Abstract
Prenatal nutritional constraint induces an altered metabolic phenotype in the offspring which in humans confers an increased risk of non-communicable disease. Feeding a protein-restricted (PR) diet to pregnant rats causes hypomethylation of specific gene promoters in the offspring and alters phenotype. We investigated how altered epigenetic regulation of the hepatic glucocorticoid receptor (GR) 110 promoter is induced in the offspring. Rats were fed a control (180g casein/kg) or a PR (90g casein/kg) diet throughout pregnancy, and chow during lactation. Offspring were killed at postnatal day 34 (n 5 per maternal dietary group). Methylation-sensitive PCR showed GR110 promoter methylation was 33% lower (P<0.001) and GR expression 84% higher (P<0.05) in the PR offspring. RT PCR showed DNA methyltransferase-1 (Dnmt1) expression was 17% lower (P<0.05) in PR offspring, while Dnmt3a/b and methyl domain binding protein-2 expression was not altered. Thus hypomethylation of the GR110 promoter may result from lower capacity to methylate hemimethylated DNA during mitosis. Histone modifications which facilitate transcription were increased at the GR110 promoter (147 to 921%, P<0.001), while those that suppress methylation were decreased (54%, P<0.01) or similar to controls. In human umbilical cord (n 15), there was a 2-fold difference between the highest and lowest level of GR1-CTotal promoter methylation. Dnmt1, but not Dnmt3a, expression predicted 49% (P = 0.003) of the variation in GR1-CTotal promoter methylation. These findings suggest that induction in the offspring of altered epigenetic regulation of the hepatic GR110 promoter, and hence metabolic phenotype, may be due to reduced Dnmt1 expression.
Keywords: Fetal programming, epigenetic, rat, glucocorticoid receptor, histones, human
Introduction
There is an increasing awareness that aspects of the prenatal environment such as maternal nutrition and stress levels provide cues that alter the phenotype of the fetus without overt reductions in fetal growth (Bateson et al. 2004). Such nutritional cues may operate within the normal range for the human population and contribute to the early origins of risk of chronic diseases such as the metabolic syndrome and cardiovascular disease (Godfrey & Barker, 2001). In rats, variations in phenotype can be induced by maternal under-nutrition (Langley & Jackson, 1994; Vickers et al. 2005) or increased intake of specific nutrients (Armitage et al. 2005). As in humans (Ravelli et al. 1998), the phenotype which is induced is dependent upon the timing of nutrient restriction during pregnancy or lactation (Remacle et al. 2004).
Induced changes to the phenotype which persist throughout the life-span are likely to involve stable alterations to the expression of the genome. The offspring of rats fed a diet with a moderate reduction in protein content (protein-restricted (PR) diet) during pregnancy show tissue-specific alterations in the expression of transcription factors which regulate a wide range of developmental and metabolic processes, specifically the glucocorticoid receptor (GR) (Bertram et al. 2001; Lillycrop et al. 2005) and peroxisomal proliferator-activated receptors (PPARs) (Burdge et al. 2004; Lillycrop et al. 2005), and changes to the expression of genes associated with fatty acid metabolism (Maloney et al. 2003; Lillycrop et al. 2005) and carbohydrate homeostasis (Burns et al. 1997; Desai et al. 1997). Little is known about how information about the availability of nutrients in the extra-uterine environment is transmitted to the offspring or how different phenotypes are induced.
The methylation of CpG dinucleotides clustered at the 5′ promoter regions of genes established during early life confers stable silencing of transcription and is critical for cell differentiation (Bird, 2001). Following fertilisation, maternal and paternal genomes undergo extensive demethylation followed by de novo methylation by the activities of DNA methyltransferases (Dnmt) 3a and 3b around the time of implantation (Bird, 2001; Reik et al. 2001). Patterns of DNA methylation are maintained through mitosis by Dnmt1 activity (Bird, 2001). Activities of Dnmt1 and DNMT3a are modified by folic acid and homocysteine (Hcyst) (James et al. 2002; Ghoshal et al. 2006). The timing of gene silencing during early development differs between genes and tissues (Grainger et al. 1983; Benvenisty et al. 1985; Gidekel et al. 2002; Hershko et al. 2003). In addition, the phenotype of an embryo can be modified by manipulation of Dnmt1 expression, and hence maintenance of patterns of DNA methylation (Biniszkiewicz et al. 2002; Stancheva & Meehan, 2000; Stancheva et al. 2001).
DNA methylation can induce transcriptional silencing either by blocking transcription factor binding and/or through the methyl CpG binding protein (MeCP2) that binds to methylated cytosines and which, in turn, recruits the histone deacetylase / histone methyl transferase (HDAC / HMT) complex to the DNA (Fuks et al. 2003). Covalent modifications to histones, such as acetylation and methylation of specific lysine residues in the N-terminal regions of histones, influence chromatin structure and hence the ability of the basal transcriptional machinery to gain access to the DNA (Turner, 2000; Strahl et al. 1999; Lachner et al. 2001; Zegerman et al. 2002; Litt et al. 2001; Nakayama et al. 2001).
Since epigenetic regulation of gene promoters which is established during development and is retained throughout the lifespan of the organism confers patterns of transcriptional expression and silencing, perturbations to such processes represent one possible molecular mechanism for induction of an altered phenotype. Feeding a PR diet to rats during pregnancy induces hypomethylation and increased expression of the GR and PPARα promoters in the liver of the adult offspring (Lillycrop et al. 2005), but was prevented by supplementation of the PR diet with folic acid. Supplementation of the PR diet with glycine or folic acid prevented induction of an altered phenotype (Jackson et al. 2002; Torrens et al. 2006). Thus 1-carbon metabolism is central to the induction of an altered phenotype in this model, which is consistent with the transient increase in plasma Hcyst, a marker of impaired 1-carbon metabolism, in early pregnancy when rats were fed a PR diet (Petrie et al. 2002).
We have tested the hypothesis that the transmission to the fetus of information regarding maternal nutrition and induction of altered DNA methylation involves modulation of Dnmt action. We investigated the effect of altered maternal protein intake during pregnancy on the epigenetic regulation of the hepatic GR promoter in the adult offspring. Specifically, we determined the effect of feeding a PR diet to pregnant rats on the methylation status and the level of histone modification at the hepatic GR promoter. In order to determine the mechanism which modifies the epigenetic regulation of the GR promoter, we investigated whether prenatal under-nutrition alters the expression of enzymes that catalyse DNA methylation de novo, methylation of hemimethylated DNA or active demethylation of DNA. As a result of our findings, we also investigated the relationship between the expression of Dnmts and the methylation of the GR promoter that is expressed in human umbilical cord (UC).
Materials and Methods
Animal procedures
Virgin Wistar rats (n 5 per dietary group) were fed isocaloric diets containing either 180g / kg casein and 1mg / kg folic acid (control), 90 g / kg casein and 1mg / kg folic acid (protein-restricted, PR). In some experiments, livers from the offspring of rats fed 90g / kg casein and 5 mg / kg folic acid were studied (Lillycrop et al. 2005) (Table 1). Dams were fed standard chow (AIN 76A) from delivery (Lillycrop et al. 2005). Litters were reduced to 8 at birth, equal numbers of males and females, and offspring were weaned onto standard chow (AIN 76A) at 28 days and were killed 6 days later. Livers were excised immediately, frozen in liquid nitrogen and stored at −80°C. One liver from each litter was selected for analysis, male to female 3:2. All animal procedures were conducted in accordance with the UK Home Office Animals (Scientific Procedures) Act (1986).
Table 1.
Composition of the diets fed during pregnancy, lactation and after weaning
| Diets fed during pregnancy | Diet fed to dams during lactation and to offspring after weaning | |||
|---|---|---|---|---|
| Control | Protein-restricted | Protein restricted, supplemented with folic acid | AIN-76A | |
| Casein (g/kg) | 180 | 90 | 90 | 200 |
| Folic acid (mg/kg) | 1 | 1 | 5 | 2 |
| Cornstarch (g/kg) | 425 | 482 | 482 | 150 |
| Sucrose (g/kg) | 213 | 243 | 243 | 500 |
| Choline chloride (g/kg) | 2 | 2 | 2 | 2 |
| DL-methionine (g/kg) | 5 | 5 | 5 | 3 |
| Vitamin mix1 (g/kg) | 5 | 5 | 5 | 5 |
| Mineral mix2 (g/kg) | 20 | 20 | 20 | 20 |
| Cellulose (g/kg) | 50 | 50 | 50 | 50 |
| Corn oil (g/kg) | 100 | 100 | 100 | 50 |
| Total metabolisable energy (MJ/kg) | 20.2 | 19.9 | 19.9 | 15.5 |
Vitamin mix: Thiamine hydrochloride 2.4 mg/kg; riboflavin 2.4 mg/kg; pyridoxine hydrochloride 2.8 mg/kg; nicotinic acid 12.0 mg/kg; D-calcium pantothenate 6.4 mg/kg; biotin 0.01 mg/kg; cyanocobalbumin 0.003 mg/kg; retinyl palmitate 6.4 mg/kg; DL-α-tocopherol acetate 79.9 mg/kg; cholecalciferol 1.0 g/kg; menaquinone 0.02 mg/kg.
Mineral mix: Calcium phosphate dibasic 11.3 g/kg; sodium chloride 1.7 g/kg; potassium citrate monohydrate 5.0 g/kg; potassium sulphate 1.2 g/kg; magnesium sulphate 0.5 g/kg; magnesium carbonate 0.1 g/kg; ferric citrate 0.1 g/kg; zinc carbonate 36.2 mg/kg; cupric carbonate 6.8 mg/kg; potassium iodate 0.2 mg/kg; sodium selenite 0.2 mg/kg; chromium potassium sulphate 12.5 mg/kg.
Measurement of DNA methylation of the GR10 promoter in rat
DNA methylation was carried out essentially as described (Lillycrop et al. 2005). Genomic DNA (5μg), isolated from the livers of rats (n 5 per maternal dietary group) was incubated with the methylation sensitive restriction endonucleases AciI and HpaII, as instructed by the manufacturer (New England Biolabs, Hitchin, Hertfordshire, UK). The resulting DNA was amplified using real time PCR, which was performed in a total volume of 25μl with SYBR® Green Jumpstart Ready Mix (Sigma, Poole, Dorset, UK) as described by the manufacturer. Cycle parameters were 55°C for 5 minutes, 95°C for 10 minutes then, 40 cycles of 95°C for 30s, 60°C for 1 minute and 72°C for 1 minute. Single bands of the correct size were verified by gel electrophoresis. Primers were designed to amplify the CpG island spanning the GR110 promoter used in rat liver (McCormick et al. 2000). Primer sequences are listed in table 2. The promoter region from the rat PPARγ2 promoter which does not contain AciI or HpaII cleavage sites was used as an internal control (Lillycrop et al. 2005). All Ct values were normalised to the internal control and each sample analysed in duplicate.
Table 2.
PCR primers for analysis of mRNA expression and promoter methylation
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Methylation-sensitive PCR | ||
| GR 110 | TCCTCCATTTTTGCGAGCTC | CCACCGCAGCCAGATAAAC |
| PPAR-γ2 | GTCTCTGCTCTGGTAATTC | AAGGCTTGTGGTCATTGAG |
| hGR1-CTotal | ATTTTGCGAGCTCGTGTCTG | CGCAGCCGAGATAAACAACT |
| hPPAR-α | CGGAGTTTATGAGGCCATATTC | AGGGAGATATCACTGTCATCCAG |
| mRNA expression | ||
| GR | TGACTTCCTTCTCCGTGACA | GGAGAATCCTCTGCTGCTTG |
| Dnmt 1 | QIAGEN Quantitect® primer assay QT00493577 | |
| Dnmt 3a | QIAGEN Quantitect® primer assay QT00429380 | |
| Dnmt 3b | QIAGEN Quantitect® primer assay QT00436002 | |
| MeCP2 | CAGCTCCAACAGGATTCCATGGT | TGATGTCTCTGCTTTGCCTGCCT |
| MDB2 | GGCAAGAGCGATGTCTACTA | CTGGACCGACTCCTTGAAGA |
| Cyclophilin | TTGGGTCGCGTCTGCTTCGA | GCCAGGACCTGTATGCTTCA |
| hGR 1-CTotal | GCTCCTCTGCCAGAGTTGAT | CAGTGGATGCTGAACTCTTGG |
Dnmt1, DNA methyltransferase-1; Dnmt3a, DNA methyltransferase-3a; Dnmt3b, DNA methyltransferase-3b; GR110, glucocorticoid receptor 110 promoter, hGR1-CTotal, human GR1-CTotal promoter; hPPARα, human peroxisomal proliferator-activated receptor-α; MeCP2; methyl cytosine binding protein-2; MDB2; methyl binding domain binding protein-2.
Measurement of mRNA expression in rat
Total RNA was isolated from liver (n 5 per maternal dietary group) samples using Tri® Reagent (Sigma) as described by the manufacturers. cDNA was prepared as described (Lillycrop et al. 2005) and amplified using real time PCR (Harris et al. 2002) which was performed in a total volume of 25μl with SYBR® Green Jumpstart Ready Mix (Sigma) as described by the manufacturer. PCR primers are listed in table 2. Samples were analyzed in duplicate and the expression of GR110 promoter transcript, Dnmt1, Dnmt3a, Dnmt3b, MeCP2 and methyl binding domain binding protein (MBD)-2 was normalised to the housekeeping gene cyclophilin (Bustin, 2000). Dnmt1, 3a and 3b were measured using primer kits from QIAGEN Ltd UK (Crawley, Sussex, UK). Cycle parameters were 55°C for 5 minutes, 95°C for 10 minutes then, 40 cycles of 95°C for 30s, 60°C (cyclophilin, phoephoenolpyruvate carboxykinase (PEPCK), Dnmt1, Dnmt 3a, Dnmt3b, MeCP2 and MBD2) or 65°C (GR) for 1 minute and 72°C for 1 minute. Single bands of the correct size were verified by gel electrophoresis (data not shown).
Measurement of histone acetylation and methylation at the GR promoter in rat
Histone modification, and MeCP2 and Dnmt1 binding at the GR110 promoter was analysed by ChIP assay (Boyd & Farnham, 1999). 100mg of liver tissue (n 5 per maternal dietary group, 1 per litter) was ground in liquid nitrogen and fixed with formaldehyde (1% v/v) for 10 minutes. The chromatin was sonicated to yield DNA fragments of 100-400bp in length. The sonicated chromatin was quantified on the basis of DNA content at A260nm. The chromatin equivalent of 40μg DNA was used in each immunoprecipitation. The sonicated supernatant was diluted 10-fold in ChIP dilution buffer (0.01% (w/v) SDS, 1.1% (v/v) Triton X-100, 1.2mM EDTA, 16.7mM NaCl, 20mM Tris-HCl pH 8.1) and pre-cleared with salmon sperm DNA/protein A agarose (50% (w/v) slurry). Pre-cleared chromatin was then incubated overnight with 2μg of antibodyat 4°C. Anti β-galactosidase (c-20) antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, California, USA). Anti-acetyl Histone H3 (06-599), anti-acetyl H4 (06-866), di-methylated H3 K4 (07-030), di-methylated H3 K9 (07-441) from Upstate Biotechnology (Dundee, Scotland) and tri-methylated H3K9 (ab8898), Dnmt1 (ab5208) and MeCP2 (ab3752) from Abcam (Cambridge, UK). The immunocomplexes were collected by the addition of salmon sperm DNA/protein A agarose slurry, washed with 0.1% (w/v) SDS, 1% (v/v) Triton X-100, 2 mM EDTA, Tris-HCl pH8.1, 150 mM NaCl, Tris-HCl pH 8.1 and then with 0.1% (w/v) SDS, 1% (v/v)Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl pH 8.1 followed by 0.25 M LiCl, 1% (v/v) NP-40, 1% (w/v) sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.1 and twice with 10 mM Tris-HCl pH 7.5, 1mM EDTA). DNA was eluted by the addition of a solution of 1% (w/v) SDS and 0.1M NaHCO3, the cross-links reversed, and the DNA purified. The ChIP precipitated DNA and the input DNA were subjected to real-time PCR using SYBR® Green Jumpstart Ready Mix (Sigma). The PCR primers used for the GR110 promoter were 5′ CGTCTTGTTCCACCCACT 3′ and 5′ CCTTGCAGTTGCCGACAG. All values were normalised with respect to the input DNA and expressed as a percent of the control group. To demonstrate the specificity of the immunoprecipitation reactions an antibody directed against β-galactodiase and a negative antibody control were carried out in each experiment (data not shown).
Human umbilical cord samples
These studies utilised samples of UC from a stratified random sample of 15 term infants in the Birthright Fetal Growth Rates Study. This study of nutrition during pregnancy and fetal growth recruited Caucasian women with singleton pregnancies and known menstrual dates who registered in early pregnancy with participating obstetricians at the Princess Anne Maternity Hospital, Southampton UK (Godfrey et al. 2001). Samples were stored at −80°C. Birthweights of the infants were in the normal range (range 2665 to 4430 g, mean 3426 g). Collection and analysis of human UC samples was carried out with written informed consent from all subjects and under IRB approval from the Southampton and South West Hampshire Joint Research Ethics Committee.
DNA methyltransferase mRNA expression in human umbilical cord
Total RNA was isolated from human UC using Tri® Reagent (Sigma, Poole, Dorset, UK) as described by the manufacturers. cDNA was prepared as described (Lillycrop et al. 2005) and amplified using real-time RT PCR, which was performed in a total volume of 25μl with SYBR® Green Jumpstart Ready Mix (Sigma) as described by the manufacturer. Human Dnmt1 shows 80% identity to rat Dnmt1 and human Dnmt3a shows 91% identity to rat Dnmt3a. There was 100% identity between rat and human Dnmt1 and rat and human Dnmt3a within the primer sequences. Therefore, we used the same RT PCR primers to measure Dnmt1 and Dnmt3a expression in human UC and rat liver. PCR primers are listed in table 2. Samples were analyzed in duplicate and the expression of Dnmt1 and Dnmt3a was normalised to the housekeeping gene cyclophilin (Bustin, 2000). Cycle parameters were 94°C for 2 minutes then, 40 cycles of 95°C for 30s, 60°C (cyclophilin and Dnmt1) or 62°C (Dnmt3a) for 1 minute and 72°C for 1 minute.
Measurement of DNA methylation of the GR 1-CTotal promoter in human umbilical cord
To confirm that GR1-CTotal promoter is expressed in human umbilical cord, total RNA was isolated from UC and human blood using Tri® Reagent (Sigma) as described by the manufacturers. The size of the GR1-CTotal promoter RT PCR transcript from UC on agarose gel electrophoresis was compared to the blood reference transcript (Turner & Muller, 2005) (Figure 4 A). PCR primers are listed in table 2. Cycle parameters were 94°C for 2 minutes then, 40 cycles of 95°C for 30s, 62°C for 1 minute and 72°C for 1 minute.
For analysis of GR1-CTotal promoter methylation, genomic DNA (400ng) was incubated with the methylation sensitive restriction endonucleases AciI and HpaII as instructed by the manufacturer (New England Biolabs, Hitchin, Hertfordshire, UK). The resulting DNA was amplified using real time PCR, which was performed in a total volume of 25μl with SYBR® Green Jumpstart Ready Mix (Sigma) as described by the manufacturer. A fragment of the human PPARα exon 7 which does not contain AciI or HpaII cleavage sites was used as an internal control gene. Primers were designed to amplify the CpG island spanning the GR1-CTotal promoter (Turner & Muller, 2005). Primer sequences are listed in table 2. Cycle parameters were 94°C for 2 minutes then, 40 cycles of 95°C for 30s, 59.3°C (GR 1-CTotal) or 66°C (PPARα) for 1 minute and 72°C for 1 minute. All Ct values were normalised to the internal control and each sample analysed in duplicate. Single bands of the correct size were verified by gel electrophoresis (not shown).
Statistical analysis
Data are presented as mean (SEM). Statistical comparisons were by Student’s unpaired t-test or 1-way ANOVA with Bonferroni’s post hoc analysis as indicated in the text. The relationship between DNA methylation and gene expression in human UC was determined by calculation of Pearson’s correlation coefficient.
Results
GR promoter methylation, and expression of GR and PEPCK in rat liver
Hepatic GR110 promoter methylation was 33% lower (Table 3) at postnatal day 34 in the offspring of dams fed a PR diet compared with offspring of control group. Hypomethylation of the GR110 promoter was associated with higher mRNA expression of GR110 (84%). The expression of the GR target gene PEPCK was also increased by 16% in the offspring of the PR group (Table 3).
Table 3.
Glucocorticoid receptor promoter methylation, and glucocorticoid receptor and phosphoenolpyruvate carboxykinase mRNA expression in the liver of day 34 offspring
| Relative to the control group (%) | |||||
|---|---|---|---|---|---|
| Control | PR | ||||
| Mean | SEM | Mean | SEM | P | |
| GR110 methylation | 100.0 | 3.5 | 67.3 | 1.5 | <0.001 |
| GR110 mRNA expression | 100.0 | 22.0 | 184.0 | 11.0 | <0.05 |
| PEPCK mRNA expression | 100.0 | 2.3 | 116.0 | 4.0 | <0.05 |
Values are mean (SEM) results of RT PCR analysis for n 5 samples/maternal dietary group. Statistical comparison was by Student’s unpaired t-test.
GR110, glucocorticoid receptor 110 promoter; PEPCK, phosphoenolpyruvate carboxykinase.
MeCP2 expression in rat liver
Having shown GR hypomethylation in PR offspring, we then investigated whether the expression and recruitment of MeCP2 to the GR110 promoter was also altered in response to maternal diet. MeCP2 mRNA expression was 29% lower in the liver of the PR offspring versus controls (Table 4). Binding of MeCP2 to the GR promoter in the liver of control and PR offspring was assessed using a chromatin immunoprecipitation (ChIP) assay. Binding of MeCP2 to the GR1 10 promoter was reduced by 43% (P<0.05) in PR offspring compared to control offspring (Table 4). No signal was obtained when chromatin was precipitated with an anti- β-galactosidase antibody or with a negative antibody control. This demonstrates that the immunoprecipitation reactions were specific.
Table 4.
MeCP2 mRNA expression, and binding of MeCP2 and histone modifications at the glucocorticoid receptor promoter
| Relative to the control group (%) | |||||
|---|---|---|---|---|---|
| Control | PR | ||||
| Mean | SEM | Mean | SEM | P | |
| MeCP2 mRNA expression and binding | |||||
| MeCP2 mRNA expression | 100.0 | 9.3 | 71.4 | 7.3 | <0.05 |
| MeCP2 binding | 100 | 2 | 57 | 2 | <0.05 |
| Histone modifications | |||||
| H3K9 acetylation | 100 | 12 | 274 | 20 | <0.0001 |
| H4K9 acetylation | 100 | 18 | 402 | 33 | <0.0001 |
| H3K9 methylation | 100 | 20 | 1025 | 87 | <0.0001 |
| H3K9 dimethylation | 100 | 22 | 19 | 16 | <0.001 |
| H3K9 trimethylation | 100 | 60 | 103 | 11 | NS |
Values are mean (SEM) for n 5 samples/maternal dietary group. Statistical comparison was by Student’s unpaired t-test. Expression of methyl cytosine binding protein (MeCP)-2 expression and binding of MeCP2 at the glucocorticoid receptor 110 promoter in liver from 34 day-old offspring of rats fed either a control or protein-restricted (PR) diet during pregnancy. Binding of MeCP2 and the extent of histone modifications at the GR110 promoter were determined by chromatin immunoprecipitation assay
Analysis of histone modifications at the hepatic GR110 promoter in rats
In order to determine whether the hypomethylation of the GR110 promoter, and reduced expression and binding of MeCP2 were associated with altered covalent modifications of histones bound to the GR promoter, ChIP assays were used to measure the acetylation and methylation of specific histone lysine residues at the GR110 promoter. The level of histone modifications which facilitate transcription was higher at the hepatic GR110 promoter in the PR versus control offspring, namely, acetylation of H3K9 (174%) and H4K9 (302%), and methylation of H3K4 (925%) (all P<0.001) (Table 4). Di-methylation and tri-methylation of H3K9, which are associated with suppression of transcription, were 54% lower (P<0.01) or not statistically significantly different, respectively, in the PR offspring versus controls at the GR110 promoter (Table 4).
Analysis of the expression of genes which regulate DNA methylation in the liver of rats
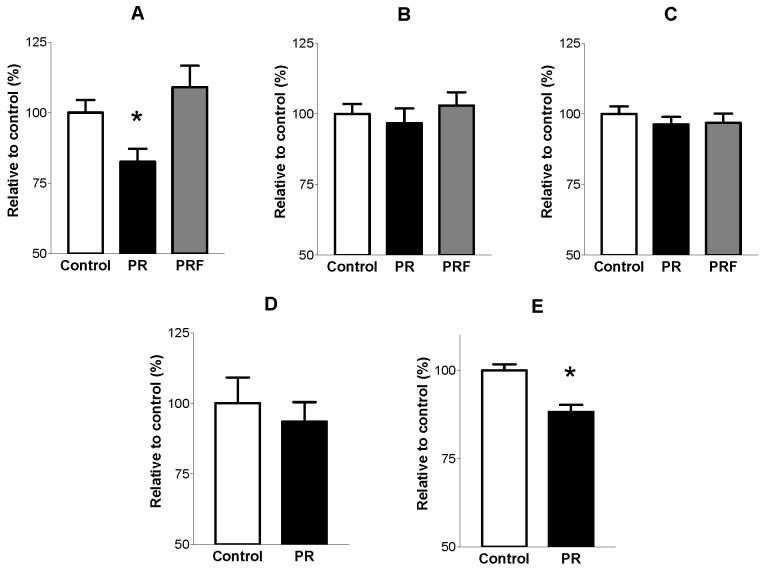
There are three possible mechanisms underlying hypomethylation of the GR110 promoter: a) impaired methylation de novo by Dnmt 3a and 3b, b) failure to maintain CpG methylation through mitosis by Dnmt1 or c) active demethylation by the putative DNA demethylase MBD2 (Detich et al. 2002). To examine which of these mechanisms may be responsible for the hypomethylation of GR, the expression of Dnmt1, Dnmt3a/b and the demethylase MBD2 were measured using real time RT-PCR. Expression of Dnmt1 mRNA was 17% lower (P<0.05) in PR versus control offspring (Figure 1 A). There were no significant differences between PR and control offspring in the expression of Dnmt 3a and 3b, or MBD2 (Figure 1 B-D). ChIP assays using an anti-Dnmt1 antibody showed that binding of Dnmt1 at the GR110 promoter was significantly lower (12%, P<0.05) in the PR offspring compared to controls (Figure 1 E).
Fig. 1.
mRNA expression of DNA methyltransferases (Dnmt (s)) in liver from 34 day-old offspring of rats fed either a control or protein-restricted (PR), or the PR diet supplemented with folic acid (PRF) diet during pregnancy. Data from RT PCR analysis are mean (SEM) relative to the control group (n 5 per maternal dietary group). (A) Dnmt1 expression. (B) Dnmt3a expression. (C) Dnmt3b expression. Statistical comparisons were by 1-Way ANOVA with Bonferroni’s post hoc analysis. 1-Way ANOVA showed a significant difference (P<0.05) between groups for Dnmt1, but not for the other DNA methyltransferases. *P<0.05 compared to the control group. (D) Expression of the methyl binding domain protein-2 from 34 day-old offspring of rats fed either a control or PR diet. Data are mean (SEM) relative to the control group (n 5 per maternal dietary group). Statistical analysis was by Student’s unpaired t-test. (E) Binding of Dnmt1 at the glucocorticoid receptor 110 promoter in liver from 34 day-old offspring of rats fed either a control or PR diet measured by chromatin immunoprecipitation assay. Data are mean (SEM) relative to the control group (n 5 per maternal dietary group). Statistical analysis was by Student’s unpaired t-test. *P < 0.05 compared to the control group.
Since 1-carbon metabolism appears to be involved centrally for the induction of an altered phenotype by variations in maternal protein intake during pregnancy (Petrie et al. 2002; Torrens et al. 2004), and hypomethylation of the hepatic GR is prevented by supplementation of the PR diet with folic acid (Lillycrop et al. 2005) we next investigated the effect of supplementation of the PR diet with 5-fold more folic acid than was present in the control or PR diet. Supplementing the PR diet with folic acid prevented reduced Dnmt1 expression compared to the control group (Figure 1 A), but did not alter Dnmt 3a or 3b expression (Figures 1 B, C).
Analysis of GR promoter methylation and Dnmt1 expression in human umbilical cord
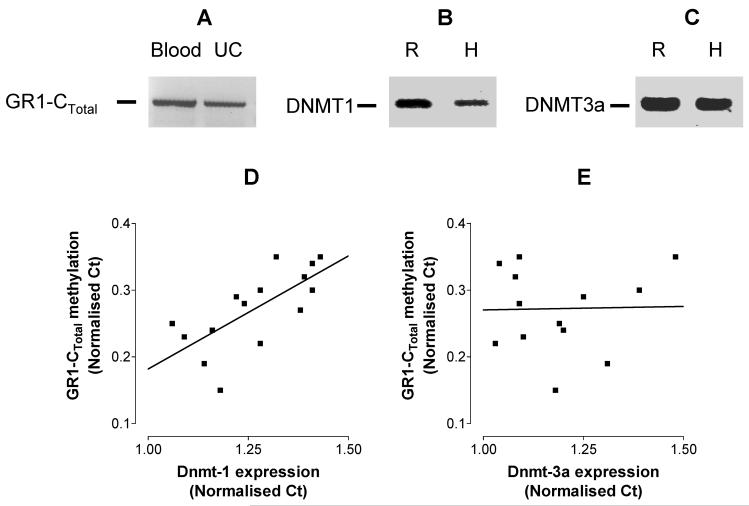
Since Dnmt1 mRNA expression appeared to be related to the level of methylation of the hepatic GR promoter, we investigated whether Dnmt1 expression was also related to the level of GR methylation in fetal human tissue. The human GR1-CTotal promoter, like the rat GR110 promoter, contains a CpG island which spans the 5′ untranslated region of the gene and which contains 11 different heterogeneous non-coding first exons (Turner & Muller, 2005). In the human fetal UC samples, we examined the methylation status of the GR1-CTotal promoter, which shows 70.6% homology with the rat GR110 promoter (Turner & Muller, 2005). We showed that transcripts from the GR1-CTotal promoter are expressed in human UC (Figure 2 A). Using a methylation sensitive restriction enzyme PCR assay we found that there was variation in the level of methylation of the GR 1Ctotal promoter between human fetal cord samples such that the highest level of methylation was approximately 2-fold greater than the lowest (Figure 2 D).
Fig. 2.
Methylation status and mRNA expression of glucocorticoid receptor (GR) 1-CTotal promoter and mRNA expression of DNA methyltransferase (Dnmt)-1 and Dnmt3a in human umbilical cord (UC). (A) Expression of the transcript of the GR1-CTotal in human UC by real-time RT-PCR using adult human blood as reference. The same primers were used to detect (B) Dnmt1 and (C) Dnmt3a in human (H) UC as used with rat (R) liver. Transcripts of appropriate size were identified by analysis of the RTPCR products by agarose gel electrophoresis. The methylation status of the GR1-CTotal promoter and the mRNA expression of Dnmt1 and Dnmt3a was measured in human UC samples from 15 pregnancies. The association between GR1-CTotal promoter methylation and Dnmt expression was analysed by calculation of Pearson’s correlation coefficient. (D) The relationship between Dnmt1 expression and GR1-CTotal promoter methylation, r = 0.70, P = 0.03. (E) The relationship between Dnmt3a expression and GR1-CTotal promoter methylation, r = 0.001, P = 0.89).
Single bands of the correct size for the PCR products of the Dnmt1 and Dnmt3a primers in human and rat tissue were verified by gel electrophoresis (Figure 2 B, C). Dnmt1 expression predicted 49% (P = 0.003) of the variation in GR1-CTotal methylation (Figure 2 D), while Dnmt3a expression was not related to GR1-CTotal methylation (Figure 2 E).
Discussion
The results of this study show for the first time that hypomethylation and increased expression of the GR110 promoter induced in the liver of the offspring of rats fed a PR diet during pregnancy are associated with reduced Dnmt1 expression and altered covalent modifications to histones at the GR promoter.
Feeding a PR diet to pregnant rats increases hepatic gluconeogenesis in the adult offspring (Burns et al. 1997; Desai et al. 1997), which reflects increased expression of hepatic GR and PEPCK expression (Bertram et al. 2001, Lillycrop et al. 2005). This is consistent with increased corticosteroid activity in the offspring of rats fed a PR diet during pregnancy (Langley-Evans et al. 1996). Our current and previous findings (Lillycrop et al. 2005) show that increased hepatic GR expression in the PR offspring is associated with hypomethylation of the GR110 promoter. The livers of these rats also show hypomethylation of the PPARα promoter and increased expression of its target gene acyl-CoA oxidase (AOX) (Lillycrop et al. 2005). Together these findings suggest that feeding a PR diet during pregnancy induces in the liver of the offspring a metabolic phenotype characterised by altered regulation of energy balance, with increased capacity for gluconeogenesis and peroxisomal fatty acid β-oxidation. Moreover, since promoter methylation status may be modified at different stages of development in a gene and tissue –specific manner (Grainger et al. 1983; Benvenisty et al. 1985; Gidekel et al. 2002; Hershko et al. 2003), this suggests a mechanism by which timing of nutritional constraint may influence the induced phenotype (Ravelli et al. 1998; Remacle et al. 2004).
The stable silencing of genes by DNA methylation is critical for cellular differentiation (Bird, 2001; Hershko et al. 2003) and for the developmental regulation of the activities of metabolic pathways (Grainger et al. 1983; Benvenisty et al. 1985). Induction of hypomethylation of the hepatic GR promoter in the PR offspring may occur either by impaired methylation de novo during cell differentiation in the early embryo, failure to maintain DNA methylation during mitosis or active demethylation. Failure to maintain methylation or active demethylation may result in the activation of genes not normally expressed in adult tissues, or accelerated or more extensive demethylation of genes that are induced during tissue maturation. Our findings show that, while the expression of Dnmt3a and 3b and the demethylase MDB2 were not altered, Dnmt1 expression was significantly lower in the liver of the PR offspring compared to controls. Thus hypomethylation of the GR110 promoter, and in turn altered regulation of hepatic glucose metabolism, may be induced by reduced capacity to methylate hemimethylated DNA during mitosis rather than failure of DNA methylation de novo or active demethylation. This conclusion is supported by lower binding of Dnmt1 at the GR110 promoter in the PR offspring. These findings agree with those showing induction of DNA hypomethylation and altered phenotype by depletion of xDnmt1 in Xenopus embryos (Stancheva & Meehan 2000; Stancheva et al. 2001) and promoter demethylation by Dnmt1 knockdown (Leu et al. 2003).
Altered 1-carbon metabolism plays a central role in phenotype induction in this model (Jackson et al. 2002; Petrie et al. 2002; Torrens et al. 2006; Lillycrop et al. 2005). Our findings show that lowering of Dnmt1 expression by the PR diet was prevented by maternal folic acid supplementation, while expression of Dnmt3a was unaltered. Hence Dnmt1 expression in the offspring is modified by maternal folic acid intake, which is consistent with the modulation of Dnmt1 expression in adult rats by folic acid intake (Ghoshal et al. 2006) and the inhibition of Dnmt1 and induction of DNA hypomethylation by hyperhomocysteinemia (James et al. 2002). Thus, the effects of differences in maternal folic acid intake, and in turn capacity for metabolism of methyl groups, during pregnancy could explain the induction or prevention of GR110 hypomethylation in the liver of the offspring. These findings also emphasise the importance of adequate dietary intake of folic acid during pregnancy for optimal fetal development.
One possible mechanism for induction of an altered metabolic phenotype in the liver of the offspring of rats fed a PR during pregnancy is impairment of 1-carbon metabolism leading to down regulation of Dnmt1 expression and progressive loss of methyl groups from the GR110 promoter. Reduced expression of Dnmt1 may be expected to result in decreased methylation of all promoters containing CpG dinucleotides. However, studies in cells lacking Dnmt1 show only a 20% decrease in genomic methylation and no changes to the methylation status of specific genes (Rhee et al. 2000). This suggests Dnmt1 activity is targeted to specific genes, possibly through binding to transcription factors such as E2F1 (Robertson et al. 2000). Thus because binding of Dnmt1 to the GR promoter was reduced in addition to an overall reduction in Dnmt1 expression, the proposed pathway is consistent with gene-specific hypomethylation in the liver in the offspring of dams fed a PR diet (Lillycrop et al. 2005). Dnmt1 activity is also required for progression through mitosis (Milutinovic et al. 2003). Thus it is possible that suppression of Dnmt1 activity or expression by altered 1-carbon metabolism in the preimplantation period could also account for the reduction in cell number during the early development in this model (Kwong et al. 2000).
The specific link between reduced protein intake and altered 1-carbon metabolism cannot be deduced from the present data. However, we offer two possible explanations. First, it is possible that it may result from decreased availability of glycine, leading to altered flux of methyl groups between different metabolic fates. Secondly, increased maternal corticosteroid activity (Langley-Evans et al. 1996), possibly as a result of stress induced by constrained nutrient availability, may reduce folic acid availability (Terzolo et al. 2004). The second mechanism could explain how maternal corticosteroid blockade prevents induction of hypertension in the PR offspring (Langley-Evans et al. 2004) and as well as prevention of altered phenotype by folic acid administration (Torrens et al. 2006; Lillycrop et al. 2005).
Analysis of GR1-CTotal in human UC samples showed for the first time that among individuals within the normal birthweight range there is considerable variation in the methylation status of a gene expressed in human fetal tissue. Dnmt1 expression predicted 49% of the variation in methylation of the GR1-C Total promoter expressed in human UC. Because the UC were frozen, we were unable to dissect and measure Dnmt1 expression and GR1-C Total methylation in specific tissues. However, these data suggest that methylation of the GR1-C Total promoter in human UC is associated with the capacity of Dnmt1 to maintain methylation of CpG dinucleotides rather than capacity for DNA methylation de novo. The extent to which maternal diet during pregnancy determines Dnmt1 expression cannot be examined within this small group of infants. However, these findings are consistent with our observations in the rat and so suggest the hypothesis that induction of different phenotypes in humans by prenatal nutrition may involve variations in Dnmt1 expression and, in turn, DNA methylation.
Covalent modifications of specific residues in the N-terminal domain of histones also confer epigenetic regulation of transcription (Turner, 2000). Such modifications are closely linked to promoter methylation as methylated CpG nucleotides are required for binding of MeCP2 and recruitment of the HDAC/HMT complex. Binding of MeCP2/HDAC/HMT to methylated CpGs causes histone deacetylation and methylation of specific lysine resides leading to suppression of transcription. We found higher levels of histone modifications which facilitate transcription, while modifications that suppress transcription were reduced, at the GR110 promoter in the offspring of rats fed the PR diet than controls. One possible explanation is that hypomethylation of the GR110 promoter reduced binding of MeCP2 and, in turn, the HDAC/HMT complex. If so, this suggests lower Dnmt1 expression is the primary process in inducing increased GR110 expression, and altered histone modifications are a secondary effect. This may be exacerbated by lower hepatic MeCP2 expression in these offspring. Such changes to the regulation of transcription of the GR110 promoter are consistent with the higher level of transcription in the PR offspring.
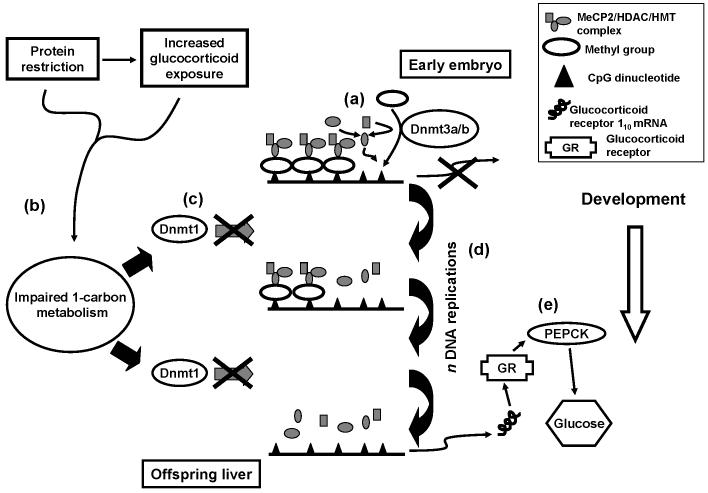
We therefore propose a mechanism for the induction of an altered metabolic phenotype in the offspring of rats fed a PR diet during pregnancy based upon our findings for the GR110 promoter (Figure 3). Promoter methylation is induced in a gene-specific manner during the development of the early embryo by the activities of Dnmt 3a and 3b. Impaired 1-carbon metabolism, either a direct result of constrained maternal nutrition or by increased corticosteroid activity, reduces Dnmt1 expression resulting in progressive hypomethylation of specific genes during successive mitotic cycles. The hypomethylated GR110 promoter would result in the reduced binding of the MeCP2/HDAC/HMT complex which facilitates persistence of histone modifications that permit transcription. In turn, gluconeogenesis increases due to up-regulation of PEPCK expression by the action of the GR. The pathway may also explain hypomethylation of PPARα and increased expression of AOX in the liver of the PR offspring (Lillycrop et al. 2005). If this pathway acts primarily by altering the epigenetic regulation of specific transcription factors, then changes in the activities of a number of metabolic pathways may be induced. However, the proposed pathway does not exclude the possibility that other genes may also be hypomethylated by this process. This pathway also explains how administration of glucocorticoids during pregnancy induces persistent changes to gluconeogenic enzymes in the offspring as a result of increasing GR expression (Nyirenda et al. 1998).
Fig. 3.
A diagram of the proposed pathway for induction of altered glucose homeostasis in the offspring of rats fed a protein-restricted (PR) diet during pregnancy. A full explanation is given in the text. (a) Glucocorticoid receptor (GR) expression is silenced in the early embryo in cells destined to become hepatocytes by the activities of DNA methyltransferases (Dnmt) 3a and 3b. (b) In the offspring of rats fed a PR diet, 1-carbon metabolism is impaired either as a direct consequence of the restricted diet or by increased glucocorticoid exposure. This down-regulates Dnmt1 expression (c). Lower Dnmt1 expression results in a impaired capacity to methylate hemimethylated DNA during mitosis (d). After sequential mitotic cycles, this results in hypomethylation of the GR promoter and loss of epigenetic memory of gene silencing, such that, in the liver of the adult offspring, expression of GR is induced in cells which do not express GR in control animals. The increased expression of GR is facilitated by lower binding and expression of methyl CpG binding protein (MeCP)-2 and reduced recruitment of the histone deactylase (HDAC) / histone methyltransferase (HMT) complex, resulting in higher levels of histone modifications which permit transcription. (e) This results in increased phosphoenolpyruvate carboxykinase (PEPCK) expression and increased gluconeogenesis.
In conclusion, the present findings suggest a pathway for induction of an altered phenotype or fetal programming. In humans this may provide a basis for interventions in early life to reduce risk of later metabolic disease.
Acknowledgements
GCB and MAH are supported by the British Heart Foundation. Collection of human umbilical cord specimens was supported by the Medical Research Council.
References
- Armitage JA, Lakasing L, Taylor PD, Balachandran AA, Jensen RI, Dekou V, Ashton N, Nyengaard JR, Poston L. Developmental programming of aortic and renal structure in offspring of rats fed fat-rich diets in pregnancy. J Physiol. 2005;565:171–184. doi: 10.1113/jphysiol.2005.084947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE. Developmental plasticity and human health. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- Benvenisty N, Mencher D, Meyuhas O, Razin A, Reshef L. Sequential changes in DNA methylation patterns of the rat phosphoenolpyruvate carboxykinase gene during development. Proc Natl Acad. Sci U S A. 1985;82:267–271. doi: 10.1073/pnas.82.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram C, Trowern AR, Copin N, Jackson AA, Whorwood CB. The maternal diet during pregnancy programs altered expression of the glucocorticoid receptor and type 2 11beta-hydroxysteroid dehydrogenase: potential molecular mechanisms underlying the programming of hypertension in utero. Endocrino. 2001;142:2841–2853. doi: 10.1210/endo.142.7.8238. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2001;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mo. Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Farnham P. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Bio. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge GC, Phillips ES, Dunn RL, Jackson AA, Lillycrop KA. Effect of reduced maternal protein consumption during pregnancy in the rat on plasma lipid concentrations and expression of peroxisomal proliferator-activated receptors in the liver and adipose tissue of the offspring. Nutr Res. 2004;24:639–646. [Google Scholar]
- Burns SP, Desai M, Cohen RD, Hales CN, Iles RA, Germain JP, Going TC, Bailey RA. Gluconeogenesis, glucose handling, and structural changes in livers of the adult offspring of rats partially deprived of protein during pregnancy and lactation. J Clin Invest. 1997;100:1768–1774. doi: 10.1172/JCI119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mo. Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Desai M, Byrne CD, Meeran K, Martenz ND, Bloom SR, Hales CN. Regulation of hepatic enzymes and insulin levels in offspring of rat dams fed a reduced-protein diet. Am J Physio. 1997;273:G899–G904. doi: 10.1152/ajpgi.1997.273.4.G899. [DOI] [PubMed] [Google Scholar]
- Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Bio. Chem. 2002;277:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, Huang Y, Young D, Jacob ST. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522–1527. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Bio. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4:611–624. doi: 10.1079/phn2001145. [DOI] [PubMed] [Google Scholar]
- Godfrey K, Walker-Bone K, Robinson S, Taylor P, Shore S, Wheeler T, Cooper C. Neonatal bone mass: influence of parental birthweight, maternal smoking, body composition, and activity during pregnancy. J Bone Min Res. 2001;16:1694–1703. doi: 10.1359/jbmr.2001.16.9.1694. [DOI] [PubMed] [Google Scholar]
- Grainger RM, Hazard-Leonards RM, Samaha F, Hougan LM, Lesk MR, Thomsen GH. Is hypomethylation linked to activation of delta-crystallin genes during lens development? Nature. 1983;306:88–91. doi: 10.1038/306088a0. [DOI] [PubMed] [Google Scholar]
- Harris RG, White E, Phillips ES, Lillycrop KA. The expression of the developmentally regulated proto-oncogene Pax-3 is modulated by N-Myc. J Bio. Chem. 2002;277:34815–34825. doi: 10.1074/jbc.M109609200. [DOI] [PubMed] [Google Scholar]
- Hershko AY, Kafri T, Fainsod A, Razin A. Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene. 2003;302:65–72. doi: 10.1016/s0378111902010910. [DOI] [PubMed] [Google Scholar]
- Jackson AA, Dunn RL, Marchand MC, Langley-Evans SC. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin. Sci. (Lond.) 2002;103:633–639. doi: 10.1042/cs1030633. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002;132(Suppl):2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development. 2000;127:4195–4202. doi: 10.1242/dev.127.19.4195. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Langley SC, Jackson AA. Increased systolic blood pressure in adult rats induced by fetal exposure to maternal low protein diets. Clin Sc. (Lond.) 1994;86:217–222. doi: 10.1042/cs0860217. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC, Gardner DS, Jackson AA. Maternal protein restriction influences the programming of the rat hypothalamic-pituitary-adrenal axis. J. Nutr. 1996;126:1578–1585. doi: 10.1093/jn/126.6.1578. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC. Hypertension induced by foetal exposure to a maternal low-protein diet, in the rat, is prevented by pharmacological blockade of maternal glucocorticoid synthesis. J Hypertens. 1997;15:537–544. doi: 10.1097/00004872-199715050-00010. [DOI] [PubMed] [Google Scholar]
- Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, Huang TH. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res. 2003;63:6110–6115. [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Maloney CA, Gosby AK, Phuyal JL, Denyer GS, Bryson JM, Caterson ID. Site-specific changes in the expression of fat-partitioning genes in weanling rats exposed to a low-protein diet in utero. Obes Res. 2003;11:461–468. doi: 10.1038/oby.2003.63. [DOI] [PubMed] [Google Scholar]
- McCormick JA, Lyons V, Jacobson MD, Noble J, Diorio J, Nyirenda M, Weaver S, Ester W, Yau JL, Meaney MJ, Seckl JR, Chapman KE. 5′-heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol Endocrinol. 2000;14:506–517. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Niveleau A, Szyf M. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem. 2003;278:14985–14995. doi: 10.1074/jbc.M213219200. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nyirenda MJ, Lindsay RS, Kenyon CJ, Burchell A, Seckl JR. Glucocorticoid exposure in late gestation permanently programs rat hepatic phosphoenolpyruvate carboxykinase and glucocorticoid receptor expression and causes glucose intolerance in adult offspring. J Clin Invest. 1998;101:2174–2181. doi: 10.1172/JCI1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie L, Duthie SJ, Rees WD, McConnell JM. Serum concentrations of homocysteine are elevated during early pregnancy in rodent models of fetal programming. Br J Nutr. 2002;88:471–477. doi: 10.1079/BJN2002695. [DOI] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Remacle C, Bieswal F, Reusens B. Programming of obesity and cardiovascular disease. Int J Obes Relat Metab Disord. 2004;28(Suppl 3):S46–S53. doi: 10.1038/sj.ijo.0802800. [DOI] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. Dnmt1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzolo M, Allasino B, Bosio S, Brusa E, Daffara F, Ventura M, Aroasio E, Sacchetto G, Reimondo G, Angeli A, Camaschella C. Hyperhomocysteinemia in patients with Cushing’s syndrome. J Clin Endocrinol Metab. 2004;89:3745–3751. doi: 10.1210/jc.2004-0079. [DOI] [PubMed] [Google Scholar]
- Torrens C, Brawley L, Anthony FW, Dance CS, Dunn R, Jackson AA, Poston L, Hanson MA. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Turner JD, Muller CP. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol. 2005;35:283–292. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Vickers MH, Gluckman PD, Coveny AH, Hofman PL, Cutfield WS, Gertler A, Breier BH, Harris M. Neonatal leptin treatment reverses developmental programming. Endocrinol. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]