Abstract
Tumor suppressor p53 is a well-characterized transcription factor that binds DNA. More enigmatic are the RNA-binding properties of p53 and their physiological relevance. We used three sensitive co-immunoprecipitation methods in an attempt to detect RNAs that tightly associate with p53 in cultured human cells. Although recombinant p53 protein binds RNA in a sequence-nonspecific mode, we do not detect specific in vivo RNA binding by p53. These results suggest that RNA binding is prevented by post-translational p53 modifications. A ribonucleoprotein (not p53) is purified by multiple IgG monoclonal antibodies (including anti-p53 antibodies) from both p53 +/+ and p53 null cells. Caution is therefore required in interpreting RNA co-immunoprecipitation experiments. Though not formally excluded, these results do not support models in which p53 binds specific RNA partners in vivo.
Introduction
The p53 tumor suppressor protein has been extensively characterized as a DNA-binding transcription factor. Most cellular responses to p53 are mediated by targets of p53 transcriptional activity [1]. p53 integrates cellular stress signals ranging from DNA damage to nutrient deprivation [2], playing a role in cell cycle arrest, DNA damage repair, and apoptosis. p53 is nonfunctional in most human cancers [3].
The domain structure and functions of p53 have been established (Fig. 1A). A peculiar observation is RNA binding by the C-terminus of p53 (Fig. 1A; [4]). Although many RNA-binding proteins are characterized by RNA-recognition motifs (RRMs; [5]), the interaction between p53 and RNA involves the C-terminal 30 amino acids of p53 [6, 7], which do not form a RRM but are disordered in solution [7, 8] and do not reorganize upon RNA binding [7, 9]. p53, like the HIV-1 nucleocapsid protein, has been termed an RNA chaperone [10] that may facilitate RNA folding [11] without ATP hydrolysis [11–13].
Fig. 1.
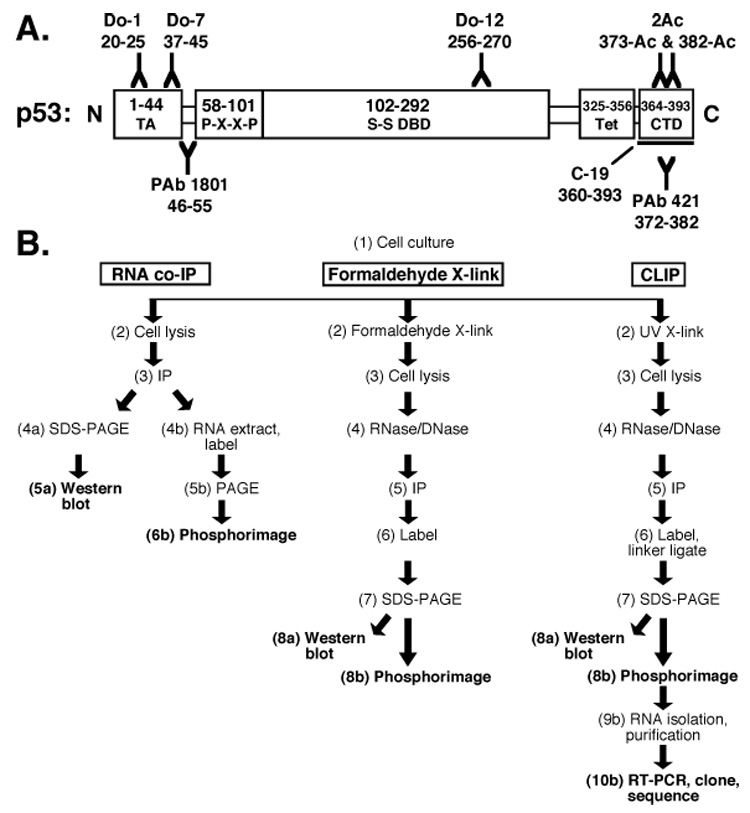
Protocols to study protein-RNA interactions. (A) p53 domains and antibodies. TA: transactivation domain, P-X-X-P: proline-rich domain, S-S DBD: sequence-specific DNA-binding domain, Tet: tetramerization domain, CTD: regulatory C-terminal domain. (B) In vivo RNA identification protocols.
Several in vitro observations motivated a search for in vivo RNA partners of p53. According to early reports, mouse p53 can be purified from SV40-transformed BALB/c cells in a covalent complex with 5.8S rRNA [14, 15]. Mosner et al. reported non-covalent interactions between p53 and the 5’-UTR of its own mRNA transcript [16], and others suggested that p53 can bind the 5'-UTR of Cdk4 mRNA [17]. Similarly, Galy et al. reported that recombinant p53 can bind the 5’-UTR of the FGF-2 mRNA [18]. In contrast, reports of p53 RNA-RNA annealing activity [13] and sequence-nonspecific RNA binding [4, 6, 7, 19] suggested that p53 binds RNA promiscuously. Acetylation of four p53 C-terminal lysine residues eliminates detectable RNA binding in vitro [7], as it does for sequence-nonspecific DNA binding [8].
We wished to test the hypothesis that p53 associates with specific target RNAs in living cells. A prior study of p53-RNA interactions in human cells suggested that RNA is co-immunoprecipitated with p53 using Do-1 and Do-7 antibodies, which recognize N-terminal p53 epitopes (Fig. 1A,B; [19]). Recovered RNAs were detected by radiolabeling but were not cloned [19]. A disadvantage of such RNA co-immunoprecipitations is the potential redistribution of complexes after cell lysis [20]. Nonspecific RNA contamination of immunoprecipitations is also a concern. High-salt washes might enhance stringency but may disrupt both specific and nonspecific RNA-protein interactions [7]. One approach to increase stringency involves in vivo covalent cross-linking with a chemical agent such as formaldehyde, which captures in vivo protein-RNA interactions rapidly and reversibly (Fig. 1B, center; [21]).
The UV-cross-linking and immunoprecipitation (CLIP) protocol was recently reported to facilitate the detection and cloning of protein-bound RNAs in vivo (Fig. 1B, right; [22]). CLIP involves the brief UV irradiation of live cells followed by extract preparation for immunoprecipitation (Fig. 1B, right; [22]). Selection among the many commercial antibodies specific for p53 (Fig. 1A) can improve the stringency and specificity of this method. The lysate used for immunoprecipitation is treated with nucleases to eliminate DNA contamination, generating protein-bound RNA tags. Linker oligonucleotides are ligated to the radiolabeled RNA tags, and the protein-RNA complexes are separated by SDS-PAGE. RNA tags are purified, amplified, cloned, and sequenced (Fig. 1B, right).
Our previous studies of RNA binding by p53 in yeast and in vitro demonstrated that p53 with incomplete post-translational modifications binds RNA without apparent sequence- or structure-specificity [4, 6, 7]. It is possible that a partially-modified form of p53 could exist in vivo with one or more specific RNA partners. RNA co-immunoprecipitation with p53 was undertaken to test the hypothesis that RNA binding partners for p53 can be cloned from human cells.
Experimental Procedures
Cell culture and antibodies
MCF-7 (TP53 +/+) human breast cancer cells were grown in Dulbecco's modified Eagle's medium, PC-3 (tp53 −/−) human prostate cancer cells were grown in McCoy’s 5a medium, and HCT116 human colorectal carcinoma cells (both p53 WT and tp53 −/−; [23]) were grown in RPMI medium. Media were supplemented with 10% fetal bovine serum.
Anti-p53 PAb 1801 and PAb 421, and anti-c-myc Ab1 antibodies were purchased from EMD Biosciences, anti-HA HA.11 antibody from Covance, anti-p53 (PAb 246, C-19, Do-1, HRP-conjugated Do-1, and Do-7) antibodies from Santa Cruz Biotechnology, 2Ac (p53 Ac373/Ac382) from Upstate, and anti-p53 Do-12 from Chemicon International.
RNA co-immunoprecipitation
Untreated cells (MCF-7, PC-3, HCT116, and HCT116 tp53−/− were lysed in 500 µl lysis buffer (50 mM Tris [pH 8.0], 1 mM EDTA, 120 mM NaCl, 10% glycerol, 0.5% NP-40). 10% of the resulting clarified whole cell lysate (WCL) was saved for Western blotting.
WCL was mixed with 50 µl protein G Dynabeads, pre-bound with anti-p53 Do-7 antibody for 2 h at 4 °C. One tenth of the supernatant was saved for Western blot analysis, and the beads were washed and split. Half was eluted in 1:1 wash buffer:LDS sample loading buffer (Invitrogen) and loaded onto a Novex 10% Bis-Tris gel (Invitrogen) for SDS-PAGE for Western analysis. The remaining half was extracted with TRIZOL reagent (Invitrogen) to isolate RNA. All RNA samples were treated with calf intestinal alkaline phosphatase (CIP; New England BioLabs) and radiolabeled using polynucleotide kinase (PNK). Samples were analyzed by electrophoresis in 4–12% Bis-Tris SDS PAGE gels (1 h, 185 V) and 6% (1:19) denaturing PAGE gels (1 h, 500V).
Formaldehyde cross-linking immunoprecipitation
A protocol was adapted from a prior report [21]. Cells were resuspended in PBS containing 1% formaldehyde (37% stock, Mallinckrodt). Samples were incubated with rotation for 10 min at room temperature. Reactions were quenched with 0.25 M glycine for 5 min at room temperature. Cell pellets were resuspended in 500 µl RIPA buffer (50 mM Tris [pH 7.5], 1% NP-40, 0.5% deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl). Samples were sonicated and the resulting lysate was treated with RQ1 DNase (Promega) for 5 min at 37 °C, and then diluted RNase A (USB) was added for an additional 10 min. The lysate was centrifuged at 14 000 RPM in a benchtop centrifuge for 10 min at 4 °C. The supernatant (clarified WCL) was recovered, and 10% was removed for Western analysis. The remaining supernatant was incubated for 2 h at 4 °C with 50 µl protein G Dynabeads pre-bound to Do-7 antibody. A sample of the cleared supernatant was retained for Western analysis, and the beads were washed three times in RIPA buffer, then twice in PNK buffer (50 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 0.5% NP-40). Beads were resuspended in 40 µl PNK reaction mix [4 µl 10X PNK buffer (New England BioLabs), 1 µl [γ-32P] ATP (GE), 2 µl T4 PNK enzyme (10 U/µl; New England BioLabs), 32 µl water] and incubated at 37 °C for 10 min. Samples were washed three times in 200 µl PNK buffer and eluted from beads in a mixture of 15 µl PNK buffer and 15 µl LDS sample buffer (Invitrogen) with heating at 70 °C. Ten µl of eluted sample were loaded into each lane of a Novex NuPAGE 10% Bis-Tris gel (Invitrogen). RNA-protein complexes were transferred to nitrocellulose (BA-85; S&S) at 4 °C.
Cross-linking immunoprecipitation
CLIP was performed as previously described [22]. Cells were irradiated on ice with 200 mJ/cm² in a Stratalinker 1800 (Stratagene). Where noted, samples were treated with CIP (1 U/µl; Roche). Reactions were performed in PNK buffer [pH 7.4]. Western analysis was performed by standard methods.
Results and Discussion
RNA co-immunoprecipitation with anti-p53 antibody Do-7
Anti-p53 antibodies were used to precipitate p53 from cells (Fig. 1B, left). Tested cell lines do (MCF-7, HCT116 +/+) or do not (PC-3, HCT116 −/−) express p53 [23]. Unstimulated levels of p53 were tested, as induction methods induce p53 post-translational modifications [24]. The high steady-state p53 levels that accumulate in some model cell lines are presumably tolerated because p53 is not properly activated for transcriptional function [25, 26]. The co-immunoprecipitation protocol required mild conditions to preserve binding to immobilized antibody Do-7 Fig. 1A; [27].
Western blots of input WCL identified p53 at ~50 kDa in parental HCT116 (TP53 +/+) (Fig. 2A, lane 2) but not tp53 −/− HCT116 cells (Fig. 2A, lane 1). p53 was detected in MCF-7 cells (Fig. 2A, lane 4), but not in PC-3 cells (Fig. 2A, lane 3). p53 proteins in MCF-7 and HCT116 cell lines displayed slightly different electrophoretic mobilities, suggesting differences in post-translational modifications (Fig. 2A, compare lanes 2 and 4). p53 was immunoprecipitated with Do-7 antibody from HCT116 (TP53 +/+) and MCF-7 cells (Fig. 2A, lanes 6 and 8) but not from HCT116 cells (tp53 −/−) or PC-3 cells (Fig. 2A, lanes 5 and 7).
Fig. 2.
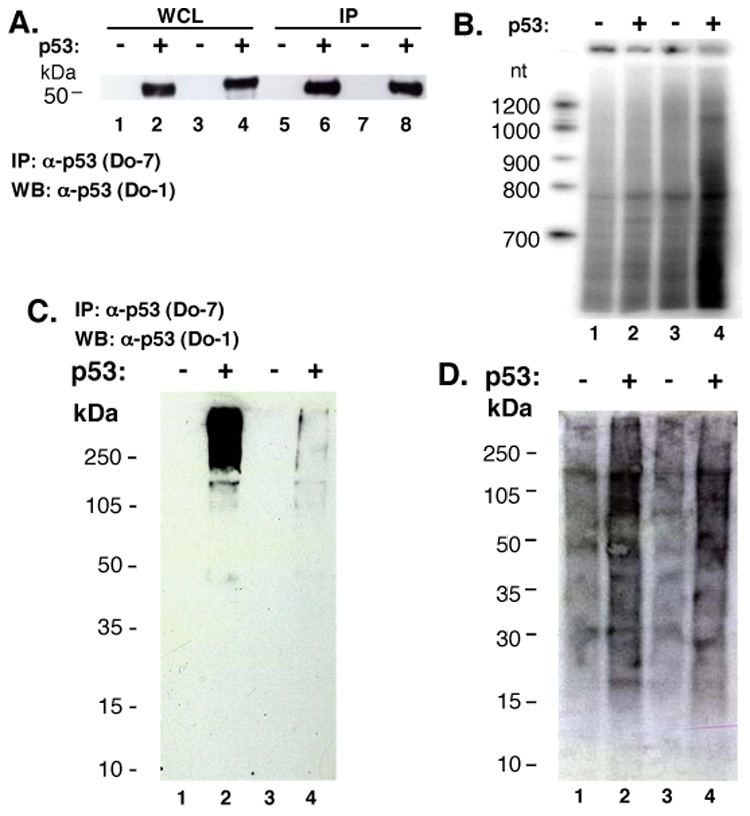
RNA is purified from cell lysates by anti-p53 antibodies regardless of p53 status. (A) Western analysis of p53. Preparations of whole cell lysate (WCL; 10% of total) [HCT116 (tp53 −/−) lane 1; HCT116 (TP53 +/+) lane 2; PC-3 (tp53 −/−) lane 3; MCF-7 (TP53 +/+) lane 4, after IP with anti-p53 Do-7 (lanes 5–8, respectively). (B) Co-immunoprecipitated RNA after [γ32P]-ATP labeling: HCT116 (tp53 −/−) lane 1; HCT116 (TP53 +/+) lane 2; PC-3 (tp53 −/−) lane 3; MCF-7 (TP53 +/+) lane 4. Formaldehyde cross-linked whole cell lysate: [HCT116 (tp53 −/−) lane 1; HCT116 (TP53 +/+) lane 2; PC-3 (tp53 −/−) lane 3; MCF-7 (TP53 +/+) lane 4], subjected to IP with anti-p53 Do-7 followed by RNA radiolabeling. (C) Western blot with anti-p53 antibody Do-1. (D) Membrane from panel (C) exposed to film.
RNAs that co-immunoprecipitated with p53 were extracted, radiolabeled, denatured, and separated on a polyacrylamide gel (Fig. 1A, left; Fig. 2B). The resulting signal was sensitive to RNase (data not shown). Unexpectedly, Do-7 antibody immunoprecipitated a wide range of RNA species from all four cell lines (Fig. 2B), even those lacking p53 expression. The pattern of labeled RNA species was indistinguishable for all cases (Fig. 2B, lanes 1–4). Because a p53-associated RNA signal might be occluded by the strong nonspecific RNA signal, we increased the wash stringency by the use of cross-linking in subsequent experiments.
RNA co-immunoprecipitation after formaldehyde cross-linking
We used formaldehyde to cross-link protein-RNA complexes in live cells prior to immunoprecipitation [21]. Cells were treated with an optimized concentration of formaldehyde before rigorous lysis and immunoprecipitation (Fig. 1B, center). Samples were treated with [γ-32P] ATP and T4 PNK to radiolabel bound RNA fragments before elution from beads (Fig. 1B, center).
p53 protein was detected exclusively in the expected cell types as a high molecular weight complex, consistent with chemical cross-linking before immunoprecipitation (Fig. 2C, lanes 2 and 4). As previously observed without cross-linking, RNA was co-immunoprecipitated from all four cell lines regardless of p53 expression. Although more RNA was obtained from the cell lines that express p53 (Fig. 2D, lanes 2 and 4), radiolabeled RNA species appeared throughout the gel, not comigrating with p53 as would have been expected for RNA species covalently linked to p53. The pattern of radiolabeled RNAs was indistinguishable among the four samples, again indicating that unique RNAs were not recovered in a p53-dependent manner.
RNA co-immunoprecipitation after CLIP
The CLIP technique [22] was applied to further enhance the specificity of RNA cross-linking and co-purification with p53. CLIP cross-links only RNAs in close p53 contact (Fig. 1B, right). Application of CLIP also assumes that UV treatment does not induce changes in p53 that prevent RNA binding. We compared MCF-7 (TP53 +/+) breast cancer cells with PC-3 (tp53 −/−) prostate cancer cells as a negative control. Similar results were obtained with HCT116 TP53 +/+ and tp53 −/− cell lines.
Anti-p53 antibodies recognizing N-terminal epitopes were selected to monitor potential RNA cross-linking at the p53 C-terminus. (Fig. 1A; [7]), while N-terminal anti-p53 antibodies Do-7 and PAb 1801 were used to directly compare protein and RNA signals immunoprecipitated from UV-cross-linked MCF-7 and PC-3 WCL (Fig. 4A–B). p53 was efficiently immunoprecipitated from MCF-7 cells (Fig. 3A, lanes M), and not from p53-null PC-3 cells (Fig. 3A, lanes P). The overexposed Western blot shows a ladder (~60–150 kDa) of ubiquitinated p53 isoforms (Fig. 3A, lanes M), which further accumulate upon proteasome inhibitor MG132 treatment (data not shown). Also visible below 50 kDa is a smaller p53 species, possibly a p53 isoform [28], and a Do-7 cross-reacting, non-p53 species was detected at ~250 kDa (Fig. 3A, lanes M and P, Do-7).
Fig. 4.
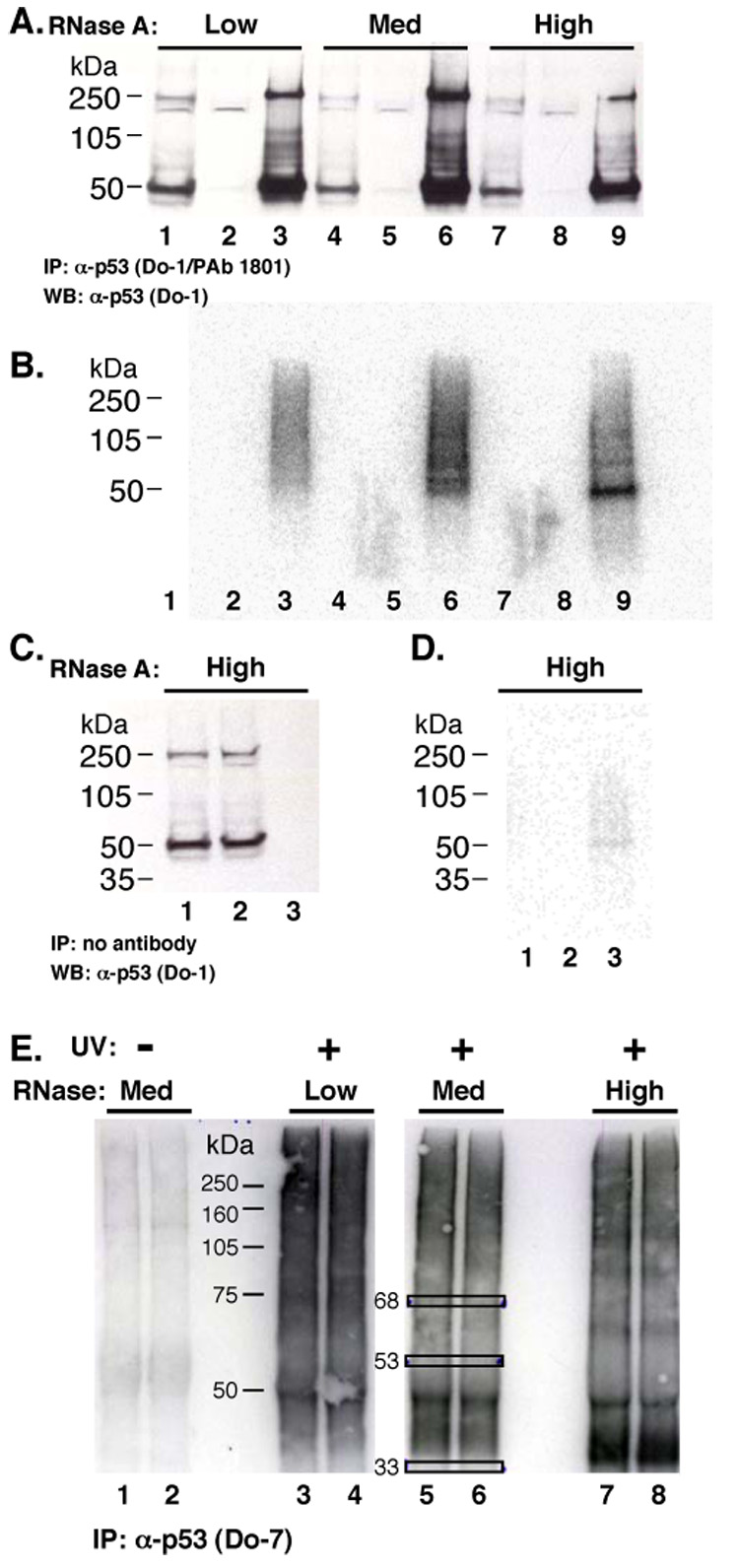
CLIP analysis of ribonucleoprotein “X”. (A) Western blot. MCF-7 cell lysate treated with RNase A. Lanes 1, 4 and 7: 10% of total WCL. Lanes 2, 5, and 8: cleared supernatant after IP with antibodies Do-1 and PAb 1801. Lanes 3, 6, and 9: IP material. (B) RNA analysis after p53 CLIP, RNAse treatment, and RNA labeling. Western blot from (A) exposed to a phosphorimager screen. (C) Mock IP without antibody, followed by processing, and Western blot. Whole cell lysate (lane 1), post-IP supernatant (lane 2) and IP sample (lane 3). (D) Phosphor image of membrane (C). (E) MCF-7 cell lysate treated with UV irradiation and RNase A, as indicated, subjected to IP by Do-7, and labeled. RNA samples of ~68, ~53, and ~33 kDa (boxes) were excised and cloned.
Fig. 3.
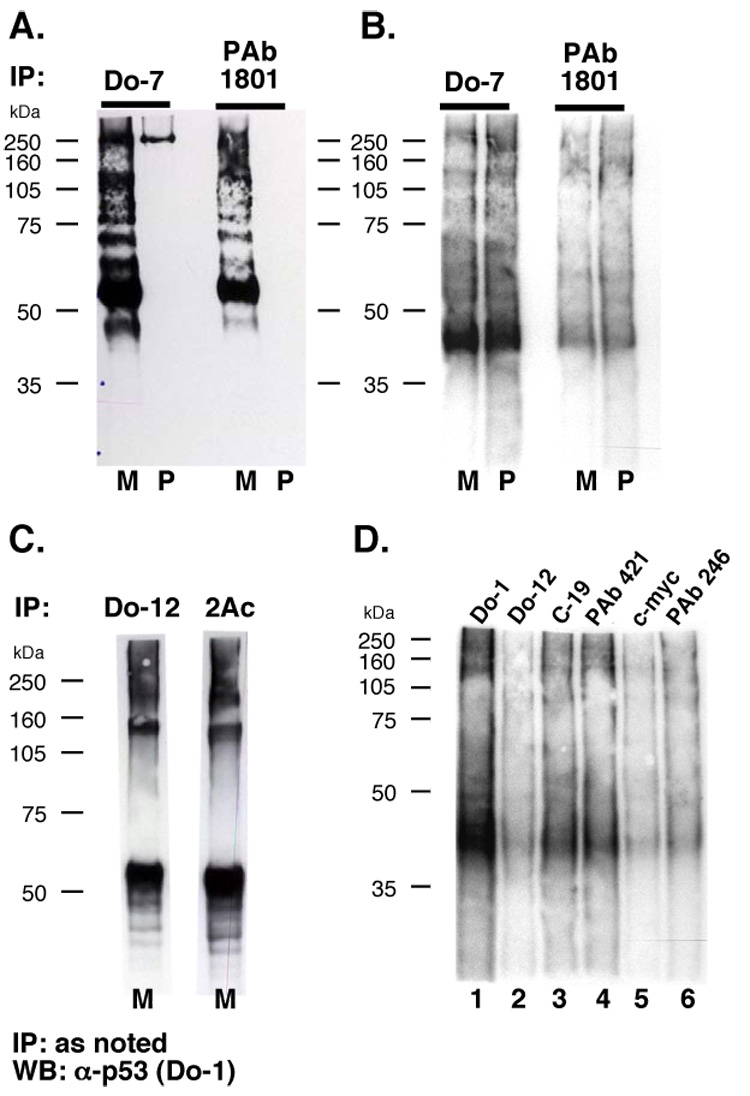
UV-cross-linked RNA is purified from p53-null cells by anti-p53 antibodies. UV-cross-linked MCF-7 (TP53 +/+) cell lysate (M) or PC-3 (tp53 −/−) cell lysate (P) treated with RNase A and cross-linked RNA fragments were labeled before IP. (A) Western detection of p53. (B) Membrane from (A) exposed to film to visualize labeled RNA. (C) CLIP with anti-p53 antibodies. MCF-7 cell lysate treated with RNase A and subjected to IP with the indicated antibodies and Western detection of p53. (D) Radiolabeled RNA IP from p53-null PC-3 cells.
Strikingly, the yield of radiolabeled RNA was not p53-dependent, being similar after immunoprecipitation of UV-cross-linked extract from TP53 +/+ and tp53 −/− cell lines with either antibody (Fig. 3B, compare lanes M and P). In addition, the migration of the major radiolabeled ribonucleoprotein species suggested a molecular weight lower than p53 (Fig. 3A–B, ~41 kDa vs. ~53 kDa). The p53-independent RNA recovery was consistent with the data in Fig. 2. We conclude that at least one ribonucleoprotein (not p53) is detected by anti-p53 antibodies in these experiments. This ribonucleoprotein “X” is evidently co-immunoprecipitated by antibodies of the IgG class, in a manner independent of their epitope specificity.
Prior in vitro experiments suggested that C-terminally acetylated p53 is incapable of interacting with RNA [7]. We assayed the relative amount of acetylated p53 in MCF-7 cells (Fig. 3C). We compared p53 immunoprecipitation by Do-12, an antibody with an internal p53 epitope in a non-acetylated region of the protein (Fig. 1), to immunoprecipitation by 2Ac, an antibody that specifically recognizes p53 acetylation on K373 and K382. Comparison of the yield of these immunoprecipitation reactions showed that the majority of p53 in MCF-7 cells has at least one C-terminal acetylated lysine (Fig. 3C).
To explore the basis for ribonucleoprotein “X” immunoprecipitation by anti-p53 antibodies, we applied the CLIP protocol to p53-null PC-3 cells (Fig. 3D). Several anti-p53 antibodies were tested (Fig. 1A). Like Do-7 and PAb 1801, Do-1 binds an N-terminal p53 epitope, Do-12 binds an internal epitope (residues 256–270) [29], and C-19 and PAb 421 (C-terminal epitopes) were included [30]. Antibodies against mouse p53 and unrelated proteins (c-myc and influenza HA) were tested. Remarkably, although no p53 protein can be immunoprecipitated from these p53-null cells (Fig. 3A), the pattern of RNA immunoprecipitation was comparable for all antibodies (Fig. 3D). This result shows that ribonucleoprotein “X” is a ubiquitous contaminant of immunoprecipitations including antibodies not specific for p53 (Fig. 3D). Because ribonucleoprotein “X” is not purified by beads alone (Fig. 4C–D), these results suggest that mouse IgG2b antibody constant region most strongly immunoprecipitates ribonucleoprotein “X”.
CLIP allows cloning of RNA sequences [31]. We cloned RNA tags from ribonucleoprotein “X” to determine what RNAs might be misinterpreted as p53 partners. An RNase A titration was performed to optimize RNA tag length (Fig. 4 A–B; [22]). “Low,” “medium,” and “high” RNase A treatments were employed (Fig. 4 A–B). At the highest RNase A treatment, the radiolabeled ribonucleoprotein migrated in a manner consistent with a molecular weight smaller than 50 kDa (Fig. 4B, lane 9). Panels C and D of Fig. 4 show results of a control experiment, where an antibody-free mock immunoprecipitation after high RNase A treatment (Fig. 4C) yielded little RNA (Fig. 4D, lane 3).
The CLIP protocol was applied to ribonucleoprotein “X” as shown in Fig. 4E. RNA recovery was dependent upon UV-cross-linking (Fig. 4E, compare lane 1 with 2–4). The indicated bands (Fig. 4E, 9 boxes) were excised and RNA tags were cloned. Nineteen sequence tags from 45 identified RNAs are listed in Table 1. No RNA sequences were obtained from the negative control sample at 33 kDa (Fig. 4E, lane 3). All of the tags were human sequences. Seven of the cloned tags could have originated from multiple human chromosomes (not shown). Excluded tags (19/45) corresponded to chromosomal regions with no putative genes. All of the remaining identified tags were located in non-coding regions of pre-mRNAs, with the majority residing near the center of introns. One tag mapped to a 5’-UTR.
Table 1.
CLIP tag sequences from ribonucleoprotein “X”
| kDa1 | Tag2 | Chr.3 | Gene4 |
|---|---|---|---|
| 53 | AAGTTACCACAGGGATAACTGGCTT | 1 | HFM1 |
| 53 | AATGGTGCCTATGCACCCATC | 5 | CTNNA |
| 68 | AAAACATCAGATTGTGAATCTGACAACAGAGGCTT | 5 | PCBD2 |
| 53 | AAGCTCCTCCTTTCCTTTCC | 6 | SOD2ps |
| 53 | AAATCACTGAGCCCCTCGGAGCCTC | 7 | PODXL |
| 53 | ATGTTGGTGTTATAGGTACCAGAAAGAGCTCT | 8 | SNX16 |
| 53 | AGAAGATGAAGAGAGGCTGAGTCACT | 8 | KIAA0196 |
| 68 | AATGTCCACCTGGATCTGCCCTGT | 9 | ALG2 |
| 68 | CGAGAAAGCCCCGGCTAACTAC | 9 | Q5VZ16 |
| 53 | ATTTGGTTTTACAGAAAAACTTTGTCAACCCCCGTTCTGAGGTTCTT | 10 | CACNB2 |
| 68 | AAAGGTGAGGAAGTTGGGTTCC | 10 | OPTN |
| 68 | ACTTGTTGTGGTTAGGGCCAGTTT | 11 | FCHSD2 |
| 53 | AAGAGAGGACTAGTGATCTGAAA | 12 | CRADD |
| 53 | AAAGGATGTGCAGGACTTGTTCA | 12 | NAP1L1 |
| 68 | ATTTTGAAGGCTTGTGTGGCAAAT | 14 | PRKCH |
| 68 | ATTAGATACCCTGGTAGTCCACGCCGTAAACGATGAGTGTTAG | 15 | PNAS-20 |
| 68 | AATTTGGGCTGTGAGGGACAGAGAAT | 17 | RAI1 |
| 68 | AATTCTGTCCCTTGTGTGTCTGGCATAAGGGAGNTGAAAGT | 21 | DSCAM |
| 68 | AACGATCTGGGCGCTGTCTCAACGG | 21 | GT335 |
Molecular weight of tagged complex
Tag sequence
Chromosomal location
Gene
RNA tags associated with ribonucleoprotein “X” implicate an interesting collection of genes, any one of which could have been mistakenly interpreted as a target for direct p53 regulation (Table 1). The yield of immunoprecipitated ribonucleoprotein “X” was insufficient for protein detection or mass spectrometric identification.
We report that no RNA partners are purified with p53 from the human cell lines tested. Rather, nonspecific RNA contamination of p53 immunoprecipitations is problematic and dependent on the isotype of the immunoprecipitating antibody. This RNA contamination can easily be misinterpreted as evidence for specific p53-RNA interactions. Whereas unmodified or partially post-translationally modified p53 proteins from E. coli [19], yeast [6], insect cells [7, 13, 16–18], or reticulocyte lysate [16–18] are prone to nonspecific RNA binding, p53 protein is not complexed with RNA when purified from the tested human cells. Perhaps p53 post-translational modifications prevent p53-RNA interactions in vivo. Together with other recent results [6, 7], this work illuminates a historically mysterious aspect of p53 biology. The physiological significance of RNA binding by p53 remains obscure [4].
Acknowledgements
We thank R. Darnell, A. Mele, J. Ule, N. Becker, D. Smith, F. Couch, I. Ivanovic, and F. Secreto. Supported by the Mayo Foundation and NIH Grant GM68128.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 2.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 3.Baroni TE, Wang T, Qian H, Dearth LR, Truong LN, Zeng J, Denes AE, Chen SW, Brachmann RK. A global suppressor motif for p53 cancer mutants. Proc. Natl. Acad. Sci. U S A. 2004;101:4930–4935. doi: 10.1073/pnas.0401162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley J-K, Maher LJ., 3rd p53-RNA interactions: new clues in an old mystery. RNA. 2007 doi: 10.1261/rna.673407. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall KB. RNA-protein interactions. Curr Opin Struct Biol. 2002;12:283–288. doi: 10.1016/s0959-440x(02)00323-8. [DOI] [PubMed] [Google Scholar]
- 6.Riley KJ, Cassiday LA, Kumar A, Maher LJ., 3rd Recognition of RNA by the p53 tumor suppressor protein in the yeast three-hybrid system. RNA. 2006;12:620–630. doi: 10.1261/rna.2286706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley KJ, Ramirez-Alvarado M, Maher LJ., 3rd RNA-p53 interactions in vitro. Biochemistry. 2007;46:2480–2487. doi: 10.1021/bi061480v. [DOI] [PubMed] [Google Scholar]
- 8.Friedler A, Veprintsev DB, Freund SM, von Glos KI, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure (Camb.) 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J. Mol. Biol. 1997;266:677–687. doi: 10.1006/jmbi.1996.0813. [DOI] [PubMed] [Google Scholar]
- 10.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 11.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. Disordered RNA chaperone proteins: from functions to disease. Cell Mol Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontoura BM, Sorokina EA, David E, Carroll RB. p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samad A, Carroll RB. The tumor suppressor p53 is bound to RNA by a stable covalent linkage. Mol Cell Biol. 1991;11:1598–1606. doi: 10.1128/mcb.11.3.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller SJ, Suthiphongchai T, Zambetti GP, Ewen ME. p53 binds selectively to the 5' untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol. 2000;20:8420–8431. doi: 10.1128/mcb.20.22.8420-8431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galy B, Creancier L, Prado-Lourenco L, Prats AC, Prats H. p53 directs conformational change and translation initiation blockade of human fibroblast growth factor 2 mRNA. Oncogene. 2001;20:4613–4620. doi: 10.1038/sj.onc.1204630. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Yoshida T, Itoh H, Kohno K. Binding of RNA to p53 regulates its oligomerization and DNA-binding activity. Oncogene. 2004;23:4371–4379. doi: 10.1038/sj.onc.1207583. [DOI] [PubMed] [Google Scholar]
- 20.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. Rna. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 22.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou YH, Chung PH, Sun TP, Shieh SY. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell. 2005;16:1684–1695. doi: 10.1091/mbc.E04-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergamaschi D, Samuels Y, O'Neil NJ, Trigiante G, Crook T, Hsieh JK, O'Connor DJ, Zhong S, Campargue I, Tomlinson ML, Kuwabara PE, Lu X. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet. 2003;33:162–167. doi: 10.1038/ng1070. [DOI] [PubMed] [Google Scholar]
- 26.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 28.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 30.Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Biol. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]


