Summary
Int protein has two classes of binding sites within the phage att site: the arm-type recognition sequences are found in three specific sites that are distant from the region of strand exchange; the junction-type recognition sequences occur as inverted pairs around the crossover region in both attP and attB. During recombination between attP and attB each of the four DNA strands is cut at a homologous position within each of the junction-type Int binding sites. In all four junction-type sites Int protein interacts primarily with the same face of the DNA helix, as determined by those purine nitrogens that are protected against methylation by dimethylsulfate. Efficient secondary attachment sites for λ contain sequences with partial homology to the junction-type binding sites. In addition, the sequence between, but not part of, the two junction-type sites (the overlap region) is strongly conserved in secondary att sites. Thus, in the vicinity of strand exchange, attP and a recombining partner, such as attB, are very similar; each comprises two junction-type Int recognition sites and an overlap (crossover) region.
Introduction
The λ genome encodes a site-specific recombination system that is utilized for integration of the phage genome into that of its host, E. coli, during establishment of lysogeny, and for excision upon phage induction. The components of this system have been well defined genetically, and considerable progress has been made in their biochemical characterization (for reviews see Gottesman, 1981; Nash, 1981a, 1981b; Weisberg and Landy, 1983).
Integrative recombination between the specific phage and bacterial sites attP and attB requires the phage-encoded Int protein (reviewed in Nash, 1981a) and the E. coli–encoded integration host factor (Miller et al., 1979; Miller and Friedman, 1980; Nash and Robertson, 1981). Excisive recombination between the prophage sites attL and attR requires the phage-encoded Xis protein in addition to Int and integration host factor. Purified Int protein (40,000 MW; Kikuchi and Nash, 1979a) is a specific DNA-binding protein (Ross et al., 1979; Hsu et al., 1980), as is the purified integration host factor (Nash and Robertson, 1981; N. Craig and H. Nash, unpublished data).
The recombination crossover occurs within a short region of sequence homology between the two sites, the 15 bp common core (Landy and Ross, 1977). The points of exchange of the two strands are staggered by 7 bp (Mizuuchi et al., 1981; Craig and Nash, personal communication). The attP and attB sites are very different in both size and protein binding characteristics (Hsu et al., 1980). The attP site, referred to as the “donor,” is quite large, spanning approximately 235 bp from −150 to +82 (0 is the center of the common core sequence) (Hsu et al., 1980; Mizuuchi and Mizuuchi, 1980). In contrast, the bacterial site, referred to as the “recipient,” is small, spanning from −11 to+11 (Mizuuchi et al., 1981).
Four specific binding sites for purified Int have been defined within attP in nuclease protection experiments. The boundaries of the phage att site coincide with the boundaries of the outermost of these binding sites (Ross et al., 1979; Hsu et al., 1980). At least three of these sites can be protected in the absence of any of the other sites (Ross and Landy, 1982). The sequences of the four sites, and of the various specific non-att binding sites previously reported, fall into two distinct classes, the arm type and the core type. These different classes may be recognized by distinct DNA-binding “domains” of Int (Ross and Landy, 1982).
Though it has been shown that the isolated core region interacts with Int independently of arm sites, and that the arm-type consensus sequence is not found in the core regions (Ross and Landy, 1982), the interaction of Int with the two cores has been less well understood. Both the attP and the attB core regions interact with Int, yet their nuclease protection patterns differ significantly in size and in position with respect to the common core. We present data from studies of Int interaction with non-att DNA, from cloned subsections of the attP core, and from methylation protection experiments, all of which indicate that the common core region of each att site comprises two binding sites. The two binding sites in each core region are situated in inverted orientation at the core–arm junctions, and we refer to them as “junction-type” sites to distinguish them from the arm-type sites previously characterized (Ross and Landy, 1982). The four junction-type binding sites are not identical in sequence, and differ in apparent affinity for Int.
Results
Int has been shown to interact specifically with two distinct types of DNA sequence: the arm-type, for which a consensus sequence has been defined through interaction of Int with non-att DNA (Ross and Landy, 1982); and a sequence in the core region that is identified and characterized in this report. Significant differences in protection patterns of the attP and attB core regions have indicated that the 15 bp core sequence per se does not constitute an Int recognition sequence (Ross et al., 1979). The attB core protected region is shorter in length (15–20 bp vs. 30–35 bp for attP), and is centered over the left core–arm junction rather than being symmetrically disposed with respect to the core. To rule out the possible influence on attB core protection patterns of a second Int binding site near the core, we have carried out footprint experiments with a much smaller subcloned attB containing only sequence from −17 to+21 (pMM291; Mizuuchi et al., 1981). The protection pattern of this fully functional attB core region is indistinguishable from that reported previously (data not shown).
A Core-like Int-Protected Sequence in Non-att DNA
A consensus recognition sequence for arm-type sites was established by analysis of specific Int-protected regions in non-att DNA (Ross and Landy, 1982). We have also observed Int binding to non-att sequences that show no homology to the seven arm-type non-att sites described previously. One of these sites (Figure 1A) is a 30 bp protected region (+140 to +170) situated within the Int structural gene beyond the functional limit (+82) of attP (Hsu et al., 1980). This protected sequence, which will be referred to as the int155 site, contains two subsections of homology to the attP core region sequence (see Figure 2). These two homologies are situated in attP at the core–arm junctions, in regions protected by Int against neocarzinostatin (NCS) or DNAase I digestion. There is no homology between the central core sequence (part of which has been shown to be sensitive to NCS digestion in the presence of Int; Ross et al., 1982) and the int155 sequence. The sequences that are homologous between the core region and int155 sites constitute in each case an imperfect inverted repeat sequence (Figure 2). The int155 sequence appears to be a lower affinity site than is the core. On a restriction fragment carrying both sites, a slightly higher Int concentration is required to observe protection of the int155 site (data not shown).
Figure 1. Int Protection of Non-att Sequences Homologous to the attP Core Region.
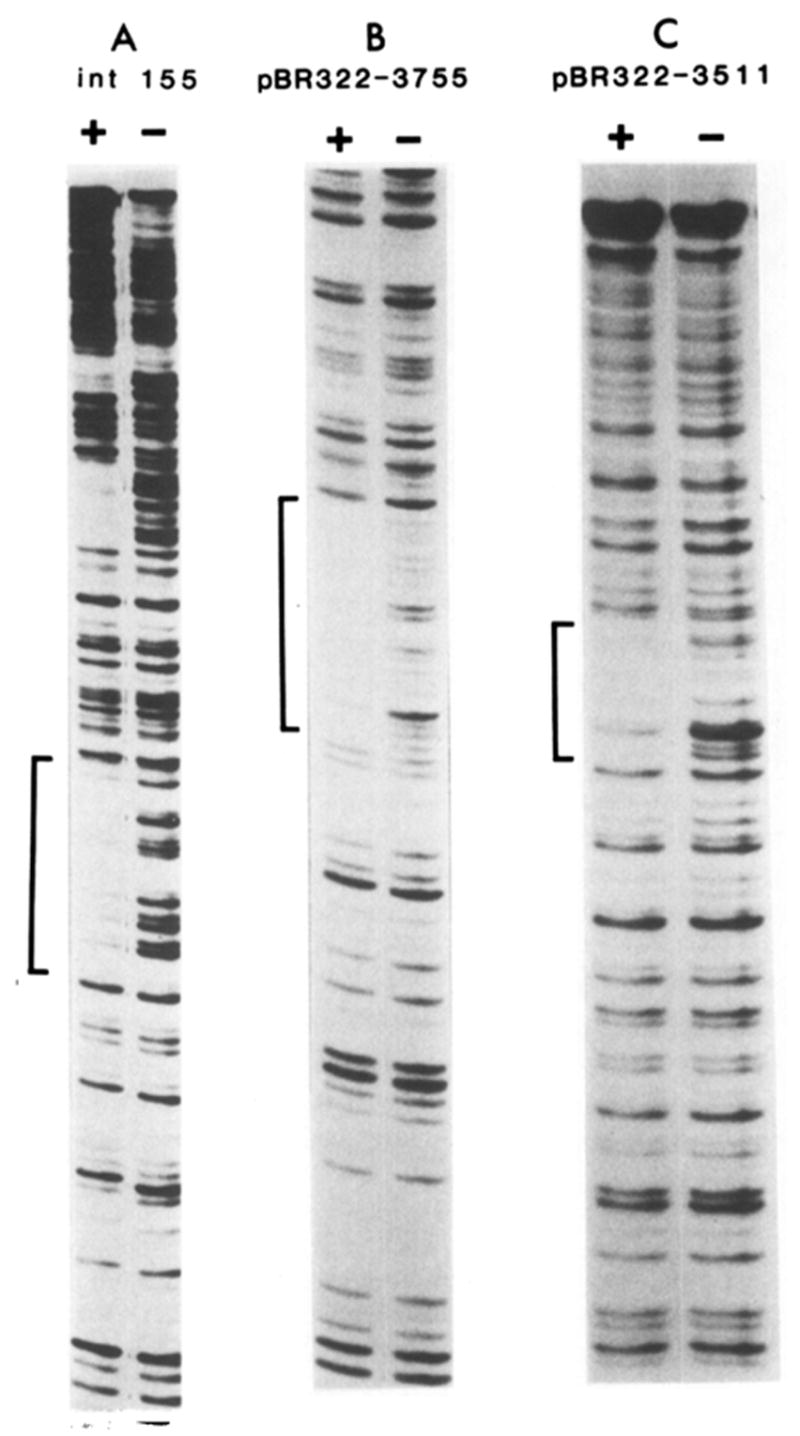
(A) The attP-containing fragment *Bam(+242)–Hind III(−250), from pWR1 (Hsu et al., 1980), partially digested with NCS in the presence (+) or absence (−) of Int. (* denotes the 5′-labeled end.) The P′ arm site and common core protected regions are visible further up the gel beyond the int155 site, which is marked by a bracket. Sequence of the 30 bp protected region at the int155 site is compared with that of the attP core region in Figure 2. (B) Fragment *Pst(3608)–Hinc II(3906) from pBR322 partially digested with DNAase I in the presence (+) or absence (−) of Int. (C) Fragment *Hinf(3361)–Pst(3608) from pBR322, partially digested with DNAase I in the presence (+) or absence (−) of Int. Sequences of the protected regions in (B) and (C) are illustrated in Figure 3A.
Figure 2. Comparison of Int-Protected Sequences in the attP Core Region and the int155 Site.
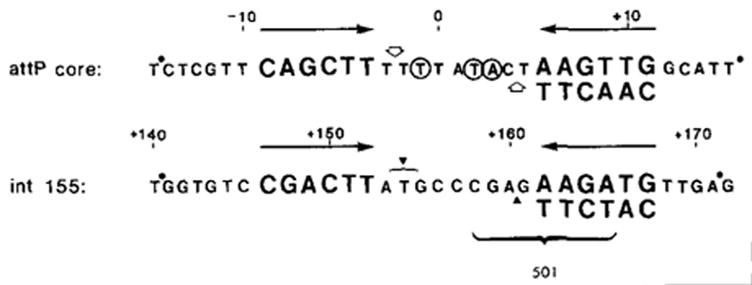
Sequences have been previously reported (Landy and Ross, 1977). The regions of homology between the two sites, which constitute in each case an imperfect inverted repeat (→←), are represented in large letters. Approximate boundaries of NCS footprint protection are indicated (●). Sites in the central attP core that are sensitive to NCS are circled (Ross et al., 1982). No such sites are seen in the int155 region, but would not be expected since NCS is an A+T-specific reagent. (
 ) Sites cut by Int during recombination (Mizuuchi et al., 1981; N. Craig and H. Nash, personal communication). The endpoints of xin-promoted deletions (▼) (Gritzmacher and Weisberg, personal communication) and the 8 bp region containing the att2501 deletion endpoint (Ross et al., 1982) are indicated within the int155 site.
) Sites cut by Int during recombination (Mizuuchi et al., 1981; N. Craig and H. Nash, personal communication). The endpoints of xin-promoted deletions (▼) (Gritzmacher and Weisberg, personal communication) and the 8 bp region containing the att2501 deletion endpoint (Ross et al., 1982) are indicated within the int155 site.
Two Binding Sites at Core
As described above, the attB core protected sequence corresponds to the left half of the attP core protected region. A sequence centered within the attB protected region shares partial sequence homology with the elements of symmetry in the attP core region at the two core–arm junctions, as noted above and in Figure 2. These attP junctions are also homologous to the int155 site discussed above (Figure 2). The size and position of the attB Int-protected region suggests the possibility that each core–arm junction region may define a separate recognition site. The larger attP core and int155 protected regions would reflect two adjacent sites situated in inverted orientation.
To determine whether each attP core–arm junction represents a separate binding site for Int, sequences that resemble each junction were identified in pBR322. Two sequences like that of the right junction, CAACTT (at pBR322 positions 3511 and 3755), and four like the left junction, CAGCTT (see below), were found. Each of these six sites was isolated on a single end-labeled restriction fragment (see Experimental Procedures and Figure 1) and was tested for Int binding by DNAase and NCS footprinting.
Of the six sites, the two CAACTT sites were clearly protected by Int against digestion (Figures 1B and 1C). Footprint protection of an additional non-att site with the sequence CAACTT was also observed in sequence adjacent to attB (+38 to +52; Ross et al., 1979). This site, referred to as “bio” (see Figure 3), occurs in a region known not to be required for attB function (Mizuuchi et al., 1981). The extent of sequence protected in each of the three cases was approximately 15–20 bp, roughly half of the full attP core site protected region, and comparable in size to the attB site footprint. The sequence CAACTTNNT is found in each protected region (Figure 3A). This consensus sequence is also found in its entirety at the right attP core–arm junction (Figure 3B).
Figure 3. Sequences of Int-Protected Junction-type Sites.

All sequences are written 5′ to 3′. (A) Non-att junction-type sites. Protection profiles of pBR322 3511 and 3755 are shown in Figures 1B and 1C, and of “bio” in Ross et al. (1979). Sequences are aligned according to homology, and the numerical coordinates given are from the published sequences (“bio”—Landy and Ross, 1977; Ross et al., 1979; pBR322—Sutcliffe, 1979). Homologous positions are presented in large type. Approximate boundaries of footprint protection are indicated for DNAase I (○) and NCS (●). Definition of very precise boundaries is not possible because not all positions are good substrates for the reagents used (NCS is A+T-specific, see D’Andrea and Hazeltine, 1978). (B) Sequences of both left and right core–arm junctions from attP and attB aligned according to homology among them and homology with the non-att sites in (A). Numbering is from Landy and Ross (1977) and protection boundaries are from Ross et al. (1979). Boundaries of the right side of the two attP sites incorporate data from NCS-sensitive central core positions (Ross et al., 1982). (C) Sequences from the left and right sides of the int155 site. Regions of homology to other sites in (A) and (B) are in large letters. The pairs of sites in (B) and (C) are present in inverted orientation and with identical spacing in the attP, attB and int155 sites (see Figures 2 and 5).
In contrast to the clearly defined Int interaction with the three CAACTTNNT non-att sites, Int protection of the four sequences from pBR322 that resemble the left attP junction (CAGCTT) was either very partial (two sites, at 14 and 2133) or undetectable (at sites 1087 and 2781) (data not shown). CAGCTT sequences may therefore be lower affinity sites. Alternatively, additional sequence in the left core–arm junction may be essential for binding. For example, in the left junction region, the T at −1 is in an analogous position to the T in the final position of the sequence shared by the three Int-protected non-att sites and the right junction (Figure 3B). None of the four pBR322 sites has this T. To address this question the attP left junction region was subcloned and will be described below.
A sequence like that found in the attB left junction, CTGCTT, was also tested for Int binding (not shown). The absence of footprint protection at this site (+189 to +194 within the Int structural gene; data not shown) supports the idea that additional sequence information is required for binding strong enough to be detectable by this assay (see Experimental Procedures). Specific interaction at junction sites that is undetectable by footprinting can be seen in the methylation protection experiments to be described below.
Methylation Protection
The interaction of Int with single junction-type sites in non-att DNA, as described above, indicates that the attP core region probably comprises two such sites. However, the lack of clear interaction with non-att sequences resembling either the left attP junction or the left attB junction raises the possibility that additional sequence positions may be critical for binding, as well as the interesting possibility that the attP and attB junction binding sites might differ in the precise mode of their interaction with Int.
To evaluate these possibilities, and to compare attB binding with attP binding, we have carried out methylation protection experiments. Proximity of a protein to purine positions N7 of guanine (in the major groove) or N3 of adenine (in the minor groove) results in protection of these positions against methylation by dimethylsulfate (DMS) (Gilbert et al., 1976).
Int partially protects several purine positions (both adenine and guanine) on each strand in the attP core region (Figures 4A, 4B, and 5) at Int concentrations comparable to those required for footprint protection. These protected positions are centered around, and include a majority of the purines in, the consensus recognition sequence defined by Int binding to non-att sites (Figure 3). Five of six purines in the left junction sequence CAGCTT, and four of six in the right junction sequence CAACTT, are protected (Figure 5). Three of the protected positions at each junction are homologous between the two sites (as marked in Figure 5). Several additional positions show either protection or an enhanced level of methylation. Two positions within sequence that is homologous between the two sites are protected in one site (−1, −4), but not in the other (+3, +6). Three protected or enhanced positions on each side are in regions of nonhomology between the two sites (−13, −3, −2 and +4, +12, +13). The four adenines in the center of the common core region (0 to +3) show methylation that is not affected by Int binding. The experiments in Figures 4A and 4B were carried out with a fragment containing only the attP core site, but no arm sites. The methylation pattern of the top strand of the attP core has also been examined on a restriction fragment containing both the core region and the P′ arm binding site (from pPH56; −90 to +242) (data not shown). No differences in the positions or relative extent of methylation protection or enhancement were observed. Different preparations of Int also gave the same results. The distribution of protected purine positions on a three-dimensional model of attP core region sequence will be discussed below.
Figure 4. Int Protection of the attP and attB Core Regions against Methylation by DMS.

The attP core region fragment from pMJB11 (containing only the core Int binding site, but no arm binding sites, see Experimental Procedures) was 5′-labeled in the lower strand (Bam site at +46) (A), or the upper strand (Hind III site at −70) (B), and methylated in the presence (+) or absence (−) of Int (see Experimental Procedures). An attB core region fragment from pMM291 (containing attB sequence from −17 to +21; Mizuuchi et al., 1981) was 5′-labeled in the lower strand (Hha site at 4257 in pBR322) (C), or the upper strand (Hga I site at 390 in pBR322) (D), and methylated in the presence (+) or absence (−) of Int. C+T chemical sequence markers were prepared by procedures of Maxam and Gilbert (1980). Samples were electrophoresed in 10% acrylamide–8 M urea gels (see Ross et al., 1979, 1982). Positions protected by Int against methylation (*). Smaller (*) indicates slight degree of protection. Positions of enhanced methylation in the presence of Int (→). The outermost affected positions in each site are numbered; see Figure 5 for identification of other protected positions.
Figure 5. attP and attB Core Region Sequences Showing Methylation Protection or Enhancement in the Presence of Int.
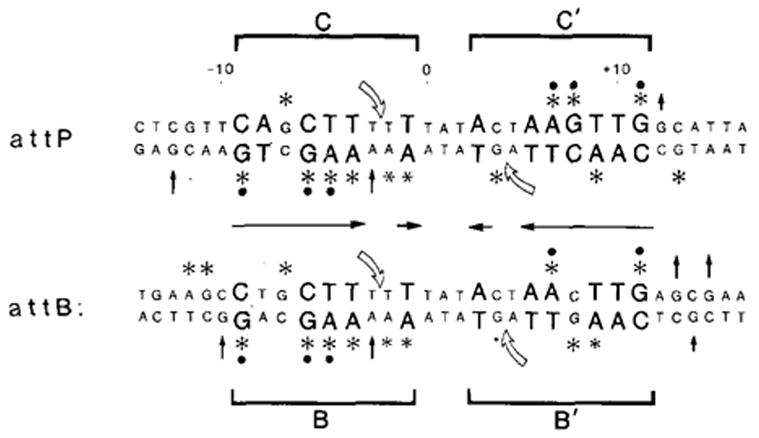
(*) Positions protected against methylation. Smaller asterisk indicates slight degree of protection. (↑) Positions showing enhanced levels of methylation. Sequences were reported previously (Landy and Ross, 1977). 0 is the center of the 15 bp common core sequence. Positions in larger letters are regions of homology between the left and right core–arm junctions (situated in inverted orientation (→←) and conserved in non-att junction-type binding sites (Figure 3). (
 ) Positions cut by Int during recombination (Mizuuchi et al., 1981; Craig and Nash, personal communication). The four junction sites are referred to as C and C′ (for core) in attP and B and B′ in attB. Protected positions that are conserved in three or four of the sites (●).
) Positions cut by Int during recombination (Mizuuchi et al., 1981; Craig and Nash, personal communication). The four junction sites are referred to as C and C′ (for core) in attP and B and B′ in attB. Protected positions that are conserved in three or four of the sites (●).
attB–Int Interaction
Methylation protection experiments were also carried out with an attB-containing fragment in order to assess whether Int interaction with the partially homologous left attB junction sequence (CTGCTT) resembles the attP junction interaction. In addition, it was of interest to evaluate whether possible weaker interactions with the right attB junction sequence might be detectable by this assay. This right junction also shares some homology to the attP junctions (CAAGTT; see Figure 3) but gives no clear Int footprint (Ross et al., 1979).
At the left attB junction, the protection or enhancement of eight positions is comparable to that seen at the homologous positions in the left attP junction (Figure 5; −9 and −7 through −1). In addition, three positions of nonhomology with attP show altered levels of methylation (Figure 5; −10, −11, −12). These effects are seen in the Int concentration range required for footprinting.
At higher Int concentrations (where footprint protection is obscured by the extensive inhibition of DNAase I or NCS digestion by nonspecifically bound Int), methylation protection experiments yielded two further results of considerable interest. Specific protection or enhancement of positions at the right attB junction was seen. At positions of homology to the right attP junction the same protection effects were found (+4, +7, +9, +11; though +4 may be less extensively protected than in attP). In addition, the nonhomologous positions +8, +13, +14, and +15 are affected. The Int concentration required to observe these effects was approximately 4-fold that required for protection at the left attB junction.
The attB core–Int interactions can be viewed as essentially paralleling those at the attP core; each core region has two junction binding sites. However, in contrast to attP, where under the conditions used, both left and right core sites fill at the same concentration (data not shown), the two sites in the attB core region require different Int concentrations to fill.
DNAase footprinting carried out at high Int concentration in the presence of methylating agent, DMS, indicates that nonspecific binding occurs under these conditions, and is not abolished, for example, by methylation effects on Int protein (data not shown). It appears, therefore, that nonspecific Int binding does not result in any selective effects on methylation, and also that the overall rate of methylation in nonspecifically protected regions is not significantly reduced.
Separated attP Junction Binding Sites
Int binding to non-att regions with the sequence CAACTTNNT, and the presence of this sequence at the right attP core junction, indicate that this consensus sequence contains the essential features of a single junction-type Int binding site. As predicted from these results, the subcloned right attP junction site in plasmid pWR221 shows partial footprint protection, and the same specific sites of methylation protection in the strand tested (upper) as are seen in the right junction side of the full core site (Figures 5 and 6; see Experimental Procedures for clone construction). The site in pWR221 gives only partial footprint protection, and is thus interpreted to be of lower affinity than the non-att sites tested. This may be due to the lack in pWR221 of the conserved T in the final consensus position, which is found in the otherwise homologous strong non-att sites (Figures 1 and 3), and in the full attP core.
Figure 6. Subcloned Left (pWR243) and Right (pWR221, pPH411) attP Core–Junction Binding Sites (C and C′).
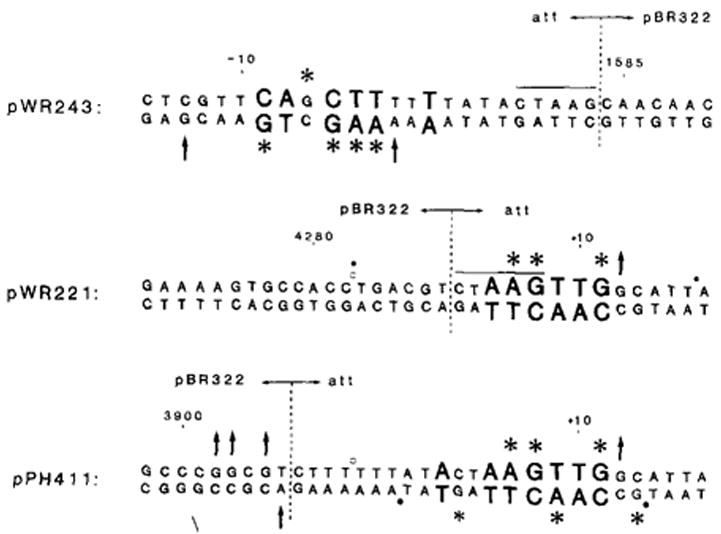
Sequence conserved among junction-type binding sites (see Figure 3) is represented in larger letters. Purine positions protected by Int against methylation by DMS (*), or showing enhanced levels of methylation in the presence of Int (↑), are indicated (compare with Figure 5). Only the upper strand of pWR221 was tested. Boundaries enclosing sequences that are protected by Int in DNAase I (○) or NCS (●) footprint protection experiments are indicated. pWR243 shows no footprint protection under the conditions employed (see text). Protection of pWR221 is partial. See Experimental Procedures for clone constructions. The newly created junctions of att and pBR322 sequence are indicated (
 ). In pWR243 and pWR221, these new junctions occur at the right or left ends of the Dde site (CTAAG). In pPH411, the new junction was created at the Hinc II site in pBR322 (3906) and the Alu site in att (−6); no new restriction site was generated, and one C was deleted during fusion formation. Numbering refers to att sequence (Figure 5) or to pBR322 sequence (Sutcliffe, 1979).
). In pWR243 and pWR221, these new junctions occur at the right or left ends of the Dde site (CTAAG). In pPH411, the new junction was created at the Hinc II site in pBR322 (3906) and the Alu site in att (−6); no new restriction site was generated, and one C was deleted during fusion formation. Numbering refers to att sequence (Figure 5) or to pBR322 sequence (Sutcliffe, 1979).
The lack of clear interaction with non-att sequences resembling the left attP junction could be attributable either to important attP sequence positions not found in the non-att sequences tested or to differences in affinity of the two attP junction sites for Int.
To determine whether the actual left attP junction region could interact with Int to yield a detectable footprint when separated from the right junction site, a clone containing the region of interest was constructed. The cloning strategy utilized a Dde I restriction site located in the right side of the core, and is described in Experimental Procedures. The sequence of the subcloned left junction region on plasmid pWR243 is shown in Figure 6. In addition to the left junction (including the final T in the consensus sequence) it contains the entire 15 bp core sequence, but lacks most of the right junction binding site. This site was tested in both footprinting and methylation protection assays. Though no clear footprint protection was detectable (not shown), specific purine positions were protected against methylation in both strands (Figure 6). The positions affected are the same as those seen at the left junction in the full core site (Figure 5). At the right (altered) side of the pWR243 core, no protection or enhancement was detected on either strand.
Thus the left attP junction does constitute a specific Int binding site: like the right attB junction, it is detectable only in methylation protection experiments. A second subcloned attP right junction site in plasmid pPH411 (see Experimental Procedures) gives a distinct 15–20 bp footprint, as well as comparable methylation protection patterns in the right junction sequence (see Figure 6). However, its left junction region is quite similar in sequence to the attP left junction. Though this altered left side exhibits neither footprint protection nor comparable methylation protection patterns, it does show four positions of enhanced methylation. This clone is thus less clearly interpretable as containing only a single junction site.
Discussion
Two Int Binding Sites at Each Core
Utilizing the interaction of purified Int protein with several non-att DNA sequences, we have determined that a minimal consensus sequence for core-type Int binding sites is CAACTTNNT. In footprint experiments with three independent non-att fragments this sequence gives rise to protected regions roughly half the size of the attP core protected region. In attP this sequence forms the right element of a region of 2-fold rotational symmetry centered at +1 in the core. All of the data presented here indicate that the attP core comprises two similar but nonidentical Int binding sites, situated in inverted orientation at the core–arm junctions. The proximity of Int to each of these symmetry elements in attP is demonstrated in the methylation protection experiments discussed below. It is not certain whether the smallest entity binding to each of these sites is an Int monomer or a multimer. However, we favor the former since the consensus does not have elements of 2-fold symmetry.
Using the methylation protection technique, we have established that attB also has this same general structure: two binding sites situated in inverted orientation at the core–arm junctions. The specific sequence positions affected in the attB methylation experiments parallel those found in attP, indicating that the mode of Int interaction with the two core regions is similar (see Figure 5).
In previous experiments in which footprint protection techniques were used, only the left (stronger) attB core binding site could be detected (Ross et al., 1979). With the methylation protection assays described here, specific Int interactions can be visualized at higher concentrations of Int than can be used in footprinting, since nonspecific Int interactions do not inhibit methylation by DMS (see Experimental Procedures). Methylation protection was previously reported to discriminate between the specific and nonspecific interactions of a proteolytic fragment of lac repressor when operator specificity was not detectable in filter binding assays (Ogata and Gilbert, 1978).
Strong support for Int recognition of core–arm junction sequences, and for the lack of critical involvement of central core sequences in recognition, is also provided by Int protection of the 30 bp int155 site (Figures 1 and 2). This region contains two sequences, homologous to core junctions, that are inverted and spaced just like those of the attP and attB core sites. There is virtually no homology between the A+T-rich central att core sequence and the G+C-rich central sequence in the int155 site (see Figure 2). These observations are all the more compelling since under some conditions the int155 site is functional in site-specific recombination (in xin-promoted deletions, Gritzmacher and Weisberg, unpublished data; and probably in formation of the att2501 deletion, Ross et al., 1982).
The Four Core Binding Sites Differ
To facilitate discussion of the four junction binding sites, we identify them as the C and C′ sites (for core) at the left and right attP junctions, and the B and B′ sites at the left and right attB junctions (Figure 5). These four sites all differ slightly from one another. The C′ site contains the consensus sequence CAACTTNNT. In sites C, B, and B′ there is at least one mismatch with the consensus (positions 2, 3, or 4; see Figure 3).
It is likely that these sequence differences among the four sites and possibly the differences in the two or three positions adjacent to the consensus which show methylation effects (see Figure 5) are at least partially responsible for the observed differences in their relative affinities for Int. Of the two sites in attB, the left site (B, detectable in nuclease protection experiments; Ross et al., 1979) has a higher affinity for Int. The right site (B′) interaction can be detected only at higher Int concentrations by using the methylation protection assay. In methylation protection experiments carried out under these binding conditions there appears to be roughly a 4-fold difference in the concentration of Int required to detect the two sites (data not shown).
In the attP core, under the conditions used, both sides of attP fill at the same Int concentration. However, when the two attP sites are physically separated, the right binding site appears to differ from the left side. Binding to the right site (C′, in pWR221, Figure 6) or its consensus sequence in non-att DNA (Figure 3), is detectable in footprint assays, whereas the separated left site (C, in pWR243) is detectable only in the methylation protection assays. Thus the affinity for the left site may be enhanced when present together with the right site, suggesting a possible cooperative interaction at these sites. This apparent cooperativity, however, is not sufficient to make the weak right site of attB (B′) visible in footprint protection experiments. (The right and left attB sites have not yet been tested separately.)
It is difficult to determine the degree of recognition flexibility for each position in the consensus sequence, CAACTTNNT. However, this sequence must include the essential features for recognition of a junction-type site. Of the single junction-type sequences in non-att DNA analyzed for Int interaction by nuclease protection, only those sites containing the precise sequence CAACTTNNT gave clear protection patterns (pBR322 sites 3511 and 3755; bio; see Figure 3). Variants of this sequence that are clearly protected by Int are found in close proximity to a second junction-type site (C, B, int155L and int155R). This suggests that one of the factors which may influence recognition flexibility is cooperative interaction of Int at two appropriately spaced junction sites (see above).
In addition, Int interaction may be influenced by local or long range context effects, such as base composition, microconformational states determined by neighboring sequences, or Int interaction at additional sites (junction-type or arm-type, and not necessarily adjacent or detectable). A possible example of context effects is the failure to find Int footprint protection of a CAACTTNNT sequence at the φX174 RFI origin of replication (data not shown). Local context effects might include, for instance, the two or three positions outside the consensus sequence in each core region site that are in close proximity to Int (see Figure 5). It is interesting to note that two of the sites (C′ and B) are each preceded by a GC, and that the Gs on each strand are protected or enhanced in the same ways at each site. One of the non-att sites (pBR322 3755) is also preceded by GC. Each of the other two junction sites (C and B′) is preceded by , in which some of the Gs show enhanced levels of methylation.
These kinds of effects, though they may influence the strength of Int interaction at a particular site, are unlikely to have a significant impact on our basic conclusions, which are supported by the number of independent sites having the same consensus sequence, and by considerable additional data from other types of experiments as discussed below. Context effects have also been noted in the analysis of target recognition sequences for the transposon Tn10 (Halling and Kleckner, 1982).
Our model of two Int binding sites for each core is consistent with other data pertaining to core interaction. In resection experiments with attB, removal of a portion of either of the two Int recognition elements defined in Figure 5 leads to loss of att site function in vitro (Mizuuchi et al., 1981). Of particular significance is the fact that the cut sites for recombination, at positions −2/−3 and +4/+5 (Mizuuchi et al., 1981), occupy identical positions within each of the consensus sequences (see Figure 5). It is interesting that the phosphodiester bond cut by Int connects consensus positions 7 and 8, the only two positions that are unspecified (Figures 3 and 5).
According to our model, the central portion of the core (between the two binding sites) is not recognized by Int protein, and this is also supported by data from other experiments (see also discussion of central core below). In an Int-protected core region, the central bases are accessible to digestion by the small probe NCS; it was the closer juxtaposition of these sensitive sites in the single base deletion mutant att24 that first suggested the model for two core binding sites (Ross et al., 1982).
Secondary att Sites
The most obvious feature of many of the secondary att site sequences reported to date is the conservation of positions homologous to the central attP or attB core sequence, in particular between the positions of the cuts introduced in core by Int (−2/−3 and +4/+5; Mizuuchi et al., 1981; Craig and Nash, personal communication). We have presented data indicating that five of the seven positions in this region between the staggered cuts are not part of the specified consensus critical to the Int binding (Figures 2 and 3), and that the central core sequence is accessible to NCS in the presence of Int (Ross et al., 1982). The conservation of central core sequence in secondary sites must, therefore, reflect some other important function for this region in the recombination mechanism; in this regard the role of DNA–DNA pairing is discussed below.
To determine whether secondary att sites also have junction-type Int recognition sites, we have examined the sequences found at fixed distances from the known (or probable) crossover points (Figure 7 and Table 1). Table 1 lists the known secondary sites together with measures of the degree of homology with attP and attB. The number of positions of homology with the seven conserved positions in the consensus junction-type Int recognition site on each side of the secondary site “cores” is indicated, as is the number of conserved positions in the region between the 7 bp staggered cuts (termed the overlap region).
Figure 7. Int Recognition Sequences in Secondary att Sites.

Sequence positions conserved with attP or attB, or non-aft junction-type binding sequences (Figure 3) are in large letters under the two regions marked INT. Comparison with the attP core junction sites (C and C′) is included since a subcloned attP core region functions well as a “recipient” site (attB analog) (Hsu et al., 1980; W. Bushman and A. Landy, unpublished observations). Positions of the 7 bp staggered cuts in upper (
 ) and lower (
) and lower (
 ) strands of attP and attB are indicated. The overlap region comprises the 7 bp between the cut positions. Secondary site sequences are aligned according to cut positions, as judged by sequence analysis of the secondary prophage sites (see Table 1 for references; sites are in the same order of measured or predicted efficiency as in Table 1). Many secondary sites contain several bases of continuous homology with the attP core, which may include one or both of the cut positions; in these cases, the cuts in the secondary sites are assumed to occur in the same relative positions as in the attP and attB cores. In some cases better sequence matches with the recognition site are found ±1 bp from the standard spacing (see Table 1), but are not indicated here. Sequence homologies in the overlap region are not given special representation, with the exception of the two positions (−1 and +3) that are conserved positions in the Int consensus sequence.
) strands of attP and attB are indicated. The overlap region comprises the 7 bp between the cut positions. Secondary site sequences are aligned according to cut positions, as judged by sequence analysis of the secondary prophage sites (see Table 1 for references; sites are in the same order of measured or predicted efficiency as in Table 1). Many secondary sites contain several bases of continuous homology with the attP core, which may include one or both of the cut positions; in these cases, the cuts in the secondary sites are assumed to occur in the same relative positions as in the attP and attB cores. In some cases better sequence matches with the recognition site are found ±1 bp from the standard spacing (see Table 1), but are not indicated here. Sequence homologies in the overlap region are not given special representation, with the exception of the two positions (−1 and +3) that are conserved positions in the Int consensus sequence.
Table 1.
Conserved Positions in Either of Two Int Binding Sites, or in the Overlap Region of Secondary att Sites
| Matches (of 7)
|
|||||
|---|---|---|---|---|---|
| Secondary att site
|
Eff.
|
Int Site I
|
Int Site II
|
Overlap Region
|
Ref. |
| galT-saf | 10−4 | 6 | 3, +5 | 7 | a |
| b511 ---- | –--- | 6 --- | 4 --- | 7 | b |
| pro A/B | 10−4 | 5 | 4 | 7 | c |
| trp C ---- | 10−5 --- | 3 --- | 4 --- | 6 | d |
| b2 | – | 5 | 4 | 5 | b |
| SC 2669 ---- | – --- | 4 --- | 3, +5 | 6 | e |
| galT | 10−7 | 6 | 3, +5 | 4 | f |
| thrA ---- | 10−7 --- | 5 --- | +3 --- | 4 | g |
| int 155 | – | 5 | 5 | 1 | h |
| SC 2639 ---- | – --- | 4 --- | 3 --- | 4 | e |
| rrnB | 10−10 | 4 | −3 | 5 | i |
| b522 ---- | –--- | 3 --- | −5 --- | 3 | b |
| rbt | – | 4 | 2 | 4 | j |
| SC 2667 ---- | –--- | 5 --- | 3 --- | 2 | e |
| B508 | – | 2 | 3 | 5 | b |
| SC 2666 | – | 5 | 2 | 2 | e |
The number of conserved positions, of a total of seven in each case, is indicated. The seven conserved positions in the Int recognition sequences ( ) are derived from data in Figure 3. Comparison has been made with attP (C and C′) junction sites as well as with attB (B and B′) junction sites, since the subcloned attP core region (without flanking arm sites) functions well as a “recipient” site (attB analog) (Hsu et al., 1980; W. Bushman and A. Landy, unpublished observations). The 7 bp overlap region is defined by the staggered cuts introduced by Int during recombination (Mizuuchi et al., 1981). Efficiencies indicated are summarized in Pinkham et al. (1980) or for galT saf in Weisberg et al. (1983), and are not available for the other sites. The sites are presented in an order reflecting known efficiencies, where available, and efficiencies predicted on the degree of conservation in the three regions. A series of homologous positions which is out of register by ±1 bp may be functionally significant, as suggested by the attP24 mutant, and such alignments are indicated under Int Site II, designated (+) or (−) The attP24 mutant, in which a 1 bp central core deletion brings the two binding sites closer together (Ross et al 1982), recombines with wild-type attB at a frequency of approximately 10−2, considerably higher than that of the best secondary sites (~10−4) The sources of sequence data for the sites are from the references cited as footnotes.
Ross et al (1982); this paper; Gritzmacher and Weisberg (unpublished)
Numerous positions of homology with the Int recognition sequence are observed. The flexibility inherent in Int recognition of junction-type sequence, evident by the variation in the actual att junction sites C, C′, B, and B′, should be kept in mind. It should also be noted that the efficiency of these sites relative to that of attB is extremely low by biochemical criteria (in the range of 10−4 to 10−10; Pinkham et al., 1980). Thus, at the bottom of the list, a rather poor homology with Int recognition sites and central core sequence is to be expected.
There is clear interaction of Int with the only known secondary site we have analyzed for this property, the int155 site (see Figures 1 and 2). We have suggested above that the particular orientation and spacing of two variants of the consensus sequence in this region may stabilize relatively weak Int interaction. We have not observed Int interaction with any of several one- or two-base variants of the consensus sequence found singly in non-att DNA, but cannot rule out weaker Int interactions at such sites that are not detectable in the nuclease protection assay. Interaction of a multimer (e. g., dimer) of Int at such a single variant site could lead to initiation of recombination (a single strand exchange) at this site. Int-promoted resolution of the intermediate so generated might occur at an adjacent sequence (e. g., 7 bp away) with which Int would not otherwise interact. Alternatively, though recombination at secondary sites is Int-dependent (Shimada et al., 1972), it is not known whether these events are mechanistically identical with attP × attB recombination. For example, Int interaction at a single variant site might initiate strand exchange, followed by resolution of the intermediate by other cellular recombination systems. This resolution event might occur at the same nucleotide pair, resulting in a “flush cut” rather than a “staggered cut.” A mechanism of this type could account for the strong preferential recovery of one strand of a mismatched (theoretical) heteroduplex region, such as has been observed for λ insertion at the galT secondary site (Weisberg et al., 1983).
A Model for Two Core Functions
On the basis of our data for Int binding sites at the core–arm junctions, and the significant conservation in secondary att sites of sequence falling between these binding sites (Table 1), we propose that the core region comprises two functionally distinct sequence features: Int binding sites; and a region of sequence homology with the partner site that is probably important for sequence pairing during recombination (the overlap region).
Data from a set of mutant att sites strongly support this model for two core functions (Weisberg et al., 1983). The attP safG core sequence contains three base substitutions in the central core region. This mutant recombines with wild-type attB at a reduced efficiency (10−2). However, when parallel substitutions are made in a partner att site (attB safG), restoring homology between the two sites, recombination efficiency is restored to nearly normal levels. Consistent with our definition of the consensus Int recognition site, and with the recombination properties of the saf mutants, Int binds well to the attP safG core region, as judged by nuclease protection experiments.
A Three-Dimensional Model of Int-Core Interaction
In methylation protection experiments, specific positions (N7 of guanine in the major groove and N3 of adenine in the minor groove) are identified as being in close proximity to the bound protein (Gilbert et al., 1976). The spatial distribution of the Int-protected purine positions in the attP core region is indicated on the B-form DNA model in Figure 8. The positions of cuts introduced by Int during recombination (Mizuuchi et al., 1981; N. Craig and H. Nash, personal communication) are in close proximity across the central major groove. It is along this same 180° “face” of the helix that virtually all of the Int “contact” positions are found in both the C and the C′ binding sites. Three homologous positions in each site are situated near the cut positions (−9, −6, −5 and +7, +8, +11; Figure 8). Other affected purines reflect differences between the two sites.
Figure 8. B-Form DNA Model of the attP Core Region from −15 to +15.

The helix is oriented such that the left or “C” site occupies the lower part of the figure, and the right or “C′” site the upper part. Red atoms = oxygen; yellow = phosphorus; white = hydrogen; black = carbon; blue = nitrogen. Positions of phosphodiester linkages cut by Int during recombination (Mizuuchi et al., 1981; Craig and Nash, personal communication) are indicated by green arrows, with the lower arrow point situated directly below the top strand (−2/−3) linkage, and the upper arrow point directly above the bottom strand (+4/+5) linkage (refer to Figure 5). Purine positions N7 of guanine (major groove) or N3 of adenine (minor groove), at which levels of methylation are affected by bound Int, are marked with labeled yellow disks. All but one of the affected purine positions are on the front 180° of the helix, as shown, G (−13), on the rear of the helix, which shows enhanced methylation, is not visible in this view. Enhanced positions are indicated by vertical arrows on the yellow disks. Three positions in each site, marked additionally with green spots on the yellow disks, indicate conserved sequence positions in the two sites: (−9, −6, −5, in C; and +7, +8, +11, in C′; see also Figure 5).
The data indicate that Int occupies the minor groove adjacent to each cut position, as well as the outer major grooves. In the central major groove, the only G position present (+4), and thus the only position available for study by this technique, is also protected by Int. Two central major groove pyrimidine positions, the Ts at −1 and +3, are conserved in the consensus sequence in symmetrical positions in each binding site (Figure 3). It is interesting to note that the methyl groups of these Ts are situated in the central major groove in very close proximity to the cut positions. The tempting speculation that these Ts are important in the reaction is supported by their conservation in most of the secondary att sites (see Figure 7).
The rear face of the attP helix (not shown) has only one purine showing any effect; the G at −13 at the extreme outer boundary of the left site shows enhanced levels of methylation. Four purine positions in the central minor groove on the rear of the helix (0 through +3) are not affected by Int binding.
In the attB helix (not shown), the G at +15 in the right binding site also shows enhanced methylation levels, and is situated on the rear of the helix at the outer boundary of the site. Additional positions showing enhanced levels of methylation in the presence of Int are found in the attP core helix at −3 and +12, and in attB at −3, −10, −13, and −14. Enhanced methylation levels have also been noted in other protein-DNA interactions, and these effects are probably attributable to increased DMS concentrations in locally hydrophobic areas in the environment of the protein (Johnson et al., 1978; Ogata and Gilbert, 1978; Johnsrud, 1978). However, influences of the protein on the structure and consequent reactivity of the DNA, including local helix unwinding, as suggested for RNA polymerase, may also contribute to these effects (Johnsrud, 1978).
Interactions of several other DNA-binding proteins have also been found in similar experiments to occur primarily along one face of the helix (λ repressor, Humayan et al., 1977; λ cro, Johnson et al., 1978; RNA polymerase and CAP, reviewed in Siebenlist et al., 1980). The crystal structures of two such proteins, cro and CAP, are consistent with this model for interaction, and features of symmetry in the DNA recognition sites probably reflect interaction with symmetrically disposed protein subunits (Anderson et al., 1981; McKay and Steitz, 1981).
One of the interesting questions raised by our results defining Int recognition sites in the core region is whether the observed sequence differences between the four junction sites, and their apparent differences in affinity for Int, are intrinsically significant and reflect features that are important in the detailed mechanism of strand exchange (such as the order of cutting and resealing).
The junction-type consensus sequence, , found in two copies in both the attP and attB core regions, is one of two classes of DNA sequence recognized by Int. The second class of recognition sequence, the arm-type sequence, , is found in five copies in the P and P′ arms of the attP site (Ross and Landy, 1982). (The overlined sequence positions are the most heavily conserved positions in each consensus.) We find no obvious homology between the two sequences, and have suggested that they may reflect two DNA-binding domains of Int (Ross and Landy, 1982). This model is supported by the observations that modification of Int with N-ethylmaleimide results in preferential loss of interaction with arm-type sites (Ross and Landy, 1982); that the non-att DNA sites recognized by Int fall clearly into one of the two classes; and that only attP core region sequences, and not arm sequences, are nicked by Int in in vitro assays (N. Craig and H. Nash, personal communication).
The role of Int interaction with two classes of recognition sequence, one class being found at some distance from the actual site of strand exchange, remains to be clarified. An interesting possibility, which is also consistent with the observation of “intasome” structures in the electron microscope (Better et al, 1982), is that Int monomers interacting with each of the four core junction sites interact simultaneously with arm-type “anchor” sequences. Such a complex might align the recombining core sequences and/or facilitate strand exchange.
Experimental Procedures
Preparation of Plasmid DNA and Labeled Restriction Fragments
Plasmid DNA was extracted from cells grown to OD650 0.5 in 1 × M9, 1 mM MgS04, 4 μg/ml B1, 0.5% glucose, 0.2% casamino acids, then grown for 15–18 hr with chloramphenicol added to 200 μg/ml. Pelleted cells were resuspended in 20% sucrose, 50 mM Tris-Cl (pH 8.1), and lysozyme (in 25 mM Tris-Cl [pH 8.1]) was added to 1.1 mg/ml and incubated on ice for 5 min. EDTA was added to 2 mM, followed by addition of an equal volume of 0.2% Triton X in 2.5 mM EDTA, 50 mM Tris-Cl (pH 8.1), and the mixture was incubated at 20°C for 5 min. Following centrifugation, the supernatant was adjusted to 1 M NaCl, extracted with phenol, then ethanol-precipitated. DNA was banded in a CsCl-ethidium bromide gradient, ethidium bromide was extracted with isobutanol, and the DNA was pelleted in a Spinco Ti60 rotor, at 36,000 rpm for 15–18 hr.
Restriction fragments, 32P-labeled at either the 5′ end or the 3′ end, were used in footprinting and methylation protection experiments. Restriction enzymes were from New England BioLabs. The 5′ fragment ends were labeled by using polynucleotide kinase (Boehringer Mannheim or P-L Biochemicals) and γ-32P-ATP (New England Nuclear) according to the procedures of Maxam and Gilbert (1980). The 3′ ends were labeled by using the Klenow fragment of E. coli DNA polymerase I (Boehringer). Following restriction enzyme digestion, 30 μg of plasmid DNA was phenol-extracted and ethanol-precipitated. The fragment was incubated for 30 min at 37°C in 30 μl 10 mM Tris-Cl (pH 7.9), 6 mM MgCl2, 6 mM β-mercapto-ethanol, 60 mM NaCl, with 100 μM each of the unlabeled deoxynucleotide triphosphates (Sigma), 2.5 μM α-32P-labeled deoxynucleotide triphosphate (New England Nuclear), and 2 U of Klenow fragment.
The φX174 RFI DNA was purchased from New England BioLabs. Plasmid sources included: pBR322 (Sutcliffe, 1979); pWR1, the Hind III (−250)-Bam (+242) attP-containing fragment inserted into the Hind III and Bam sites of pBR322; pMJB11 (M. Buraczynska, unpublished), attP DNA from −70 to +46 inserted into the Hind III and Bam sites of pBR322 with a Hind III linker at −70 and a Bam linker at +46; pPH56 (Hsu et al., 1980). Plasmids pWR243, pWR221, and pPH411 are described below.
Nuclease Protection Experiments
Protection experiments with DNAase I (Worthington) or NCS (gift from W. Haseltine) were carried out as described previously (Ross et al., 1979, 1982; Ross and Landy, 1982). Int protein was purified by N. Hasan according to a modification of the procedure of Kikuchi and Nash (1979a) (Hasan and Landy, unpublished data).
Construction of Clones Containing Single attP Junction-type Binding Sites
The strategy for cloning separated left and right attP junction binding sites utilized a Dde restriction site in the right side of the attP core (CTAAG, +4 to +8). To obtain each cloned site separately, the Dde fragment containing the second site was eliminated from plasmid pMJB11 (see above). To clone the left site (C) and eliminate the right site (C′), the 1245 bp Dde fragment (+4 in att to 1580 in pBR322) was removed; to clone the right site and eliminate the left site, the 175 bp Dde fragment (4290 in pBR322 to +4 in att) was removed. To remove these fragments, pMJB11 partial Dde digestion products of the correct length (lacking either of the fragments described) were gel-purified, circularized by using T4 DNA ligase (New England BioLabs), and used to transform strain HB101. Ampicillin-resistant clones were screened for a Dde profile lacking the correct fragment. The att region sequences of the clones of interest, pWR243, a left site (C) clone, and pWR221, a right site (C′) clone, were determined by the procedure of Maxam and Gilbert (1980). These clones contain precise religation of the Dde sites described (see Figure 6). A second right binding site clone, pPH411, was constructed by utilizing the Alu site at the left core-arm junction (AGCT, −8 to −5), by inserting an Alu core-containing fragment from pMJB11 (−6 to 681 in pBR322) into the Hinc II site of plasmid pKT21 (P.-L. Hsu, unpublished data).
Methylation Protection
Procedures were adapted from chemical sequence methods (Maxam and Gilbert, 1977, 1980) and methylation protection experiments (Gilbert et al., 1976). The N7 position of guanine and the N3 position of adenine were methylated at a DMS concentration affecting approximately one purine per DNA molecule, as for chemical sequencing. The presence of specifically bound protein results in either reduced or enhanced methylation levels at specific purine positions. The release of both methylated purines was accomplished by the acid-release procedure of Maxam and Gilbert (1977).
Labeled DNA fragment (approximately 5–10 ng, or 0.01 pmole) was incubated for 20 min at 20°C with a range of concentrations of purified Int (3.0 μg/ml to 0.25 μg/ml) in 200 μl 50 mM Na-cacodylate (pH 8.0), 60 mM KCl, 1 mM EDTA, 2 mg/ml BSA, 1 mM β-mercaptoethanol and 10% glycerol. Of 10.7 M DMS, (Aldrich Chemical Co.), 1.2 μl was then added, and the incubation continued at 20°C for 5 min. Then 300 μl of ice-cold phenol and 50 μl of 1 M Tris-acetate (pH 7.5), 1 M β-mercaptoethanol, 1.5 M Na-acetate. 50 mM Mg-acetate, 1 mM EDTA, and 200 μg/ml sonicated salmon-sperm DNA (Sigma) were added. Samples were vortexed and chilled immediately, and following brief centrifugation the aqueous phase was ethanol-precipitated, and the pellet was washed with 95% ethanol, dried, and resuspended in 20 μlH20. Then 5 μl of 1 M perchloric acid was added, and the samples were kept on ice water for 2 hr. Following ethanol precipitation, samples were incubated in 0.5 M piperidine, as for chemical sequence procedures (Maxam and Gilbert, 1980), and electrophoresed as described previously (Ross et al., 1979, 1982).
Partial DNAase I digests of fragments incubated with Int under these conditions (incubation buffer described above, but in 1 mM MgCl2 with no EDTA) yielded footprints comparable to those reported previously (Ross et al., 1979, 1982; Ross and Landy, 1982). Methylation reactions carried out in 1 mM MgCl2 gave results identical with those in 1 mM EDTA.
Protection against methylation by DMS was observed at Int concentrations comparable to those required for DNAase I or NCS footprint protection. The same specific sites are also selectively protected against methylation at much higher Int concentrations. At these high concentrations DNAase I or NCS digestion is completely inhibited by nonspecific Int binding. Partial DNAase I digestions carried out at high Int concentrations in the presence of DMS indicated that nonspecific binding is not altered by the presence of DMS. Thus nonspecific Int interactions do not significantly alter the rate or relative extents of methylation in nonspecific sequence regions. For this reason, specific Int interactions that are obscured in footprint experiments are detectable in methylation protection experiments.
Acknowledgments
We thank Noaman Hasan for purified Int protein, Bob Weisberg and Chris Gritzmacher for providing the results on xin-promoted deletions prior to publication, and Kiyoshi Mizuuchi for plasmid pMM291. Lina Vargas, Helen Flynn, and Eva Deignan contributed expert technical assistance. This work was supported by grants from the National Institutes of Health, PHS Al 13544, and from the National Foundation, March of Dimes, 1-543, to A. L. W. R. was supported by a National Research Service Award, NIH 5 T32 GM 07601, from the National Institute of General Medical Sciences.
References
- Anderson WF, Ohlendorf DH, Takeda Y, Matthews BW. Structure of cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981;290:754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Better M, Lu C, Williams RC, Echols H. Site-specific DNA condensation and pairing mediated by the Int protein of bacteriophage λ. Proc Nat Acad Sci USA. 1982;79:5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell K, Landy A. Structural features of X site-specific recombination at a secondary att site in ga/T. Cell. 1979;16:397–406. doi: 10.1016/0092-8674(79)90015-1. [DOI] [PubMed] [Google Scholar]
- Chapman J, Gardner JF. Secondary lambda attachment site in the threonine operon attenuator of Escherichia coli. J Bacteriol. 1981;146:1046–1054. doi: 10.1128/jb.146.3.1046-1054.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Platt T. A secondary attachment site for bacteriophage λ in trpC of E. coli. Cell. 1979;16:407–413. doi: 10.1016/0092-8674(79)90016-3. [DOI] [PubMed] [Google Scholar]
- Csordas-Toth E, Boros I, Venetianer P. Nucleotide sequence of a secondary attachment site for bacteriophage lambda on the Escherichia coli chromosome. Nucl Acids Res. 1979;7:1335–1341. doi: 10.1093/nar/7.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea AD, Haseltine W. Sequence specific cleavage of DNA by the anti-tumour antibiotics neocarzinostatin and bleomycin. Proc Nat Acad Sci USA. 1978;75:3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Maxam A, Mirzabekov A. Contact between the lac repressor and DNA revealed by methylation. In: Kjeldgaard NC, Maaløe O, editors. In Control of Ribosome Synthesis, Alfred Benson Symposium. Vol. 9. Copenhagen; Munksgaard: 1976. pp. 139–148. [Google Scholar]
- Gottesman S. Lambda site-specific recombination; the att site. Cell. 1981;25:585–586. doi: 10.1016/0092-8674(81)90165-3. [DOI] [PubMed] [Google Scholar]
- Halling SM, Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Hsu PL, Ross W, Landy A. The λ phage att site: functional limits and interaction with Int protein. Nature. 1980;285:85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humayan Z, Kleid D, Ptashne M. Sites of contact between lambda operators and lambda repressor. Nucl Acids Res. 1977;4:1595–1607. doi: 10.1093/nar/4.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Meyer B, Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc Nat Acad Sci USA. 1978;75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Nat Acad Sci USA. 1978;75:5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Nash HA. Integrative recombination of bacteriophage lambda: requirement for super twisted DNA in vivo and characterization of Int. Cold Spring Harbor Symp Quant Biol. 1979a;43:1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Nash HA. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Nat Acad Sci USA. 1979b;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Ann Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Landy A, Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977;197:1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A, Hoess RH, Bidwell K, Ross W. Site-specific recombination in bacteriophage lambda: structural features of recombining sites. Cold Spring Harbor Symp Quant Biol. 1979;43:1089–1097. doi: 10.1101/sqb.1979.043.01.121. [DOI] [PubMed] [Google Scholar]
- Loviny T, Neuberger MS, Hartley BS. Sequence of a secondary phage lambda attachment site located between the pentitol operons of Klebsiella aerogenes. Biochem J. 1981;193:631–637. doi: 10.1042/bj1930631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A, Gilbert W. A new method for sequencing DNA. Proc Nat Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Miller HI, Friedman DI. An E. coli gene product required for λ site-specific recombination. Cell. 1980;20:711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Miller HI, Kikuchi A, Nash HA, Weisberg RA, Friedman DI. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harbor Symp Quant Biol. 1979;43:1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M, Mizuuchi K. Integrative recombination of bacteriophage lambda: extent of DNA sequence involved in attachment site function. Proc Nat Acad Sci USA. 1980;77:3220–3224. doi: 10.1073/pnas.77.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K, Weisberg R, Enquist L, Mizuuchi M, Buraczynska M, Foeller C, Hsu PL, Ross W, Landy A. Structure and function of the phage lambda att site: size, Int-binding sites and location of the crossover point. Cold Spring Harbor Symp Quant Biol. 1981;45:429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Nash HA. Site-specific recombination protein of phage lambda. In: Boyer PD, editor. The Enzymes. 3. Vol. 14. New York: Academic Press; 1981a. pp. 471–480. [Google Scholar]
- Nash HA. Integration and excision of bacteriophage lambda: the mechanism of conservative site specific recombination. Ann Rev Genet. 1981b;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Nash HA, Robertson CA. Purification and properties of the E. coli protein factor required for lambda integrative recombination. J Biol Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- Ogata RT, Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Nat Acad Sci USA. 1978;75:5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham JL, Platt T, Enquist LW, Weisberg RA. The secondary attachment site for bacteriophage lambda in the pro A/B gene of Escherichia coli. J Mol Biol. 1980;144:587–592. doi: 10.1016/0022-2836(80)90339-3. [DOI] [PubMed] [Google Scholar]
- Ross W, Landy A. Int recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Nat Acad Sci. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Landy A, Kikuchi Y, Nash H. Interaction of Int protein with specific sites on λ att DNA. Cell. 1979;18:297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Shulman M, Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982;156:505–529. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- Shimada K, Weisberg RA, Gottesman ME. Prophage λ at unusual chromosomal locations I. Location of the secondary attachment sites and properties of the lysogens. J Mol Biol. 1972;63:483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Simpson RB, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion, recombination and gene expression involving the bacteriophage lambda attachment site. J Mol Biol. 1981;152:517–533. doi: 10.1016/0022-2836(81)90266-7. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harbor Symp Quant Biol. 1979;43:77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Weisberg RA, Landy A. Site-specific recombination in phage λ. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1983. in press. [Google Scholar]
- Weisberg RA, Enquist LW, Foeller C, Landy A. A role for DNA homology in site-specific recombination: the isolation and characterization of a site affinity mutant of coliphage λ. J Mol Biol. 1983 doi: 10.1016/s0022-2836(83)80151-x. in press. [DOI] [PubMed] [Google Scholar]


