shown).
Figure 5.
C600gfp (prophage negative) and C600Pgfp (prophage positive) exposed to high density of Tetrahymena. (a) Estimated bacterial densities (CFU data) at 12 and 24 h, C600Pgfp (closed triangles) and C600gfp (open triangles). (b) Number of viable C600gfp and C600Pgfp cells per food vacuole, direct counts of fluorescing cells (*p<0.05 and ****p<0.00005). (c) C600Pgfp in food vacuoles. (d) C600gfp in food vacuoles. Scale bar, 5 μm.
Additional evidence for the enhanced survival of C600P within the food vacuoles was obtained from the results of experiments using the Live/Dead BacLight viability assay. As determined by this protocol, which measures bacterial membrane integrity, a substantial fraction of C600P cells in the vacuoles were viable; they did not take up propidium iodide. On the other hand, few, if any, C600 cells failed to take up propidium iodide (data not shown).
4. Discussion
The results of our in vitro experiments with mixed cultures of E. coli bearing and not bearing the Stx-encoding prophage of E. coli O157:H7 are consistent with the hypothesis that predation by protozoa favours lysogens for this prophage. In the presence but not the absence of T. pyriformis, the carriage of the Stx-encoding prophage augments the fitness of both the laboratory strain E. coli K-12 (C600) and the clinical isolates of E. coli O157:H7 and E. coli O157:H−.
Our experiments with E. coli with prophage carrying non-functional stx loci suggest that the advantage of these lysogens in the presence of grazing protozoa can, to some extent, but not entirely, be attributed to Stx2. In the presence but not the absence of Tetrahymena, all of the clinical isolates of E. coli O157 carrying prophage with functional stx2 loci had an advantage over C600 non-lysogens as well as otherwise isogenic cells with non-functional stx2 alleles. On the other hand, one clinical isolate bearing stx1 but not stx2 also had an advantage over C600 in the presence but not the absence of Tetrahymena.
Our results indicated that some, if not all, of the advantage of E. coli with the Stx-encoding prophage in the presence of Tetrahymena can be attributed to the prophage increasing the rate of the survival of these bacteria within intact food vacuoles. While it has been suggested that build-up of toxin in the food vacuoles leads to their expulsion by the protozoa before all the bacteria are digested (Matz & Kjelleberg 2005), toxin-mediated expulsion alone would not account for the observed advantage of the prophage-bearing cells. In these mixed cultures, the vacuoles would contain both lysogens and non-lysogens for this phage. If these vacuoles are expelled before the bacteria are digested or bacteria with and without prophage survive equally well in these vacuoles, there would be no advantage associated with the carriage of these prophages. On the other hand, owing to their greater rate of survival in the vacuoles before expulsion, the relative frequency of Stx-prophage-bearing bacteria in the extra-ciliate environment at large would continue to increase as long as the ciliates grazed on these bacteria.
Under what conditions, in what habitats and to what extent predation by protozoa contributes to the persistence of Stx-encoding phages in E. coli are, at this time, unanswered questions. Is this protozoa-mediated selection for E. coli bearing Stx-encoding prophage occurring in the external environment, water or soil, or in the gastrointestinal tract of colonized bovines? The density of E. coli in the rumen of cattle, which has been estimated to be less than 105 cells g−1 (Russell et al. 2000), may be too low for the protozoa to be effective predators and thereby to select for bacteria carrying Stx-encoding prophage. If, however, the total density of bacteria in the rumen is sufficient to maintain a substantial population of grazing ciliates and possibly other even if E. coli is a small minority population, selection could still favour E. coli carrying Stx-encoding prophage. This can be seen with the simple mathematical model where we consider three populations of bacteria (Appendix A).
Bactivorous protozoa are present in the lower colon (Coleman 1964), and that too may be a habitat for protozoa-mediated selection favouring E. coli bearing Stx-encoding prophage. The bovine rectal mucosa is the primary site of E. coli O157:H7 colonization (Naylor et al. 2003). In grain-fed cattle, the densities of E. coli in the colon have been reported to be of the order of 108 cells ml−1 (Russell et al. 2000), and at E. coli densities of this magnitude, there is evidence that gastrointestinal protozoa take up these bacteria at a high rate (Coleman 1964). As mentioned above and illustrated in Appendix A, even if the density of E. coli O157:H7 in the intestine is too low of a density to sustain a population of grazing protozoa by itself, if the majority population of bacteria consumed by these protozoa is dense enough, selection mediated by predation would still favour the carriage of the Stx-encoding prophage in the E. coli O157:H7 minority.
The results of our in vitro experiments as well as other studies of bacterial predation by grazing protozoa point to the need to more extensively evaluate the contribution of this predation to the maintenance of Stx-encoding prophage in the ecology of E. coli O157:H7 and possibly E. coli O157:H7 itself. Mixed culture experiments similar to those reported here with prophage-bearing and prophage-free E. coli O157 could be performed in situ as well as in vitro with naturally occurring protozoa from the gastrointestinal tracts of cattle and other ungulates. These protozoa include species from genera Eudiplodinium, Metadinium, Polyplastron, Isotricha, Entodinium and Diplodinium (Rasmussen et al. 2005), which are all bactivorous ciliates like Tetrahymena. These experiments can also be performed in the soil and water in areas where cattle graze and where the faeces of cattle are used for fertilizer or are a probable source of contamination.
It is conceivable that the increased capacity to survive in the food vacuoles of protozoa enhances the term of survival of E. coli O157:H7 in the environment and thereby the likelihood of their transmission to humans. A number of studies have shown that bacteria ingested by protozoa are more resistant to disinfectants and biocides than free bacteria (Watson et al. 1981; Barker et al. 1992; Brandl et al. 2005). Could this be the case for the E. coli O157:H7 contaminants of bean sprouts, lettuce and spinach? Protozoa could also play a role in the transmission of E. coli O157:H7 between cattle and thereby the persistence of these bacteria in time and space. At any given time, the overall prevalence of E. coli O157:H7 on a single farm may be too low for a few individual colonized cattle shedding bacteria (Matthews et al. 2006) serving as a source for the persistence of these bacteria in the herd at large. However, anything that promotes the survival of E. coli O157:H7 in the extra-host environment could also contribute to the maintenance of these bacteria in the herd as well as their spread to other farms (Wetzel & LeJeune 2006). Moreover, if E. coli O157:H7 is like Legionella pneumophila and S. enterica, their carriage in protozoa may also make them more virulent to humans than when they are ingested as free bacteria (Cirillo et al. 1994; Rasmussen et al. 2005). Finally, it is possible that ingestion by and survival in these unicellular eukaryotes may be the selective force responsible for the evolution of the toxins and other prophage-encoded virulence factors of E. coli O157:H7. In this interpretation, protozoa are acting like what Barker and Brown refer to as ‘Trojan horses’ (Barker & Brown 1994).
To be sure, these extensions of the results of this in vitro study are, at this stage, no more than speculations. They are however speculations (hypotheses) that can be tested experimentally. We believe that these results and hypotheses have the dual virtues of being interesting to ecology, population and evolutionary biology as academic subjects and, at the same time, relevant to our understanding of the epidemiology of an infectious disease and the development of procedures for its control.
Acknowledgments
This work was supported by grants from the Wellcome Trust IPRAVE project and the US National Institutes of Health AI40662 (B.R.L.), and a National Science Foundation Graduate Research Fellowship (K.M.S.). We thank Robert Tauxe for inspiration and advice on this project and comments on the manuscript, and Mary Reynolds for advice as well as for constructing the C600 lysogens used in our initial experiments. We would also like to thank Nancy Strockbine, Herbert Schmidt and Arthur Donohue-Rolfe for generously providing strains, and Andreas Handel, Phillip Tarr and Nicholas Parrish for helpful comments while preparing the manuscript. Finally, we wish to express our gratitude to Howard Rees for assistance with confocal microscopy.
Appendix A.
In our article, we suggest that even if E. coli O157:H7 is a minority population of insufficient density to support a grazing population of protozoa, protozoa-mediated selection could still favour the ascent of Stx-encoding E. coli O157:H7. The necessary condition for this is that the dominant population(s) of bacteria, be them E. coli or not, are grazed by these protozoa and are sufficiently dense for these protozoa to grow to substantial densities. Here, we use a simple mathematical model and numerical analysis of its properties to illustrate that, at least in theory, this would obtain.
In this model, we assume that there is one population of protozoa of density P and three populations of bacteria of densities and designations B, M and T cells ml−1. The B population of bacteria is consumed by the protozoa but is not E. coli O157. The M and T populations are, respectively, prophage-free and prophage-bearing E. coli O157:H7, Tox− and Tox+. We assume that bacteria and protozoa encounter each other at random at rates proportional to their densities, the standard mass action assumption of predator–prey models (e.g. Wilson & Bossart 1971). To account for the observation that Tetrahymena stop feeding when the density of bacteria becomes too low (Watson et al. 1981), unlike standard predator–prey models, we assume that the rate at which bacteria are consumed upon contact with the protozoa increases monotonically with the density of the total population of prey (bacteria). For the consumption dynamics, we assume hyperbolic function of the sort employed by Monod (1949). The constants γB, γM and γT are the maximum rate parameters of consumption of the B, M and T bacterial population. The parameter k is the density of bacteria when the rate at which they are consumed upon contact with the protozoa is half its maximum value. We assume that the bacteria do not replicate and die only through predation by the protozoa. The protozoa do not die over the course of the experiment and only replicate by the consumption of the bacteria. A protozoan requires e bacteria to divide, the conversion efficiency (Stewart & Levin 1984). With these definitions and assumptions, the rates of change in the density of bacteria and protozoa are given by the following set of differential equations:
where N=B+M+T and ϕ(N)=N/(N+k)
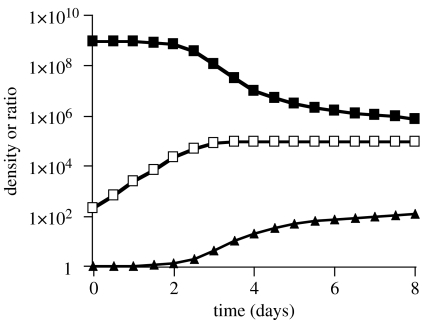
For our illustration, we use a numerical solution to these equations, a computer simulation, programed in Berkeley Madonna. Copies of this program are available on www.eclf.com. Save for the conversion efficiency e, which has been estimated to be approximately 10−4 bacteria (Watson et al. 1981) and the present study, i.e. it takes approximately 104 bacteria to produce a single protozoon, the other parameters were chosen to provide an approximate fit of Tetrahymena–E. coli observations in figure 1 of our report. In these simulations, we assume a delay of 4 h between the time the bacteria are consumed and the time the protozoa are produced.
With the chosen parameter values and initial densities of bacteria and protozoa, as with the real data in figure 1a,b in our article, the ratio of Tox+/Tox− increases when the density of protozoa increases. As the total density of the bacteria declines, the rate of ascent of the protozoa population declines and, within a few days, the density of both bacteria and protozoa levels off (figure 6).
Figure 6.
Simulation result changes in the total density of bacteria and protozoa and the ratio of prophage-bearing (Tox+) and prophage-free (Tox−) E. coli 0157:H7. Parameters γB=2×10−6, γM=2×10−6 and γT=5×10−7; e=10−4, k=10−7. The initial densities of the populations were B=109, M=T=5×104 and P=2×102 cells ml−1. Open squares, density of Tetrahymena; filled squares, density of bacteria; filled triangles, ratio (Tox+/Tox−).
References
- Barker J, Brown M.R. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- Barker J, Brown M.R, Collier P.J, Farrell I, Gilbert P. Relationship between Legionella pneumophila and Acanthamoeba polyphaga: physiological status and susceptibility to chemical inactivation. Appl. Environ. Microbiol. 1992;58:2420–2425. doi: 10.1128/aem.58.8.2420-2425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser R.E, Griffin P.M, Slutsker L. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 1999;50:355–367. doi: 10.1146/annurev.med.50.1.355. doi:10.1146/annurev.med.50.1.355 [DOI] [PubMed] [Google Scholar]
- Brandl M.T. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 2006;44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. doi:10.1146/annurev.phyto.44.070505.143359 [DOI] [PubMed] [Google Scholar]
- Brandl M.T, Rosenthal B.M, Haxo A.F, Berk S.G. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 2005;71:1562–1569. doi: 10.1128/AEM.71.3.1562-1569.2005. doi:10.1128/AEM.71.3.1562-1569.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo J.D, Falkow S, Tompkins L.S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman G.S. The metabolism of Escherichia coli and other bacteria by Entodinium caudatum. J. Gen. Microbiol. 1964;37:209–223. doi: 10.1099/00221287-37-2-209. [DOI] [PubMed] [Google Scholar]
- Cornick N.A, Booher S.L, Moon H.W. Intimin facilitates colonization by Escherichia coli O157:H7 in adult ruminants. Infect. Immun. 2002;70:2704–2707. doi: 10.1128/IAI.70.5.2704-2707.2002. doi:10.1128/IAI.70.5.2704-2707.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Nystrom E.A, Bosworth B.T, Moon H.W, O'Brien A.D. Bovine infection with Shiga-toxin producing Escherichia coli. In: Kaper J.B, O'Brien A.D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology; Washington, DC: 1998a. pp. 261–267. [Google Scholar]
- Dean-Nystrom E.A, Bosworth B.T, Moon H.W, O'Brien A.D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 1998b;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWaal C, Johnson K, Bhuiya F. Center for Science Public Interest; Washington, DC: 2006. Outbreak Alert! Closing gaps in our federal food safety net. [Google Scholar]
- Gunzer F, Bohn U, Fuchs S, Muhldorfer I, Hacker J, Tzipori S, Donohue-Rolfe A. Construction and characterization of an isogenic slt-ii deletion mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 1998;66:2337–2341. doi: 10.1128/iai.66.5.2337-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W, Hofle M.G. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 2001;35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. doi:10.1111/j.1574-6941.2001.tb00794.x [DOI] [PubMed] [Google Scholar]
- Hancock D.D, Besser T.E, Rice D.H. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices. In: Kaper J.B, O'Brien A.D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology; Washington, DC: 1998. pp. 85–91. [Google Scholar]
- Jurgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek. 2002;81:413–434. doi: 10.1023/a:1020505204959. doi:10.1023/A:1020505204959 [DOI] [PubMed] [Google Scholar]
- King C.H, Shotts E.B, Jr, Wooley R.E, Porter K.G. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 1988;54:3023–3033. doi: 10.1128/aem.54.12.3023-3033.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, McKendrick I.J, Ternent H, Gunn G.J, Synge B, Woolhouse M.E. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiol. Infect. 2006;134:131–142. doi: 10.1017/S0950268805004590. doi:10.1017/S0950268805004590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. doi:10.1016/j.tim.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Menge C, Blessenohl M, Eisenberg T, Stamm I, Baljer G. Bovine ileal intraepithelial lymphocytes represent target cells for Shiga toxin 1 from Escherichia coli. Infect. Immun. 2004;72:1896–1905. doi: 10.1128/IAI.72.4.1896-1905.2004. doi:10.1128/IAI.72.4.1896-1905.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949;3:371–394. doi:10.1146/annurev.mi.03.100149.002103 [Google Scholar]
- Naylor S.W, Low J.C, Besser T.E, Mahajan A, Gunn G.J, Pearce M.C, McKendrick I.J, Smith D.G, Gally D.L. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. doi:10.1128/IAI.71.3.1505-1512.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel J.M, Sparling P.H, Crowe C, Griffin P.M, Swerdlow D.L. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M.A, Carlson S.A, Franklin S.K, McCuddin Z.P, Wu M.T, Sharma V.K. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 2005;73:4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005. doi:10.1128/IAI.73.8.4668-4675.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.M, Sinclair J.F, Smith M.J, O'Brien A.D. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl Acad. Sci. USA. 2006;103:9667–9672. doi: 10.1073/pnas.0602359103. doi:10.1073/pnas.0602359103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J.B, Diez-Gonzalez F, Jarvis G.N. Invited review: effects of diet shifts on Escherichia coli in cattle. J Dairy Sci. 2000;83:863–873. doi: 10.3168/jds.S0022-0302(00)74950-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Cold Spring Harbour Laboratory Press; Cold Spring Harbour, NY: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Schlimme W, Baur B, Hanselmann K, Jenni B. An agarose slide method to follow the fate of bacteria within digestive vacuoles of protozoa. FEMS Microbiol. Lett. 1995;133:169–173. doi: 10.1111/j.1574-6968.1995.tb07879.x. doi:10.1111/j.1574-6968.1995.tb07879.x [DOI] [PubMed] [Google Scholar]
- Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999;65:3855–3861. doi: 10.1128/aem.65.9.3855-3861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H, Lim J.Y, Knecht H.J, Li J, Hovde C.J. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 2006;74:4685–4693. doi: 10.1128/IAI.00406-06. doi:10.1128/IAI.00406-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart F.M, Levin B.R. The population biology of bacterial viruses: why be temperate. Theor. Popul. Biol. 1984;26:93–117. doi: 10.1016/0040-5809(84)90026-1. doi:10.1016/0040-5809(84)90026-1 [DOI] [PubMed] [Google Scholar]
- Tezcan-Merdol D, Ljungstrom M, Winiecka-Krusnell J, Linder E, Engstrand L, Rhen M. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 2004;70:3706–3714. doi: 10.1128/AEM.70.6.3706-3714.2004. doi:10.1128/AEM.70.6.3706-3714.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.J, Ohtaguchi K, Fredrickson A.G. Kinetics of growth of the ciliate Tetrahymena pyriformis on Escherichia coli. J. Gen. Microbiol. 1981;122:323–333. doi: 10.1099/00221287-122-2-323. [DOI] [PubMed] [Google Scholar]
- Wetzel A.N, LeJeune J.T. Clonal dissemination of Escherichia coli O157:H7 subtypes among dairy farms in northeast Ohio. Appl. Environ. Microbiol. 2006;72:2621–2626. doi: 10.1128/AEM.72.4.2621-2626.2006. doi:10.1128/AEM.72.4.2621-2626.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte H, Wolfe D.M, Tamewitz A, Lawrence J.G. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl Acad. Sci. USA. 2004;101:10 644–10 649. doi: 10.1073/pnas.0404028101. doi:10.1073/pnas.0404028101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E.O, Bossart W.H. Sinauer; Sunderland, MA: 1971. A primer of population biology. [Google Scholar]