Abstract
There is considerable evidence for the induction of different phenotypes by variations in the early life environment, including nutrition, which in humans is associated with graded risk of metabolic disease; fetal programming. It is likely that induction of persistent changes to tissue structure and function by differences in the early life environment involves life-long alterations to the regulation of gene transcription. This view is supported by both studies of humans and animal models. The mechanism which underlies such changes to gene expression is now beginning to be understood. In this review we discuss the role of changes in the epigenetic regulation of transcription, specifically DNA methylation and covalent modification of histones, in the induction of an altered phenotype by nutritional constraint in early life. Demonstration of altered epigenetic regulation of genes in phenotype induction suggests the possibility of interventions to modify long-term disease risk associated with unbalanced nutrition in early life.
Keywords: Epigenetic, fetal programming, metabolic disease, DNA methylation, DNA methyltransferase
Introduction
Epidemiological studies (for review see Godfrey & Barker 2001) and experiments using animal models (for review see Bertram & Hanson, 2001; Armitage et al. 2004; 2005) show that both under and over nutrition during pregnancy and / or lactation induce stable alterations to the physiological and structural phenotype of the offspring. This process has been termed fetal programming, but perhaps is better described as phenotype induction because ‘programming’ has connotations of the genetic programme for development and is deterministic (Gluckman & Hanson, 2005). Induced phenotypic traits determine the ability of the organism to respond to changes in its environment within the reaction norm (Gluckman and Hanson 2004a) and are of particular interest because the health and Darwinian fitness of the organism depend on its ability to mount appropriate responses to environmental changes (Bateson et al. 2004; Gluckman & Hanson, 2004b). Developmental cues, including nutrition, which operate within the normal range for the human population induce graded changes in risk of metabolic and cardiovascular disease (Godfrey & Barker, 2001). Analogous effects have been produced in laboratory animals. Possibly the best known examples are induction of differences in blood pressures and insulin sensitivity in the offspring of rats fed different amount of protein during pregnancy (Langley & Jackson, 1994; Burns et al. 1997). It has been proposed that the change in development induced in the fetus is used to predict the postnatal environment (Gluckman & Hanson, 2004b). However, if the offspring does not predict correctly the environment experienced after birth, then it is at increased risk of developing cardiovascular and metabolic disease because its homoeostatic capacity is mismatched to that environment. The graded nature of the response of the fetus to nutritional cues from the mother potentially gives rise to a large number of phenotypes from one genotype so that at least a proportion of the offspring will be able to survive to reproductive age. A discussion of the evolutionary significance of such predictive adaptive responses is given in Gluckman et al. (2005). We now know that the phenotype of the offspring is also dependent on the timing of the nutritional cue during development, at least in humans and rats (Ravelli et al. 1998; Remacle et al. 2004).
The mechanism by which cues about nutrient availability in the postnatal environment are transmitted to the fetus and the process by which different, stable phenotypes are induced are beginning to be understood. The purpose of this review is to discuss the results of recent studies of the role of epigenetic regulation of genes in the induction of an altered fetal phenotype by maternal nutrition during pregnancy.
Phenotype induction and gene transcription
The induction of changes to the phenotype of the offspring that persist throughout the lifespan implies stable changes to gene transcription resulting in altered activities of metabolic pathways and homeostatic control processes, and in differences in the structure of tissues. The latter may result from changes in stem cell allocation to various lineages, variations in the rate and / or number of mitosis, the extent of apoptosis and / or differences in the expression of genes that encode key structural proteins. Together these processes potentially provide cellular and molecular explanations for variation between individuals in body structure and metabolic capacity (Wootton & Jackson, 1996). In women, differences in body structure and metabolic capacity induced by the environment experienced before birth may, in turn, influence the timing of their sexual maturation (Gluckman & Hanson, 2006), their reproductive success (Davies, 2006) and the birth weight of their children (Catalano & Kirwan, 2001). Thus phenotypic changes induced in one generation may be passed to more than one successive generation. We will discuss later how phenotypic traits can be transmitted between generations by altered regulation of transcription in parallel to or as an alternative to constraint in the reproductive capacity of the mother.
There are several studies which have investigated the effect of maternal nutrition during pregnancy and/ or lactation on gene expression in the offspring in animal models. These studies have used a candidate gene approach to identifying the molecular basis for changes in activities of metabolic and endocrine pathways, with a specific focus on corticosteroid activity, and carbohydrate and lipid metabolism. Feeding a protein-restricted (PR) diet to pregnant rats induced increased glucocorticoid receptor (GR) expression and reduced expression of the enzyme which inactivates corticosteroids, 11β-hydroxysteroid dehydrogenase type II, in liver, lung, kidney and brain in the offspring (Bertram et al. 2001). In the liver, increased GR activity up-regulates phosphoenolpyruvate carboxykinase (PEPCK) expression and activity, and so increases capacity for gluconeogenesis. This may contribute to the induction of insulin resistance in this model (Burns et al. 1997). Altered expression of GR has also been reported in the lung, liver, adrenal gland and kidney of the offspring of sheep fed a restricted diet during pregnancy (Whorwood et al. 2001; Brennan et al. 2005; Gnanalingham et al. 2005). Feeding a PR diet to pregnant rats also up-regulates glucokinase (GK) expression in the liver of the offspring which implies increased capacity for glucose uptake (Bogdarina et al. 2004).
Restricting maternal protein intake during pregnancy and / or lactation in rats also alters the expression of genes involved in lipid homeostasis. Expression of acetyl-CoA carboxylase and fatty acid synthase was increased in the liver of the offspring of rats fed a PR diet during pregnancy and lactation (Maloney et al. 2003). The offspring of rats fed a PR diet during pregnancy also show increased blood triacylglycerol (TAG) and non-esterified fatty acid (NEFA) concentrations (Burdge et al. 2004). Peroxisomal proliferator-activated receptor (PPAR) -α expression was increased in the liver of the offspring of rats fed a PR diet during pregnancy and was accompanied by up-regulation of its target gene acyl-CoA oxidase (AOX), while PPARγ1 expression was unchanged (Burdge et al. 2004, Lillycrop et al. 2005). In contrast, in adipose tissue the expression of the adipose specific isoform of PPARγ (PPARγ2) was reduced (Burdge et al. 2004). The different effects of maternal PR diet on PPARγ expression in liver and adipose tissue may reflect the fact that the PPARγ isoforms PPARγ1 and PPARγ2 expressed in liver and adipose tissue, respectively, are generated from different promoters. If so, this suggests a mechanism by which the process that induces altered gene expression, and hence phenotype, may result in different effects on expression of the same gene in different tissues. Increased PPARα expression would be expected to increase TAG clearance. However, increased hepatic TAG synthesis may result from increased flux of NEFA from adipose tissue as a result of reduced expression of PPARγ expression (Burdge et al. 2004) and insulin resistance (Burns et al. 1997), and so may have exceeded the capacity of fatty acid clearance pathways regulated by PPARα. Overall, the offspring of dams fed a PR diet during pregnancy show impaired lipid homeostasis.
The offspring of dams fed a PR diet during pregnancy also show lower concentration of the n-3 polyunsaturated fatty acid docosahexaenoic acid (DHA) in liver and brain which is required for the biophysical and functional properties of cell membranes, in particular in the central nervous system (Burdge et al. 2003). As DHA was not provided in the diet fed to the offspring, these rats were dependent upon synthesis from the precursor α-linolenic acid. Since the rate limiting enzyme for this conversion, Δ6-desaturase, is negatively regulated by PPARα (Tang et al. 2003), increased PPARα expression in the PR offspring may also contribute to lower DHA status.
One important consideration in understanding the mechanism responsible for phenotype induction is the interaction between any process resulting in different phenotypes, environmental cues and gene polymorphisms, in particular those located in gene promoters. It is possible that individuals with different variants of the same gene may differ in their response to the early life environment. For example, Eriksson et al. (2002) showed that increased risk of insulin resistance in adults was only associated with lower birth weight in individuals who had the Pro12Ala or Ala12Ala genotype of the PPARγ2 gene. The substitution of proline to alanine reduces the transcriptional activity of the gene. The Pro12Ala polymorphism was also only associated with increased risk of dyslipidaemia in men and women who were born at the lower end of the normal range of birth weight (Eriksson et al. 2003). Furthermore, Dennison et al. (2004) report a significant interaction between weight at 1 year of age and rate of bone loss in 61 to 73 year old adults who were homozygous for polymorphisms in promoter and coding region of the growth hormone gene. These findings demonstrate that differences in the structure of key regulatory genes can have profound effect the impact of the early life environment on disease risk in later life.
Although the number of genes studied so far is limited, stable effects of nutrient restriction on transcription have been demonstrated. Importantly, some of the genes which show altered expression following prenatal under-nutrition are transcription factors which affect multiple pathways in development and nutrient homeostasis; for example PPARs and GR. Thus by modifying the expression of a few key transcription factors, the process by which maternal nutrition alters the phenotype of the offspring may alter a large number of metabolic and developmental pathways.
Epigenetic regulation of transcription
The remainder of this review will discuss how epigenetic regulation of genes may provide a molecular mechanism for phenotype induction. The methylation of CpG dinucleotides, which are clustered at the 5' promoter regions of genes, confers stable silencing of transcription (Bird, 2001). A diagram showing a simplified mechanism for the effect of cytosine methylation on transcription is shown in Figure 1. Methylation patterns are largely established during embryogenesis or in early postnatal life. Following fertilisation, maternal and paternal genomes undergo extensive demethylation. This is followed by de novo methylation just prior to implantation (Bird, 2001; Reik et al. 2001). About 70% of CpGs are methylated, mainly in repressive heterochromatin regions and in repetitive sequences such as retrotransposable elements (Yoder et al. 1997). Promoter methylation is important for asymmetrical silencing of imprinted genes (Li et al. 1993) and retrotransposons (Walsh et al. 1998; Waterland & Jirtle, 2003). DNA methylation also plays a key role in cell differentiation by silencing the expression of specific genes during the development and differentiation of individual tissues. For example, the expression of Oct-4, a key regulator of cellular pluripotency in the early embryo, is permanently silenced by hypermethylation of its promoter around E6.5 in the mouse (Gidekel et al. 2002), while HoxA5 and HoxB5 which are required for later stages of development are not methylated and silenced until early postnatal life (Hershko et al. 2003). For some genes there also appear to be gradations of promoter demethylation associated with developmental changes in role of the gene product. The δ-crystallin II and PEPCK promoters are methylated in the early embryo but undergo progressive demethylation during fetal development, and are hypomethylated compared to the embryo and expressed in the adult (Grainger et al. 1983; Benvenisty et al. 1985). Thus changes in methylation which are associated with cell differentiation and functional changes are established at different times during development of the embryo.
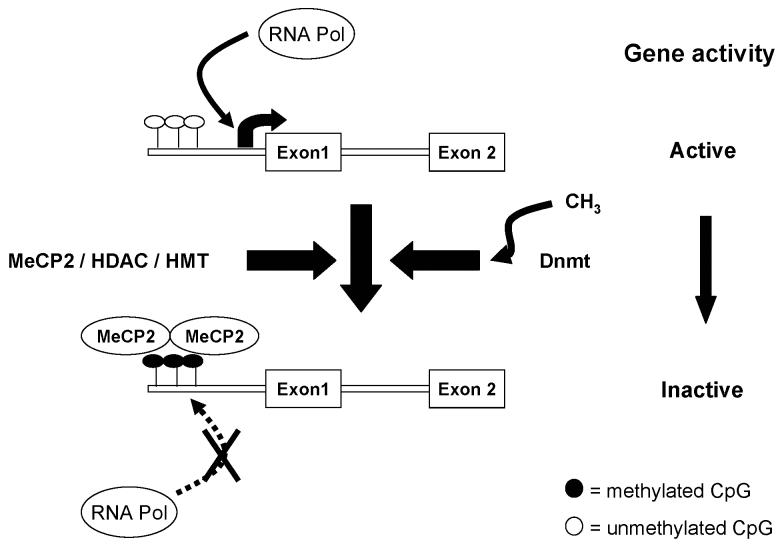
Fig. 1.
Gene silencing by DNA methylation. When CpG dinucleotides are unmethylated in the promoter, RNA polymerase (Pol) can bind and the coding region is transcribed. Methylation of CpGs by the activity of DNA methyl transferases (Dnmt) enables recruitment of methyl CpG binding protein-2 (MeCP2) which in turn recruits the histone deacetylase (HDAC)/ histone methyl transferase (HMT) complex. The HDAC/HMT complex removes acetyl groups from histones and methylates specific lysine residues. The overall effect of DNA and histone methylation is to induce long-term silencing of transcription.
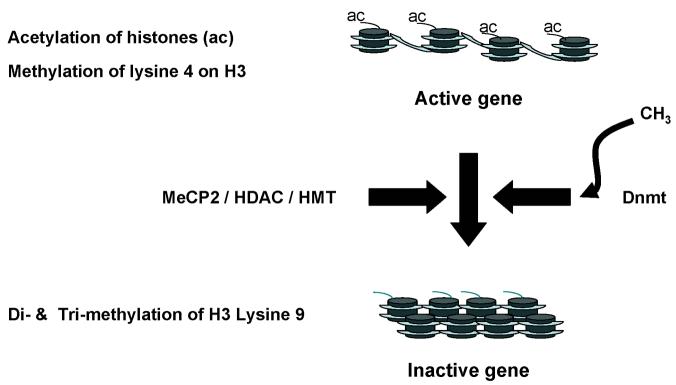
DNA methylation can induce transcriptional silencing by blocking the binding of transcription factors and/ or through promoting the binding of the methyl CpG binding protein (MeCP2). The latter binds to methylated cytosines and, in turn, recruits histone modifying complexes to the DNA (Fuks et al. 2003). MeCP2 recruits both histone deacetylases (HDACs), which remove acetyl groups from the histones, and histone methyl transferases (HMTs) which methylate lysine 9 on H3, resulting in a closed chromatin structure and transcriptional silencing (Fuks et al. 2003). Covalent modifications to histones such as acetylation and methylation influence chromatin structure and hence the ability of the transcriptional machinery to gain access to DNA (Figure 2). Acetylation which occurs at specific lysine residues in the histone tails is generally associated with transcriptional activity (Turner, 2000). In contrast, methylation of lysines has been linked to either activation or repression depending on which lysine residue is modified. Methylation of histone H3-lysine 4 (K4) promotes transcriptional activity and euchromatin (Strahl et al. 1999; Lachner et al. 2001; Zegerman et al. 2002). In contrast, di and tri -methylation of lysine 9 on histone H3 has been shown to be marker of heterochromatin in a range of organisms from yeast to mice (Litt et al. 2001; Nakayama et al. 2001).
Fig. 2.
In euchromatin, packing of histone is loose due to acetylation of lysine residues in the N-terminal domains. This facilitates transcription. Recruitment of the histone deacetylase (HDAC)/ histone methyl transferase (HMT) complex results in deacetylation and methylation of specific lysine residues histones which results in closer packing of histones and transcriptionally inactive heterochromatin.
Since epigenetic regulation of gene promoters which is established during development and is retained throughout the lifespan of the organism confers patterns of transcriptional expression and silencing, perturbations to such processes constitute a strong candidate molecular mechanism for induction of persistent alterations in phenotype.
Links between maternal nutritional and epigenetic regulation of transcription in the offspring
Two non-nutritional models have shown that the prenatal and neonatal environments modify the epigenetic regulation of specific genes. In an elegant study of the effect of maternal behaviour during suckling on the development of stress response in the offspring, Weaver et al. (2004) showed that pups raised by rat dams which showed poorer nurturing had an increased stress response. The effect was due to hypermethylation of specific CpG dinucleotides within the promoter of the GR gene in the hippocampus of the offspring. These changes were reversed in adult brain by intra-cranial administration of the histone deacetylase inhibitor Trichostatin A and L-methionine (Weaver et al. 2005; 2006). Others have shown that ligation of a uterine artery in the rat decreases p53 expression in the kidney of the offspring, associated with increased apoptosis and reduced nephron number (Pham et al. 2003). Such reports set the scene for consideration of the effect of maternal nutrition during pregnancy on the role of epigenetic regulation of transcription in determining the phenotype of the offspring.
Early nutrition and the epigenetic regulation of transposons and imprinted genes
The effects of early nutrition on the epigenetic regulation of transposable elements and on the expression of imprinted genes, in particular insulin-like growth factor (IGF) -2, has been the subject of a number of studies. The findings of these investigations will be summarised here as they are described in detail in Waterland and Jirtle (2004). Mouse embryos cultured in Whitten's medium without amino acids showed bi-allelic expression of the H19 gene, while those cultured in medium containing amino acids showed mono-allelic expression (Doherty et al. 2000). This indicates loss of methylation of an upstream regulatory region of the H19 gene in the embryos cultured in the amino acid deficient medium. Differential methylation of the IGF-2 and H19 genes also occurred when embryos were cultured with or without fetal calf serum (Khosla et al. 2001). In humans, in vitro fertilisation using the intracytoplasmic sperm injection technique is associated with increased risk of Angelman's syndrome (Cox et al. 2002; Orstavik et al. 2003) and Beckwith-Weidemann syndrome (DeBaun et al. 2003) due to loss of methylation of regulatory regions of the UBE3A, and H19 and IGF-2 genes, respectively (Cox et al. 2002; DeBaun et al. 2003). This may reflect epigenetic effects of short-term culture on the embryos prior to transfer to the mother. Such alterations to the epigenetic regulation of imprinted genes produce dramatic alterations to the phenotype of the offspring which are evident in early life and so contrast with the phenotypes induced by variations in maternal nutrition during pregnancy which are more subtle and only become clinically apparent after the neonatal period in childhood or adulthood (Gluckman & Hanson 2004b).
Differences in maternal intake of nutrients involved in 1-carbon metabolism during pregnancy in the agouti mouse induce differences in the coat colour of the offspring. Supplementation of the NIH-31 diet fed to pregnant mice with betaine, choline, folic acid and vitamin B12 shifted the distribution of coat colour of the offspring from yellow (agouti) to brown (pseudo-agouti) (Wolff et al. 1998). This shift is due to increased methylation of seven CpG islands 600 bp downstream of the Avy intracisternal-A particle insertion site which acts as a cryptic promoter directing the expression of the agouti gene (Waterland & Jirtle, 2003). Moreover, differential methylation of the seven CpG islands was associated with gradations in coat colour between agouti and pseudo-agouti. Thus maternal intake of nutrients involved in 1-carbon metabolism can induce graded changes to DNA methylation and gene expression in the offspring which persist into adulthood. However, for this process to operate in the induction of phenotypes associated with metabolic disease the effects of maternal nutrition would need to operate within the range typical for a population and, in general, do not involve retrotransposons.
The rat maternal dietary protein restriction model
Feeding a PR diet to pregnant rats is a well-established model of phenotype induction in the offspring which exhibit some of the characteristics of the metabolic syndrome in humans (Bertram & Hanson, 2001; Armitage et al. 2004). Feeding a PR diet to rats during pregnancy induces hypomethylation of the PPARα and GR promoters and increased expression of the GR and PPARα in the liver of the recently-weaned offspring (Lillycrop et al. 2005). This study shows for the first time that, in contrast to modifying the maternal intake of nutrients associated with 1-carbon metabolism (Waterland & Jirtle, 2003), stable changes to the epigenetic regulation of the expression of transcription factors, and hence a phenotype, can be induced in the offspring by modest changes to maternal intake of a macronutrient during pregnancy. The interaction between maternal protein intake and 1-carbon metabolism will be discussed below. The expression of the PPARα and GR target genes, AOX and PEPCK, respectively, was also increased which supports the suggestion that such altered epigenetic regulation of transcription factors modifies the activities of important metabolic pathways (Lillycrop et al. 2005; 2006a). Methylation of the GR and PPARα promoters was also reduced in the heart (Lillycrop et al. 2006b) and the PPARα promoter was hypomethylated in the whole umbilical cord (Burdge – unpublished observation) in the offspring of rats fed a PR diet during pregnancy (Figure 3). Thus in all the tissues examined so far methylation of PPARα and GR promoters was reduced in the offspring of dams fed a PR diet. One possible explanation is that hypomethylation of the GR and PPARα promoters was induced in the early embryo before the different cell lineages began to differentiate. Hypomethylation of the GR promoter has also been found in the offspring of mice fed a PR diet during pregnancy (Figure 3) (Slater-Jefferies – unpublished observation) which suggests that the effect of the PR diet may not be specific to one species. The fundamental role of changes in the regulation of transcription factor expression in altering the activity of pathways controlled by target genes is underlined by the observation that although GK expression was increased in the liver of the PR offspring, this was not accompanied by changes to the methylation status of the GK promoter (Bogdarina et al. 2004). Since GR activity increases GK expression, greater GK expression in the PR offspring may have been due to increased GR activity as a result of hypomethylation of the GR promoter (Lillycrop et al. 2005) rather than a direct effect of prenatal under-nutrition of GK.
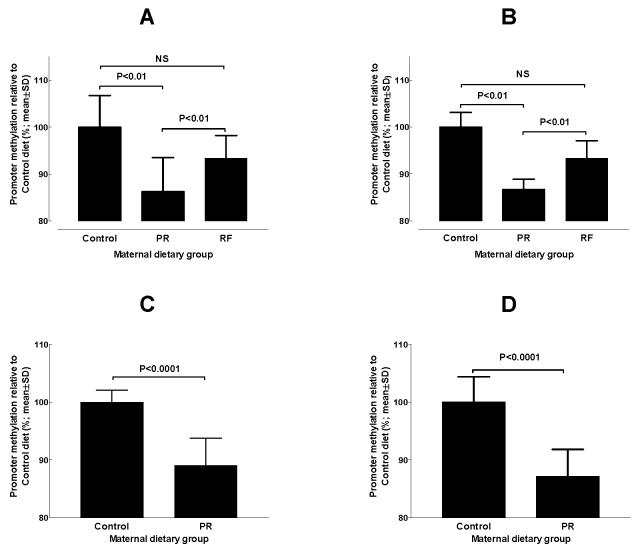
Fig. 3.
The effect of feeding a protein-restricted diet during pregnancy in rats and mice on the methylation status of promoters in the offspring. (A) PPARα promoter methylation status and (B) glucocorticoid receptor (GR) promoter methylation status in the heart of rat offspring on postnatal day 34. (C) PPARα methylation status in rat umbilical cord at post-conceptual day 20. (D) GR promoter methylation status in mouse liver at 3 months after birth. Maternal dietary groups were Control, 18% (w/w) protein; protein-restricted (PR), 9% (w/w) protein; the PR diet supplemented with 5-fold more folic acid (RF). Analysis of promoter methylation was as described by Lillycrop et al. (2005).
The precise biological meaning of a proportional difference in promoter methylation is unclear. For example, the methylation status of the GR promoter in the liver of the PR offspring is 23% lower than the control group (set at 100% for convenience) (Lillycrop et al. 2005). One possible interpretation is that the relative methylation of GR promoter between the two groups of offspring is related to a difference in the proportion of cells in which GR is transcribed. Thus the reduction in methylation status could be interpreted as the induction of GR transcription in a greater proportion of cells in the liver in relation to the control offspring. If so, this implies a metabolic shift within the tissue and a change in metabolic capacity. Although this suggestion awaits experimental investigation, it is consistent with the induction of gluconeogenesis in a subpopulation of cells in the liver of the PR offspring (Burns et al. 1997).
Hypomethylation of the GR was associated with an increase in histone modifications at the GR promoter which facilitate transcription, i.e. acetylation of histones H3 and H4 and methylation of histone H3 at lysine K4, while those that suppress gene expression were reduced or unchanged (Lillycrop et al. 2006a). While this may be primarily the result of reduced binding of the methyl CpG binding protein (MeCP) -2 to the GR promoter because of the reduced level of DNA methylation, reduced MeCP2 expression may also have contributed to higher levels of histone acetylation.
Induction of an altered phenotype (hypertension and endothelial function) in the offspring of rats fed PR diet during pregnancy was prevented by supplementation of the PR diet with glycine or folic acid (Jackson et al. 2002; Brawley et al. 2004). Hypomethylation of the hepatic GR and PPARα promoters was also prevented by addition of 5-fold more folic acid to the PR diet (Lillycrop et al. 2005). Thus 1-carbon metabolism plays a central role in the induction of an altered phenotype by maternal dietary restriction as it does in the Agouti mouse (Waterland & Jirtle, 2004). This will be discussed in more detail below.
Promoter methylation may be induced during the early development of the embryo or during the differentiation of individual tissues (Bird, 2002). Tissue differentiation is also associated with a progressive decrease in the methylation of specific genes (Grainger et al. 1983; Benvenisty et al. 1985). Such variation in the timing and direction of methylation events provides a possible mechanism by which differences in the timing of a nutritional cue during development may result in different phenotypic outcomes.
Transgenerational transmission of the induced phenotype
There are now several strands of evidence in humans and in experimental models for non-genomic transmission between generations of induced phenotypic traits associated with impaired metabolic homeostasis. Diabetes mortality was increased in men if the paternal grandfather was exposed to abundant nutrition during puberty (Pembrery et al. 2006). The daughters of women exposed to nutrient-restriction and psychological stress during pregnancy as a result of the Dutch Hunger Winter in 1944 / 5 showed decreased birthweight and increased risk of insulin resistance, while their daughters were also born with a lower birthweight (Stein & Lumey, 2000; Painter et al. 2005). These effects may be analogous to increased susceptibility to cancer in later life of the children and grandchildren of women exposed to the synthetic oestrogen and endocrine disruptor diethylstilbestrol (Newbold et al. 2006). In rats, feeding a protein-restricted diet (PRD) during pregnancy in the F0 generation resulted in elevated blood pressure and endothelial dysfunction (Torrens et al. 2002) and insulin resistance (Martin et al. 2000; Zambrano et al. 2005) in both the F1 and F2 generations, despite normal nutrition during pregnancy in the F1 generation. The adverse effects on glucose homeostasis in the F1 generation of feeding a PR diet during pregnancy in the F0 generation have been found in the F2 and F3 generation (Benyshek et al. 2006). Administration of dexamethasone to dams in late pregnancy induced increased expression of the glucocorticoid receptor (GR) and its target gene PEPCK in the liver of the F1 generation and this effect was transmitted to the F2, but not the F3, offspring by both male and female lines (Drake et al. 2005). Transmission of the effects of the F0 PR diet, but not dexamethasone exposure, to the F3 generation suggests the possibility of differences in the mechanism by which transgenerational effects are transmitted or sustained by these treatments. When the female offspring of rats fed a PR diet during pregnancy were mated and fed chow throughout pregnancy and lactation, the 80 days old F2 males showed comparable hypomethylation of the hepatic GR and PPARα promoters to 80 days old males of the F1 generation which were born to dams fed a PR diet during pregnancy (Burdge et al. 2006). This suggests that transmission of a phenotype induced in the F1 generation to the F2 generation may involve preservation of levels of DNA methylation of specific genes. It also implies that the female line is sufficient from transmission of such epigenetic information between generations and that the level of methylation of the GR and PPARα promoters in gametes must be similar to that of somatic cells and be preserved during post-fertilisation methylation, possibly by a similar mechanism to that which preserves the expression of imprinted genes (Lane et al. 2003). An alternative view is that prenatal nutritional constraint induced changes in the female which, in turn restrict the intra-uterine environment in which her offspring develop, and that the transmission of an altered phenotype between generations involves induction of changes in gene methylation de novo in each generation. If so, it may be anticipated that the magnitude of the induced effect, epigenetic or phenotypic, would differ between generations with differences in the developmental cues. However, this is not supported by the similarity in the reduction in birth size and blood glucose concentration in the F1 and F2 generations born to rat dams exposed to dexamethasone in late gestation (Drake et al. 2005), or the degree of hypomethylation of the hepatic GR and PPARα in the F1 and F2 offspring of dams fed a PR diet in pregnancy (Burdge et al. 2006). Furthermore, the observation that the effects of prenatal dexamethasone exposure are transmitted through both the male and female lines (Drake et al. 2005) also suggests that the mechanism of transmission between generations is not primarily by effects on the reproductive capacity of the female offspring. Although there is a need for substantial investigation of this process, if it occurs in humans one implication would be that nutrition of the grandmother during pregnancy may determine the capacity of critical metabolic pathways in her grandchildren and beyond.
DNA methyltransferases, 1-carbon metabolism and nutrient signalling from mother to fetus
Methylation of CpG dinucleotides de novo is catalysed by DNA methyltransferases (Dnmt) 3a and 3b, and is maintained through mitosis by gene-specific methylation of hemimethylated DNA by Dnmt1 (Bird, 2002). Over-expression of Dnmt1 results in hypermethylation of DNA and embryonic lethality (Biniszkiewicz et al. 2002), while transient depletion of xDnmt1 in Xenopus embryos induces DNA hypomethylation producing an altered phenotype (Stancheva & Meehan, 2000) and sustained depletion causes apoptosis (Stancheva et al. 2001). Thus variations in Dnmt1 expression alter the phenotype of the embryo. Dnmt1 activity is inhibited by homocysteine (Hcyst) (James et al. 2002), and Dnmt1 and 3a expression is modulated by folic acid intake in adult rats (Ghoshal et al. 2006). Changes to the activities of Dnmts as a result of altered 1-carbon metabolism represent one candidate mechanism for the transmission of information regarding maternal 1-carbon metabolism status to the fetus, for induction of modified epigenetic regulation of transcription and thus modified phenotype. However, hypomethylation of gene promoters may also be induced by impaired methylation de novo by the activities of Dnmt3a and 3b, or by active demethylation, for example by the activity of methyl binding domain (MBD) -2 (Detich et al. 2002). Feeding a PR diet to rats during pregnancy induced a reduction in Dnmt1 expression and in binding of Dnmt1 at the GR promoter (Lillycrop et al. 2006a). However, the expression of Dnmt3a, Dnmt3b and MBD2, and the binding of Dnmt3a at the GR promoter were unaltered (Lillycrop et al. 2006a). This suggests that hypomethylation of the GR promoter in the liver of the offspring, and probably other genes including PPARα, is induced by the maternal diet as a result of lower capacity to maintain patterns of cytosine methylation during mitosis.
Pregnant rats fed a PR diet show increased blood Hcyst concentration in early gestation (Petrie et al. 2002) and a trend towards a higher Hcyst concentration in late gestation (Brawley et al. 2004). Since Dnmt1 expression is negatively regulated by Hcyst and increased by folic acid, modulation of Dnmt1 expression by differences in 1-carbon metabolism may provide a link between maternal diet, and epigenetic regulation of gene expression in the fetus. This is supported by the finding that lower Dnmt1 expression induced by the PR diet was prevented by increasing the folic acid content of the PR diet (Lillycrop et al. 2006a) and is consistent with a central role for Dnmt1 in the induction of an altered phenotype (Jackson et al. 2002; Brawley et al. 2004). We suggest two possible mechanisms by which feeding a PR diet during pregnancy may alter 1-carbon metabolism. First, it is possible that decreased availability of glycine leads to altered flux of methyl groups between different metabolic fates and a constraint in the remethylation of Hcyst to methionine. Secondly, increased maternal corticosteroid levels (Langley-Evans et al. 1996), possibly as a result of stress induced by constrained nutrient availability, may reduce folic acid availability (Terzolo et al. 2004). The latter mechanism could explain how maternal corticosteroid blockade prevents induction of hypertension in the PR offspring (Langley-Evans et al. 1997) and as well as prevention of altered phenotype by folic acid administration (Brawley et al. 2004; Lillycrop et al. 2005).
Loss of Dnmt1 might be expected to result in global demethylation. However, loss of Dnmt1 leads only to a subset of genes being demethylated (Rhee et al. 2000; Jackson-Grusby et al. 2001). This indicates that Dnmt1 is targeted to specific genes, for example by binding to E2F1 binding sites (Robertson et al. 2000). Thus reduced Dnmt1 expression is consistent with hypomethylation of specific genes in the liver in the PR offspring (Lillycrop et al. 2005). Dnmt1 activity is also required for progression through mitosis (Milutinovic et al. 2003) and its expression is substantially reduced in non-proliferating cells (Suetake et al. 2001). Thus suppression of Dnmt1 activity or expression by altered 1-carbon metabolism in the preimplantation period could also account for the reduction in cell number during the early development in this model (Kwong et al. 2000). MicroRNAs have been proposed to induce DNA methylation by reduction in Dnmt1 and Dnmt3a activities (Kawasaki et al. 2005). If Dnmt1 activity was reduced in the offspring as a result of maternal under-nutrition during pregnancy then it might be anticipated that it would interfere with any process involving induction of DNA methylation involving microRNAs.
A possible pathway for the induction of an altered metabolic phenotype in the offspring by maternal under-nutrition during pregnancy
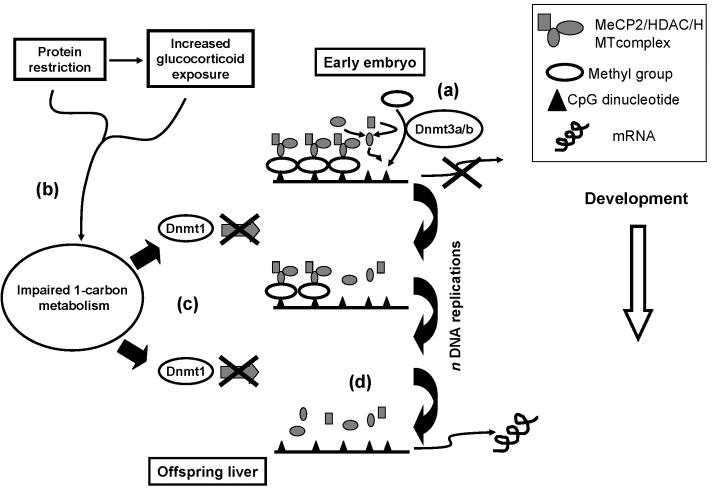
Based upon data currently available, we therefore now propose a mechanism for the induction of an altered metabolic phenotype in the offspring of rats fed a PR diet during pregnancy (Figure 4). Promoter methylation is induced in a gene-specific manner during the development of the early embryo by the activities of Dnmt 3a and 3b. Impaired 1-carbon metabolism, either a direct result of constrained maternal nutrition or by increased corticosteroid activity, reduces Dnmt1 expression resulting in progressive hypomethylation of the promoters of specific genes during subsequent mitoses. This is accompanied by reduced binding of the MeCP2/HDAC/HMT complex which facilitates persistence of histone modifications that permit transcription. This suggests how administration of glucocorticoids during pregnancy induces stable changes to the expression of gluconeogenic enzymes in the offspring as a result of increasing GR activity (Nyirenda et al. 1988). While the model explains many of the effects of maternal under-nutrition during pregnancy on the phenotype of the offspring, evidence from other nutritional challenges, such as increased nutrient abundance during pregnancy (Armitage et al. 2005), is required to test how widely the proposed pathway may operate. It is possible that over nutrition represents a stress which induces increased corticosteroid activity by increasing the demands on metabolic pathways to dispose of excess nutrients in competition with metabolic drives to deliver nutrients to the developing fetus. If so, it may be anticipated that this model may also explain how over-nutrition in pregnancy induces an altered phenotype in the offspring.
Fig. 4.
A pathway for induction of altered epigenetic regulation of the expression of specific genes in the offspring of rats fed a protein-restricted (PR) diet during pregnancy. (a) Gene expression is silenced in the early embryo by the activities of DNA methyltransferases (Dnmt) 3a and 3b. (b) In the offspring of rats fed a PR diet, 1-carbon metabolism is impaired either as a direct consequence of the restricted diet or by increased glucocorticoid exposure. This down-regulates Dnmt1 expression (c). Lower Dnmt1 expression results in impaired capacity to methylate hemimethylated DNA during mitosis (d). After sequential mitotic cycles the methylation status of the promoter is reduced and expression is induced in cells which do not express the gene in control animals. Increased gene expression is facilitated by lower binding and expression of methyl CpG binding protein (MeCP)-2 and reduced recruitment of the histone deacetylase (HDAC) / histone methyltransferase (HMT) complex, resulting in higher levels of histone modifications which permit transcription.
Epigenetic regulation of gene expression and metabolic capacity
Induction of altered epigenetic regulation of gene expression provides a molecular mechanism to explain how prenatal nutritional constraint alters the metabolic capacity of a tissue (Wootton & Jackson, 1996). However it is important to emphasise that, since transcription is regulated by a number of proteins, it is possible that although a gene promoter may show altered methylation, the effect on transcription and, in turn, metabolism may not become apparent until there is a stimulus, such as the activation of a transcription factor. For example, altered epigenetic regulation of PPARα expression may only be detected when the animal is fed a high fat diet which induces both PPARα transcription and its activation. If so, this provides a mechanism by which the effects of nutrition in early life on the epigenetic regulation of genes become apparent if there is an additional challenge from the postnatal environment. Thus the effect of induced differences in the epigenetic regulation of transcription may be regarded as altering the level at which a cell responds to environmental stress. The sum total of these cellular processes confers risk of disease on the individual.
Conclusions
Although this field of study is in its early stages, it is advancing rapidly, identification of altered epigenetic regulation of gene expression as a potential mechanism in the induction of different phenotypes by maternal nutrition during pregnancy suggests a shift in understanding of the phenomena known as fetal ‘programming’ away from detailed (although crucial) phenotypic characterisation of outcomes towards a molecular paradigm. In so doing, the processes which result in changes to the fetus in response to unbalance nutrition during pregnancy, which in humans are associated with increased disease risk in adulthood, may come to be viewed in the broader context of processes underpinned by epigenetic regulation. In this respect metabolic and cardiovascular disease may be more akin to conditions such as cancer where there is already considerable research demonstrating the role of epigenetic mechanisms. As in cancer, understanding of the underlying epigenetic processes may provide novel early markers of individuals at risk and valuable new advances in treatment and intervention.
Acknowledgements
GCB and MAH are supported by the British Heart Foundation
References
- Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol. 2004;561:355–377. doi: 10.1113/jphysiol.2004.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JA, Taylor PD, Poston L. Experimental models of developmental programming: consequences of exposure to an energy rich diet during development. J Physiol. 2005;565:3–8. doi: 10.1113/jphysiol.2004.079756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P, Barker D, Clutton-Brock T, et al. Developmental plasticity and human health. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F(3) generation) of rats malnourished during gestation and perinatal life. Diabetologia. 2006;49:1117–1119. doi: 10.1007/s00125-006-0196-5. [DOI] [PubMed] [Google Scholar]
- Benvenisty N, Mencher D, Meyuhas O, Razin A, Reshef L. Sequential changes in DNA methylation patterns of the rat phosphoenolpyruvate carboxykinase gene during development. Proc Nat. Acad Sci USA. 1985;82:267–271. doi: 10.1073/pnas.82.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram C, Trowern AR, Copin N, Jackson AA, Whorwood CB. The maternal diet during pregnancy programs altered expression of the glucocorticoid receptor and type 2 11beta-hydroxysteroid dehydrogenase: potential molecular mechanisms underlying the programming of hypertension in utero. Endocrinol. 2001;142:2841–2853. doi: 10.1210/endo.142.7.8238. [DOI] [PubMed] [Google Scholar]
- Bertram CE, Hanson MA. Animal models and programming of the metabolic syndrome. Br Med Bull. 2001;60:103–121. doi: 10.1093/bmb/60.1.103. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2001;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bogdarina I, Murphy HC, Burns SP, Clark AJ. Investigation of the role of epigenetic modification of the rat glucokinase gene in fetal programming. Life Sci. 2004;74:1407–1415. doi: 10.1016/j.lfs.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Brawley L, Torrens C, Anthony FW, Itoh S, Wheeler T, Jackson AA, Clough GF, Poston L, Hanson MA. Glycine rectifies vascular dysfunction induced by dietary protein imbalance during pregnancy. J Physiol. 2004;554:497–504. doi: 10.1113/jphysiol.2003.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan KA, Gopalakrishnan GS, Kurlak L, Rhind SM, Kyle CE, Brooks AN, Rae MT, Olson DM, Stephenson T, Symonds ME. Impact of maternal undernutrition and fetal number on glucocorticoid, growth hormone and insulin-like growth factor receptor mRNA abundance in the ovine fetal kidney. Reproduction. 2005;129:151–159. doi: 10.1530/rep.1.00229. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Delange E, Dubois L, Dunn RL, Hanson MA, Jackson AA, Calder PC. Effect of reduced maternal protein intake in pregnancy in the rat on the fatty acid composition of brain, liver, plasma, heart and lung phospholipids of the offspring after weaning. Br J Nutr. 2003;90:345–352. doi: 10.1079/bjn2003909. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Phillips ES, Dunn RL, Jackson AA, Lillycrop KA. Effect of reduced maternal protein consumption during pregnancy in the rat on plasma lipid concentrations and expression of peroxisomal proliferator–activated receptors in the liver and adipose tissue of the offspring. Nutr Res. 2004;24:639–646. [Google Scholar]
- Burdge GC, Slater-Jefferies JL, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2006 doi: 10.1017/S0007114507352392. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns SP, Desai M, Cohen RD, Hales CN, Iles RA, Germain JP, Going TC, Bailey RA. Gluconeogenesis, glucose handling, and structural changes in livers of the adult offspring of rats partially deprived of protein during pregnancy and lactation. J Clin Invest. 1997;100:1768–1774. doi: 10.1172/JCI119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano PM, Kirwan JP. Maternal factors that determine neonatal size and body fat. Curr Diab Rep. 2001;1:71–77. doi: 10.1007/s11892-001-0013-y. [DOI] [PubMed] [Google Scholar]
- Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MJ. Evidence for effects of weight on reproduction in women. Reprod Biomed Online. 2006;12:552–561. doi: 10.1016/s1472-6483(10)61180-7. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison EM, Syddall HE, Rodriguez S, Voropanov A, Day IN, Cooper C, Southampton Genetic Epidemiology Research Group Polymorphism in the growth hormone gene, weight in infancy, and adult bone mass. J Clin Endocrinol Metab. 2004;89:4898–4903. doi: 10.1210/jc.2004-0151. [DOI] [PubMed] [Google Scholar]
- Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Drake AJ, Walker BR, Seckl JR. Intergenerational consequences of fetal programming by in utero exposure to glucocorticoids in rats. Am J Physiol. 2005;288:R34–R38. doi: 10.1152/ajpregu.00106.2004. [DOI] [PubMed] [Google Scholar]
- Eriksson JG, Lindi V, Uusitupa M, Forsen TJ, Laakso M, Osmond C, Barker DJ. The effects of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-gamma2 gene on insulin sensitivity and insulin metabolism interact with size at birth. Diabetes. 2002;51:2321–2324. doi: 10.2337/diabetes.51.7.2321. [DOI] [PubMed] [Google Scholar]
- Eriksson J, Lindi V, Uusitupa M, Forsen T, Laakso M, Osmond C, Barker D. The effects of the Pro12Ala polymorphism of the PPARgamma-2 gene on lipid metabolism interact with body size at birth. Clin Genet. 2003;64:366–370. doi: 10.1034/j.1399-0004.2003.00150.x. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, Huang Y, Young D, Jacob ST. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522–1527. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. The developmental origins of the metabolic syndrome. Trends Endocrinol Metab. 2004a;15:183–187. doi: 10.1016/j.tem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004b;305:1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. The fetal matrix: Evolution, Development and Disease. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Gluckman PD, Hanson MA, Spencer HG. Predictive adaptive responses and human evolution. Trends Ecol Evol. 2005;20:527–533. doi: 10.1016/j.tree.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. Evolution, development and timing of puberty. Trends Endocrinol Metab. 2006;17:7–12. doi: 10.1016/j.tem.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Gnanalingham MG, Mostyn A, Dandrea J, Yakubu DP, Symonds ME, Stephenson T. Ontogeny and nutritional programming of uncoupling protein-2 and glucocorticoid receptor mRNA in the ovine lung. J Physiol. 2005;565:159–169. doi: 10.1113/jphysiol.2005.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4:611–624. doi: 10.1079/phn2001145. [DOI] [PubMed] [Google Scholar]
- Grainger RM, Hazard-Leonards RM, Samaha F, Hougan LM, Lesk MR, Thomsen GH. Is hypomethylation linked to activation of delta-crystallin genes during lens development? Nature. 1983;306:88–91. doi: 10.1038/306088a0. [DOI] [PubMed] [Google Scholar]
- Hershko AY, Kafri T, Fainsod A, Razin A. Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene. 2003;302:65–72. doi: 10.1016/s0378111902010910. [DOI] [PubMed] [Google Scholar]
- Jackson AA, Dunn RL, Marchand MC, Langley-Evans SC. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin Sci (Lond) 2002;103:633–639. doi: 10.1042/cs1030633. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002;(132 Suppl):2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K, Morris KV. siRNA induced transcriptional gene silencing in mammalian cells. Cell Cycle. 2005;4:442–448. doi: 10.4161/cc.4.3.1520. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development. 2000;127:4195–4202. doi: 10.1242/dev.127.19.4195. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Langley SC, Jackson AA. Increased systolic blood pressure in adult rats induced by fetal exposure to maternal low protein diets. Clin Sci (Lond) 1994;86:217–222. doi: 10.1042/cs0860217. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC, Gardner DS, Jackson AA. Maternal protein restriction influences the programming of the rat hypothalamic-pituitary-adrenal axis. J Nutr. 1996;126:1578–1585. doi: 10.1093/jn/126.6.1578. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC. Hypertension induced by foetal exposure to a maternal low-protein diet, in the rat, is prevented by pharmacological blockade of maternal glucocorticoid synthesis. J Hypertens. 1997;15:537–544. doi: 10.1097/00004872-199715050-00010. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2006a doi: 10.1017/S000711450769196X. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson AA, Burdge GC. Dietary protein restriction in the pregnant rat induces altered epigenetic regulation of the glucocorticoid receptor and PPARα in the heart of the offspring which is prevented by folic acid. Proc Nutr Soc. 2006b In Press. [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Maloney CA, Gosby AK, Phuyal JL, Denyer GS, Bryson JM, Caterson ID. Site-specific changes in the expression of fat-partitioning genes in weanling rats exposed to a low-protein diet in utero. Obes Res. 2003;11:461–468. doi: 10.1038/oby.2003.63. [DOI] [PubMed] [Google Scholar]
- Martin JF, Johnston CS, Han C-H, Benyshek DC. Nutritional origins of insulin resistance: a rat model for diabetes-prone human populations. J Nutr. 2000;130:741–744. doi: 10.1093/jn/130.4.741. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Niveleau A, Szyf M. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem. 2003;278:14985–14995. doi: 10.1074/jbc.M213219200. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinol. 2006;147(Suppl):S11–S17. doi: 10.1210/en.2005-1164. [DOI] [PubMed] [Google Scholar]
- Nyirenda MJ, Lindsay RS, Kenyon CJ, Burchell A, Seckl JR. Glucocorticoid exposure in late gestation permanently programs rat hepatic phosphoenolpyruvate carboxykinase and glucocorticoid receptor expression and causes glucose intolerance in adult offspring. J Clin Invest. 1998;101:2174–2181. doi: 10.1172/JCI1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72:218–219. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Repro Toxicol. 2005;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J, ALSPAC Study Team Sex-specific, male-line transgenerational responses in humans. Euro J Human Genetics. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Petrie L, Duthie SJ, Rees WD, McConnell JM. Serum concentrations of homocysteine are elevated during early pregnancy in rodent models of fetal programming. Br J Nutr. 2002;88:471–477. doi: 10.1079/BJN2002695. [DOI] [PubMed] [Google Scholar]
- Pham TD, MacLennan NK, Chiu CT, Laksana GS, Hsu JL, Lane RH. Uteroplacental insufficiency increases apoptosis and alters p53 gene methylation in the full-term IUGR rat kidney. Am J Physiol Regul Integr Comp Physiol. 2003;285:R962–R970. doi: 10.1152/ajpregu.00201.2003. [DOI] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- Remacle C, Bieswal F, Reusens B. Programming of obesity and cardiovascular disease. Int J Obes Relat Metab Disord. 2004;28(Suppl 3):S46–S53. doi: 10.1038/sj.ijo.0802800. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. Dnmt1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Stein AD, Lumey LH. The relationship between maternal and offspring birth weights after maternal prenatal famine exposure: the Dutch Famine Birth Cohort Study. Hum Biol. 2000;72:641–654. [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake L, Shi L, Watanabe D, Nakamura M, Tajima S. Proliferation stage-dependent expression of DNA methyltransferase (Dnmt1) in mouse small intestine. Cell Struct Funct. 2001;26:79–86. doi: 10.1247/csf.26.79. [DOI] [PubMed] [Google Scholar]
- Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human delta-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- Torrens C, Brawley L, Dance CS, Itoh S, Poston L, Hanson MA. First evidence for transgenerational vascular programming in the rat protein restriction model. J Physiol. 2002;543.P:41P–42P. [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorwood CB, Firth KM, Budge H, Symonds ME. Maternal undernutrition during early to midgestation programs tissue-specific alterations in the expression of the glucocorticoid receptor, 11beta-hydroxysteroid dehydrogenase isoforms, and type 1 angiotensin ii receptor in neonatal sheep. Endocrinology. 2001;142:2854–2864. doi: 10.1210/endo.142.7.8264. [DOI] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12:949–957. [PubMed] [Google Scholar]
- Wootton SA, Jackson AA. Influence of under-nutrition on growth, body composition and metabolic competence. In: Henry CJK, Ulijaszek SJ, editors. Long-term consequences of early ebvironment, development and the lifespan developmental perspective. Canbridge, UK: Cambridge University Press; 1996. [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Zambrano E, Martinez-Samayoa PM, Bautista CJ, Deas M, Guillen L, Rodriguez-Gonzalez GL, Guzman C, Larrea F, Nathanielsz PW. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol. 2005;566:225–236. doi: 10.1113/jphysiol.2005.086462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- Terzolo M, Allasino B, Bosio S, Brusa E, Daffara F, Ventura M, Aroasio E, Sacchetto G, Reimondo G, Angeli A, Camaschella C. Hyperhomocysteinemia in patients with Cushing's syndrome. J Clin Endocrinol Metab. 2004;89:3745–3751. doi: 10.1210/jc.2004-0079. [DOI] [PubMed] [Google Scholar]