Abstract
Unassembled (free) heavy chains appear during two stages of the class I MHC molecule's existence: immediately after translation but before assembly with peptide and β2-microglobulin, and later, upon disintegration of the heterotrimeric complex. To characterize the structures of folding and degradation intermediates of the class I heavy chain, three monoclonal antibodies have been produced that recognize epitopes along the H-2Kb heavy chain which are obscured upon proper folding and subsequent assembly with β2-microglobulin (KU1: residues 49-54; KU2: residues 23-30; KU4: residues 193-198). The Kb heavy chain is inserted into the lumen of the endoplasmic reticulum in an unfolded state reactive with KU1, KU2, and KU4. Shortly after completion of the polypeptide chain, reactivity with KU1, KU2 and KU4 is lost synchronously, suggesting that folding of the class I heavy chain is a rapid, cooperative process. Perturbation of the folding environment in intact cells with the reducing agent dithiothreitol or the trimming glucosidase inhibitor N-7-oxadecyl-deoxynojirimycin prolongs the presence of mAb-reactive Kb heavy chains. At the cell surface, a pool of free Kb heavy chains appears after 60–120 min of chase, whose subsequent degradation, but not their initial appearance, is impaired in the presence of concanamycin B, an inhibitor of vacuolar acidification. Thus, free heavy chains that arise at the cell surface are destroyed after internalization.
Peptides derived from proteins degraded in the cytosol are presented to the immune system by class I MHC molecules (1). Assembly of the class I MHC heavy chain, a type I membrane glycoprotein, with β2-microglobulin (β2m)1 and peptide occurs rapidly after cotranslational insertion of the subunits into the endoplasmic reticulum (2). En route to heterotrimer formation, the nascent heavy chain transiently associates with the endoplasmic reticulum (ER) resident proteins calnexin and calreticulin, lectins that bind to and retain incompletely assembled or misfolded proteins in the ER (3). Inhibition of the binding of calnexin and/or calreticulin to the nascent murine class I heavy chain decreases the efficiency of assembly of the latter with β2m (4) in agreement with studies on influenza hemagglutinin maturation, in which newly synthesized hemagglutinin subunits were more likely to aggregate and/or form aberrant disulfide bonds in the absence of calnexin/calreticulin association (5). After the heavy chain and β2m associate, the heterodimer binds to the peptide transporter complex TAP1–TAP2 (6, 7), an interaction that likely facilitates peptide loading of the empty class I molecule. Properly assembled class I complexes appear rapidly (<5 min) after initial completion of the polypeptide chains (8), and are subsequently transported to the plasma membrane via the secretory pathway. Once at the cell surface, class I molecules are largely stable (9), although exchange of bound β2m for bovine β2m present in serum has been observed for both murine and human class I molecules (10). Little if anything is known about how class I molecules are destroyed after they have exceeded their useful lifespan at the cell surface.
Antibody reagents that have been prepared against class I molecules can be categorized as (a) folding or assembly independent, (b) folding or assembly dependent, or (c) specific for the free heavy chain. For example, in the case of the mouse class I molecules, the antiserum p8 (11), raised against a peptide derived from the cytoplasmic tail of H-2Kb molecules, recognizes all forms of the H-2Kb heavy chain, whereas the mAb Y3 (12) binds only to properly conformed states, and the rabbit anti free-heavy chain serum binds to nonassembled material exclusively (13). Unfolded forms of class I heavy chain are likely to appear at two points during the lifetime of the molecule: early in the course of folding and assembly in the ER, and later, immediately before degradation. To characterize this population of class I heavy chains in living cells, we have prepared three mAbs against denatured Kb molecules that recognize nonassembled heavy chains exclusively. Folding and assembly studies on other glycoproteins such as the mouse class I molecule H-2Ld (14, 15), or the influenza hemagglutinin molecule (5), have utilized conformation-dependent antibodies to characterize the structure of biosynthetic intermediates; this study complements those by utilizing antibodies whose epitopes are exposed only when the protein is unfolded and has not yet associated with β2m. We show that these antibodies can be used to immunoprecipitate both assembly and degradation intermediates from pulselabeled cells. Our results suggest that the folding of the extracellular region is a rapid process, in which all of the epitopes detected by our antibodies disappear synchronously.
Materials and Methods
Cell Lines.
The Rauscher virus–transformed mouse lymphoma cell line RMA (16) was grown in RPMI-1640 supplemented with 10% fetal calf serum, l-glutamine (2 mM), penicillin (1:1000 dilution U/ml), streptomycin (100 μg/ml), sodium pyruvate (1 mM; GIBCO BRL, Gaithersburg, MD), nonessential amino acids (0.1 mM; GIBCO BRL), and β-mercaptoethanol (0.1 mM; American Bioanalytical, Natick, MA). Splenocytes from C57BL/6 mice were stimulated in the above medium supplemented with 2.5 μg/ml ConA for 48 h before labeling.
Antibodies.
The following antisera and mAbs were used: Y3 (IgG2b; recognizes the α1/α2 domain of properly conformed Kb molecules; 12), rabbit anti–mouse free heavy chain (Raf HC; recognizes non-β2m-associated mouse class I heavy chains; 13), rabbit anti-p8 (recognizes the cytoplasmic tail of Kb; prepared in our lab essentially as described 11) and rabbit anti-calnexin (recognizes the COOH-terminus of calnexin; a gift of Dr. D. Williams, University of Toronto, Canada). The conformation sensitive rabbit anti-H2 serum was provided by Dr. S. Nathenson. The mAbs introduced in this study (KU1, KU2, and KU4) were generated by immunizing mice with inclusion bodies of recombinantly expressed Kb heavy chains (a gift of Dr. S. Nathenson, Albert Einstein University, NY). Production of hybridomas was performed as described (17). All three antibodies were purified from cell culture supernatants with protein A–Sepharose (Repligen Corp., Cambridge, MA), and used in immunoprecipitations at 10 μg/ml lysate.
Western Blotting.
107 freshly isolated splenocytes from C57BL/6 (b haplotype), BALB/c (d haplotype), C3H (k haplotype), or PLJ (u haplotype) mice were lysed directly in either 2× SDS sample buffer or 1× IEF sample buffer and run on a 12.5% SDS-PAGE gel, or 1D–IEF gel (18), respectively. Before blotting, the IEF gel was washed five times (10 min each) with 300 ml of a solution of 50% (vol/vol) methanol, 5 mm Tris–HCl, pH 8.0, and 1% SDS. The transfer to nitrocellulose was performed in 25 mM Tris base, 200 mM glycine, and 20% methanol for 2.5 h at 400 mA in a Bio Rad Trans Blot Cell (Bio Rad, Hercules, CA). After a 2 h incubation in blocking buffer (phosphate-buffered saline with 10% nonfat powdered milk and 0.05% Tween-20), blots were probed with either Raf HC (1/3000 dilution) or KU2 (5 μg/ml) overnight, washed in PBS/0.05% Tween-20, and probed for 1 h with horseradish peroxidase–conjugated goat anti–rabbit or goat anti– mouse IgG (Southern Biotechnology Assoc., Birmingham, AL) before visualization with ECL (Kirkegaard and Perry Laboratories, Gaithersburg, MD).
Pulse Chase Analyses and Immunoprecipitations.
RMA cells or Con A-stimulated splenocytes were starved in RPMI-1640 medium lacking methionine and cysteine for 45 min before being pulsed with 500 μCi/ml [35S]methionine/cysteine (80:20) for the times indicated. When included, DTT (5 mM) was added 5 min before labeling, and the inhibitors N-7-oxadecyl-dNM (7-0-dec; 2 mM) and concanamycin B (Con B; 20 nM) were added during the starvation period. Labeling was terminated by adding 1 mM cold methionine/cysteine to the cell suspension. For the short (1–2 min) pulse–chase analyses (see Figs. 3 and 4), aliquots of cells (2–3 × 106) were removed at each timepoint and directly lysed in ice cold digitonin lysis buffer (0.5% digitonin [Sigma], 25 mM Hepes, pH 7.2, 10 mM CaCl2, 1 mM PMSF, 10 mM iodoacetamide) containing mAb or antiserum. After centrifugation of the cell lysates to remove nuclei and cellular debris, immune complexes were isolated after a 2-h incubation (with agitation) at 4°C by incubation with 50 μl 10% fixed Staphylococcus aureus for an additional 45 min. The S. aureus pellets were washed once in ice cold digitonin lysis buffer and then boiled for 10 min in denaturation/reduction buffer (2% SDS, 5 mM dithiothreitol, 50 mM Tris/HCl, pH 7.8, 1 mM EDTA). After one preclearing step with normal rabbit serum, the eluted Kb class I heavy chains were then reimmunoprecipitated in NP-40 lysis buffer (0.5% NP-40, 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 1 mM PMSF, 10 mM iodoacetamide) with the anti-p8 antiserum before gel analysis.
Figure 3.
The monoclonal antibodies KU1, KU2, and KU4 are specific for free heavy chains. (A) mRNAs for H-2Kb and mouse β2m were translated in vitro for 60 min, following which class I molecules were immunoprecipitated with the anti-H-2 antiserum, or the monoclonal antibodies KU1, KU2, and KU4, before analysis by SDS–PAGE. (B) RMA cells were pulsed for 1 min and chased for the times indicated. Aliquots of cells were lysed directly in digitonin lysis mix containing either Y3 (class I complexes), Raf HC (free heavy chains), KU2, or α-calnexin. Before SDS–PAGE analysis, the immunoprecipitated class I material was reimmunoprecipitated with p8.
Figure 4.
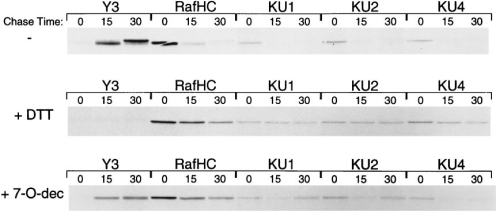
Effects of redox potential and calnexin association on the folding and assembly of the Kb molecule. RMA cells were pulsed for 1 min and chased for the times indicated. Nascent heavy chains were directly immunoprecipitated from NP-40 detergent lysates as in Fig. 3 with the antibodies indicated in each panel. The reducing agent DTT (B) was added to cells at 5 mM 5 min prior to labeling, while the glucosidase inhibitor 7-O-dec (C ) was added to cells at 2 mM during starvation (45 min prior to labeling).
For the longer (5 min) pulse–chase experiment shown in Fig. 5, aliquots of cells (Con A-stimulated splenocytes) were spun down at each timepoint, lysed in NP-40 lysis buffer, and the postnuclear supernatant precleared once with a 1:1 mixture of normal rabbit and normal mouse serum with 50 μl S. aureus before immunoprecipitation of class I heavy chains. For the pulse– chase in Fig. 6, owing to the high background commonly observed in immunoprecipitates from RMA cell lysates, we employed the direct immunoprecipitation/reimmunoprecipitation methodology described above. In vitro transcription and translation reactions were performed as described (22).
Figure 5.
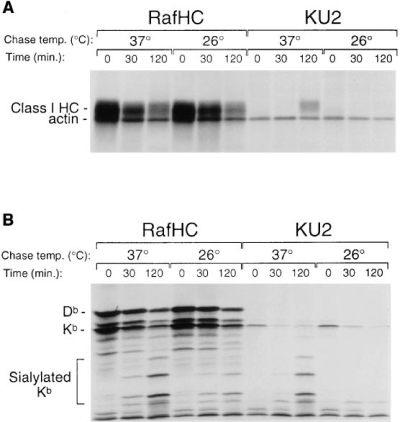
Appearance of unfolded class I heavy chains at the cell surface. Con A-stimulated splenocytes were pulse-labeled for 5 min and chased at either 37° or 26°C for the times indicated. Free (Raf HC; recognizes both Kb and Db) or KU2-reactive heavy chains (Kb alone) were immunoprecipitated from precleared lysates and resolved on SDS–PAGE (A) or 1D–IEF (B) gels.
Figure 6.
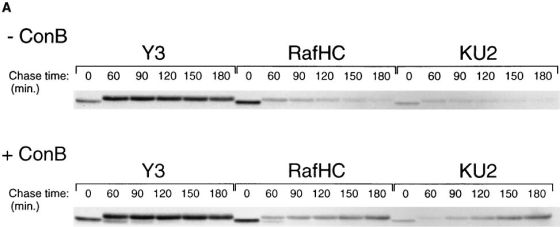
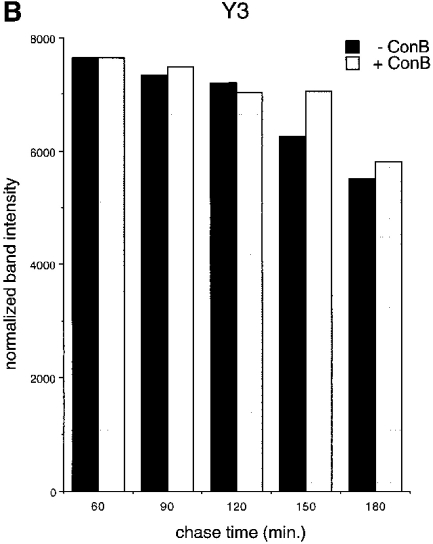
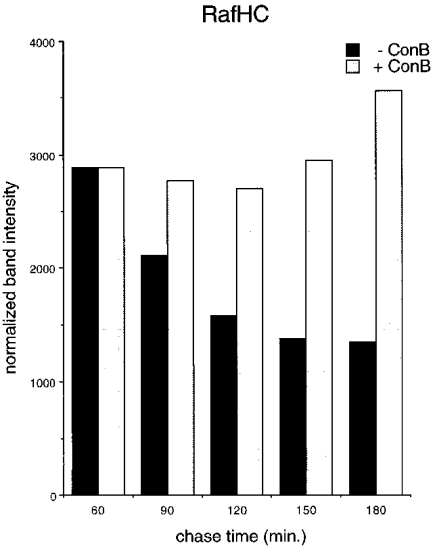
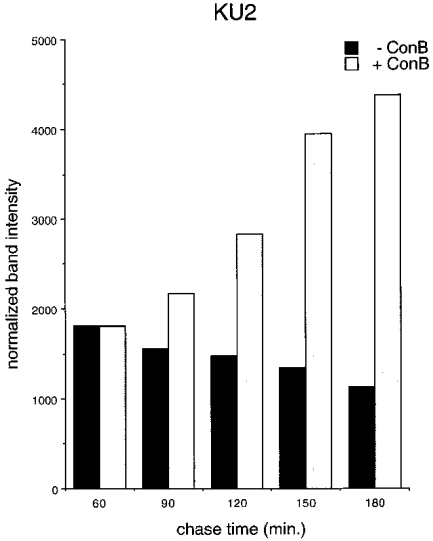
Breakdown of unfolded Kb heavy chains at the cell surface. (A) RMA cells were starved for 45 min in the absence or presence of the vacuolar H+-ATPase inhibitor Con B, pulse-labeled for 5 min, and then chased for the times indicated. Class I heavy chains were directly immunoprecipitated from NP-40 detergent lysates (no initial preclear) with the antibodies indicated in each panel, and reimmunoprecipitated with the p8 antiserum before SDS–PAGE. (B) Phosphoimager quantitation of the band densities in A. The data sets for each pulse–chase were normalized around the 60 min chase point densities.
Epitope Mapping.
The epitopes recognized by the mAbs KU1, KU2, and KU4 were determined by immunoprecipitation from a library of M13 phage displaying a random 10–amino acid insert (19). The library was constructed from a vector encoding the fd-tet phage and the tetracycline gene. Before immunoprecipitation, the phage suspension (6 × 1010 phage units/0.8 ml NP-40 lysis mix) was precleared with normal mouse serum (2 μl) added with a 1:1 suspension of protein A–Sepharose beads for 1 h at 4°C. Immunoprecipitations were performed by incubating the precleared phage suspension with 10 μg/ml mAb for 2 h at 4°C, after which the antibody–phage complexes were adsorbed onto protein A–Sepharose beads, collected by centrifugation, and washed five times in NET buffer (0.5% NP-40, 50 mM Tris– HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA). Phage were eluted from the beads with 200 μl of glycine HCl, pH 2.2, for 15 min at 4°C, neutralized with Tris–HCl (pH 9) to pH 6–8, and then grown on K91 (kanamycin-resistant) cells cultured in the presence of 20 μg/ml tetracycline. The mAb-selected phage were further purified through two additional rounds of immunoprecipitation, elution, and amplification, after which clones were picked and amplified for sequencing.
Results
Epitope Mapping and Specificity of Free Heavy Chain mAbs.
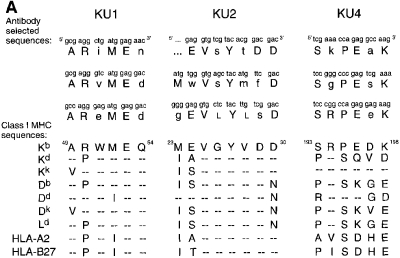
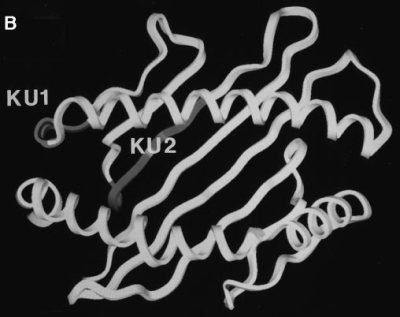
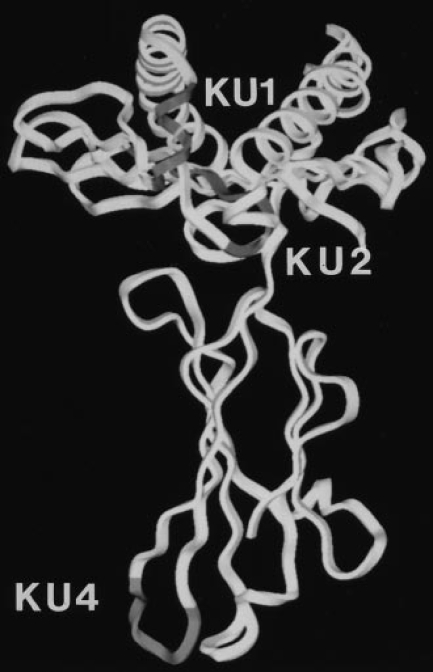
With the aim of characterizing partially folded or assembled forms of the Kb class I heavy chain, we raised three mAbs against denatured Kb molecules: KU1, KU2, and KU4. The immunogen was injected as urea-solubilized inclusion bodies obtained from E. coli, and presumably contained little coherent structure. Therefore, we expected that antibodies from this immunization would recognize unstructured epitopes along the Kb polypeptide chain. Thus, we employed an M13-based phage display library (19) encoding a random 10 amino acid insert to map the epitopes for each mAb. Three representative inserts are shown for each antibody (Fig. 1 A), although we sequenced at least 10 clones from each pool of selected phage to determine the anchor residues for each epitope. Beneath each consensus sequence is shown the corresponding Kb sequence and the sequences of other class I heavy chains in the same region (20). The location of each epitope could be established without ambiguity, and their projection onto the fully assembled Kb molecule is shown in Fig. 1 B (21). The epitope recognized by KU1 (residues 49–54) is normally folded into a 310 helix that immediately precedes the long α helix in the α1 domain which forms one side of the peptide-binding groove. The involvement of four out of five consecutive residues in generation of the KU1 epitope argues in favor of recognition of this stretch in nonhelical configuration and, conversely, folding of this region should conceal the KU1 epitope. KU2 binds to an epitope that spans part of the second β strand and connecting loop sequence in the α1 domain (residues 23–30); this region contains residues that directly contact β2m (Y27, E32). The KU4 epitope maps to the first β strand in the α3 domain (residues 193–198) and is adjacent to residues that contact β2m (R202, W204) as well as one of the cysteines (C203) that participates in the α3 disulfide bond. Note that the residues recognized by each antibody, as identified by phage display, are buried or rearranged upon folding of the heavy chain and assembly with β2m.
Figure 1.
Epitope mapping of the monoclonal antibodies KU1, KU2, and KU4. (A) Three representative phage insert sequences selected by each antibody are shown, with translation, above the Kb sequence that is predicted to contain the epitope. Residues in the phage sequences that match the actual Kb sequence are shown in capital letters. For comparison, sequences of other class I heavy chains are shown below each predicted epitope. (B) Ribbon diagrams of the properly conformed Kb heavy chain, highlighting in grey the locations of the predicted epitopes.
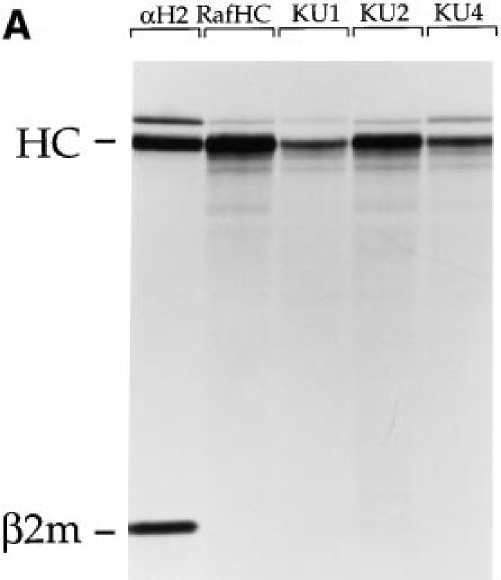
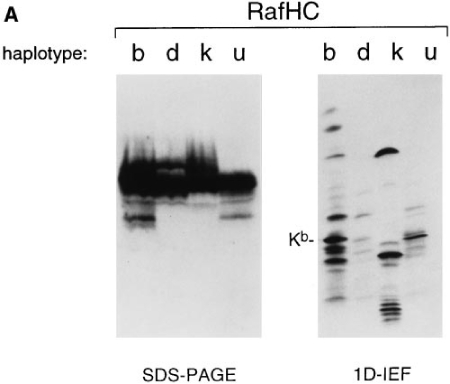
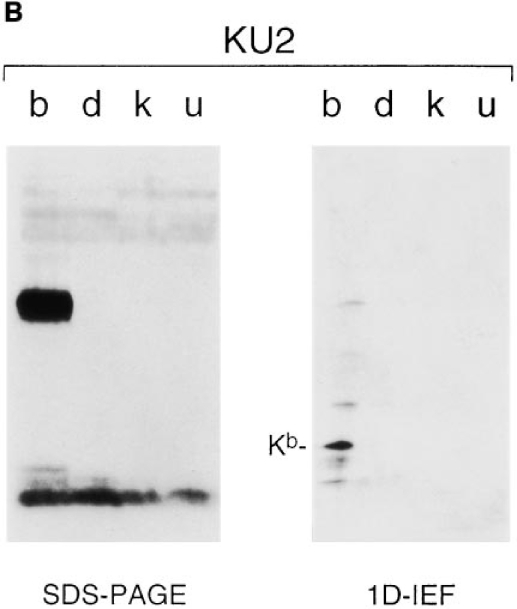
The results of the epitope mapping described above and sequence comparisons for class I heavy chains in that region suggested that KU1 and KU2 would recognize Kb exclusively, and that KU4 would recognize both Kb and Kk molecules. To probe directly the specificity of the antibodies, Western blots were performed on splenocyte extracts prepared from mice of b, d, k, and u haplotype resolved on SDS-PAGE and 1D–IEF gels (Fig. 2). As a control, we blotted with the rabbit anti-free heavy serum (Raf HC), which recognizes most murine class I heavy chains (13). As anticipated, KU2 recognized Kb heavy chains exclusively (Fig. 2 B), whereas Raf HC decorated all of the class I material present (Fig. 2 A). Neither KU1 nor KU4 blotted as efficiently as KU2, although the specificity for each was as expected based on the epitope mapping (data not shown). Further confirmation of the predicted epitopes for the antibodies was obtained from immunoprecipitations performed on Kb COOH-terminal truncation fragments translated in vitro (KU1, KU2, and KU4 all react with translation products containing Kb residues 1–204; only KU1 and KU2 react with fragments containing residues 1–74; data not shown).
Figure 2.
Western blot analysis of diverse class I material. Splenocyte extracts from H-2 b, d, k, and u haplotype mice were resolved on SDS– PAGE or 1D–IEF gels and blotted with either RafHC (A) or KU2 (B).
The mAbs KU1, KU2, and KU4 Recognize Unfolded Forms of the Kb Heavy Chain.
Having mapped the binding sites for each monoclonal antibody, we next sought to determine at which points these epitopes would be exposed during the life of the Kb heavy chain. Little if any detectable material could be immunoprecipitated with the antibodies from RMA cells labeled for 60 min (data not shown), suggesting that, while unfolded forms of the heavy chain might appear transiently during both biogenesis and degradation, they were too unstable to accumulate to appreciable levels at steady state. We analyzed reactivity of KU1, KU2, and KU4 on Kb molecules translated in vitro under conditions that support folding and assembly (22). We observed that none of the mAbs recognize any β2m containing forms of heavy chains, whereas the conformation sensitive rabbit anti-H2 antiserum clearly does (Fig. 3 A). The stronger reactivity of KU2, compared with KU1 and KU4, is demonstrated in this experiment, although the behavior of KU1, KU2, and KU4, in terms of loss of reactivity with Kb heavy chains, appears indistinguishable (see also Figs. 3 B and 4). Because of the stronger reactivity of KU2, much of the remainder of the experiments was performed with this antibody.
We first chose to examine the early events in the assembly of the Kb molecule, by performing a series of brief pulse chase experiments on RMA cells with pulse times of 1 min in order to resolve the rapid early stages of Kb folding and calnexin association. To minimize the lag time between cell lysis and exposure to antibody, aliquots of cells removed from the chase mixture were added directly to lysis buffer containing the relevant antibody (1° antibody) and immune complexes were recovered (8). The crude immunoprecipitates were then denatured completely by exposure to 2% SDS at 100°C, and reimmunoprecipitated with the p8 antiserum, which recognizes all full-length Kb heavy chains. The kinetics of assembly of the Kb molecule are evident from the pulse–chase experiment depicted in Fig. 3 B: properly conformed class I complexes (Y3 immunoprecipitates) form within 5 min, while, conversely, free heavy chains (Raf HC) are present immediately following completion of the 1-min pulse, but are largely assembled beyond 5 min of chase. KU2 also binds to newly synthesized heavy chains extracted within the first 5 min of synthesis, suggesting that the α1 domain β-strand recognized by the antibody remains accessible until the heavy chain has assembled with β2m. Although the recovery of calnexin-bound heavy chains was inefficient, notwithstanding the use of digitonin lysis buffer, the peak of heavy chains coimmunoprecipitated with calnexin occurs around 2–5 min after the 1-min pulse, slightly later than the appearance of the Raf HC and KU2-reactive material. This observed lag suggests that calnexin may not be stably bound to the newly synthesized heavy chains until 1–2 min after completion of the Kb polypeptide chain. Although folding clearly is an early and rapid process, and is likely to start on the nascent chain (5), it is completed posttranslationally.
Both KU1 and KU4 could also immunoprecipitate newly synthesized class I heavy chains within the first few minutes after completion of the polypeptide chain (Fig. 4 A). To determine whether disulfide bond formation is required for the folding of the regions surrounding the epitopes recognized by the antibodies, we performed a short pulse–chase experiment in the presence of the reducing agent dithiothreitol (DTT; 5 mM), which has been shown to prevent the formation of disulfide bonds in living cells (Fig. 4 B) (23). Under these conditions, no properly conformed Kb molecules are detectable, even after 30 min of chase (Y3). Unassembled Kb heavy chains persist throughout the chase (RafHC ), and all three monoclonal antibodies show prolonged reactivity, suggesting that, in the absence of proper disulfide bond formation, the folding of three distinct regions of the class I heavy chain is impeded.
To investigate the role of calnexin/calreticulin in the early folding events of the class I heavy chain, we performed a short pulse–chase experiment in the presence of the glucosidase inhibitor 7-O-dec (Fig. 4 C) (24). By blocking the trimming of terminal glucose residues on the N-linked oligosaccharides of the class I heavy chains, 7-O-dec effectively prevents the association of calnexin/calreticulin with the latter (25). The formation of properly conformed class I complexes (Y3) still occurs in the absence of calnexin/calreticulin binding, consistent with recent studies performed on the folding of influenza hemagglutinin (5), although assembly of free heavy chains with β2 is clearly less complete over the chase period (Raf HC ). All three mAbs react with class I heavy chains immediately after their synthesis, and by 15 min, much of the reactivity has been lost. However, unfolded Kb heavy chains reappear by 30 min, most likely from the disintegration of unstable class I complexes that were formed in the absence of the quality control normally provided by calnexin/calreticulin.
Kb Molecules Unfold at the Cell Surface and Are Degraded in an Acidic Compartment.
To follow the fate of free Kb heavy chains at the cell surface, we performed a pulse– chase experiment on Con A-stimulated splenocytes (prepared from C57BL/6 mice) with a chase time of 2 h, and resolved the Raf HC and KU2-reactive class I material on SDS (Fig. 5 A) and 1D–IEF (Fig. 5 B) gels. The chase was performed at 26°C as well as 37°C, because at the former temperature, the stability of class I molecules has been shown to be markedly enhanced. IEF allows the visualization of sialylated class I heavy chains. Addition of sialic acids to the class I glycan occurs in the trans-Golgi network, and immediately precedes surface deposition. For the description of the results, we shall equate sialylation with cell surface expression. At the onset of the chase, free heavy chains are observed exclusively in the ER (compare SDS and 1D–IEF 0 min timepoints), but appear at the cell surface by 30 min, most likely from disintegration of unstable class I complexes. Fewer free heavy chains are detectable during the 26°C chase, consistent with the increased stability of class I complexes at this temperature.
KU2-reactive heavy chains appear in abundance late in the chase, following the loss of β2m from unstable class I molecules at the cell surface (compare KU2 immunoprecipitates, SDS and IEF, 120 min timepoints). At the lower temperature, free heavy chains that accumulate at the cell surface (see Raf HC, 26° chase, IEF ) maintain a conformation that is not recognized by the KU2 antibody (compare Raf HC and KU2, 120 min timepoints, 26°C chase). Thus, the free heavy chain is capable of retaining the conformation of properly folded molecules, even after β2m dissociation, consistent with studies on the H-2Ld heavy chain (26), as well as with our previous observations (13). The KU2 antibody only recognizes a fraction of the free heavy chains that have been retained in the ER at either temperature (compare Raf HC and KU2, IEF gels); clearly, most of these free heavy chains are maintained in a conformation or complex that obscures the epitope. It is unlikely that calnexin is responsible for this effect, as the noncovalent complex between calnexin and the class I heavy chain does not survive exposure to NP-40 (27). Because Raf HC-reactive Kb heavy chains persist at timepoints where little reactivity is observed with the mAb KU2, we infer that the latter is the more stringent tool for monitoring folding, whereas Raf HC reports on both folding and assembly: association of Kb heavy chains with β2m obscures the Raf HC reactivity. We note that intact cells can be stained with the mAbs as revealed by cytofluorimetry, and that such staining is reduced for cells maintained at 26°C (data not shown), a condition known to prevent unfolding of otherwise labile Kb molecules (28).
By what means are class I complexes at the cell surface degraded? If breakdown occurs in lysosomal compartments, pH neutralization of these compartments might inhibit the dissociation of the heavy chain from the class I molecule, or degradation of the latter, or both. To address this question, we pulse-labeled RMA cells in the presence or absence of Con B, a fungal macrolide antibiotic that inhibits vacuolar H+ ATPases (29), and followed the pools of conformed (Y3), free (Raf HC ), or unfolded (KU2) Kb molecules over the 3-h chase period (Fig. 6). Owing to the higher background typically seen in immunoprecipitations from briefly pulse-labeled RMA cells, we utilized the direct immunoprecipitation/reimmunoprecipitation methodology as described for the short pulse–chase experiments in Figs. 3 and 4. This technique also results in improved recovery of nascent heavy chains that are only reactive with KU2 immediately following their deposition in the ER. Heavy chains that dissociate at the cell surface in untreated cells are largely degraded after 3 h of chase (Fig. 6 A). In contrast, when cells are treated with Con B, the destruction of both Raf HC- and KU2-reactive heavy chains is inhibited. Quantitation by phosphoimaging (Fig. 6 C ) reveals that the decay of properly formed complexes (Y3 panel) is similar in the presence and absence of the inhibitor, suggesting that degradation, but not disintegration, of the class I complex is dependent on lysosomal function. Neuraminidase digestion of intact cells chased in the presence of Con B demonstrates that after 3 h, about half of the sialylated KU2-reactive heavy chains are cell surface disposed, while the remainder resides within the cell, presumably within lysosomes (data not shown).
Discussion
The pool of class I molecules within the cell is comprised of both partial (i.e., monomeric and heterodimeric) and complete (i.e., heterotrimeric) complexes. Intermediate forms of the class I molecule are most evident during the initial folding and assembly processes that occur in the ER, as well as during the disintegration of the complex at the cell surface. The three mAbs introduced in this study have allowed us to characterize in part the structure of non-β2massociated free heavy chains as they appear in the course of the lifetime of the class I molecule. These reagents recognize epitopes on the class I heavy chains that are buried upon folding and assembly with β2m, and thus can be used to monitor the local conformation of the respective subdomains surrounding each epitope.
Newly synthesized class I heavy chains interact with the ER resident chaperones calnexin and calreticulin rapidly after synthesis (30, 3). We show that three distinct epitopes, recognized by KU1, KU2, and KU4, persist after synthesis for at least 5 min, during which period the heavy chains are associated with calnexin (Fig. 3). We have been unable to observe any differential loss of reactivity over time with the three antibodies in the course of folding and assembly of the class I heavy chain, in contrast with recent studies performed on the much larger glycoprotein, influenza hemagglutinin, which have delineated a number of discrete folding intermediates (5). If the calnexin–calreticulin interaction is abrogated by incubation of cells with the glucosidase inhibitor 7-O-dec, then a pool of mAb-reactive heavy chains reappears later in the chase, demonstrating that, even though properly conformed molecules are formed in the absence of calnexin–calreticulin association, many of the heavy chains do not fold productively, in agreement with results from the influenza hemagglutinin system (25) as well as with a recent study on the murine class I molecules H-2Kb and Db (4). In the presence of the reducing agent DTT, the folding of the heavy chains is blocked at an early stage, before the packing of the epitopes recognized by each of the three mAbs. However, since calnexin contains disulfide bonds that are essential for its function (31), addition of DTT most likely perturbs the association between the class I heavy chain and calnexin as well, and thus heavy chains synthesized under reducing conditions must attempt to fold both without disulfide bonds and calnexin.
Once the class I molecule has been properly assembled, it proceeds to the cell surface via the secretory pathway. However, soon after deposition at the plasma membrane a fraction of the class I complexes dissociates, as evidenced by the appearance of sialylated free heavy chains 60–120 min after synthesis. In contrast with the pool of intracellular free heavy chains that persist throughout the chase, the cell surface-disposed free heavy chains are also reactive with the KU2 mAb, and thus are unfolded to a greater extent. One explanation for this rapid disintegration of a portion of class I molecules is that complexes that have bound low affinity peptides might attain a transport-competent structure, but then lose peptide once they arrive at the cell surface. This fraction may be stabilized by suitable peptide ligands added extracellularly (32) (33), but should no peptides be available, the complex is more likely to resolve into the separate subunits. Treatment of cells with Con B, a vacuolar proton pump inhibitor, prevents the subsequent destruction of sialylated free heavy chains, implying that degradation occurs intracellularly in an acidic compartment. While generally difficult to detect, internalization of class I molecules is unlikely to be restricted to free heavy chains, and thus, this pathway may provide an opportunity for empty class I molecules that are not degraded to bind peptides derived from exogenous antigens (34).
Acknowledgments
We would like to thank Dr. D. Williams for the rabbit anti-calnexin antiserum, and Dr. S. Nathenson for the purified soluble H-2Kb and the rabbit anti-H2 antiserum.
Footnotes
This work was supported by National Institutes of Health grants R01-AI33456-01 and R01-AI07463-17.
1 Abbreviations used in this paper: β2m, β2-microglobulin; HA, hemagglutinin; Raf HC, rabbit anti-free haevy serum.
References
- 1.Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell JW, Bennink JR. Cell biology of antigen processing and presentation to MHC class I molecule– restricted T lymphocytes. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 3.Williams DB, Watts TH. Molecular chaperones in antigen presentation. Curr Opin Immunol. 1995;7:77–84. doi: 10.1016/0952-7915(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 4.Vassilakos A, Cohen-Doyle MF, Peterson PA, Jackson MR, Williams DB. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO (Eur Mol Biol Organ) J. 1996;15:1495–1506. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Helenius J, Braakman I, Helenius A. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/β2-microglobulin complexes associate with TAP transporters before peptide binding. Nature (Lond) 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 7.Suh W, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science (Wash DC) 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 8.Neefjes JJ, Hammerling GJ, Momburg F. Folding and assembly of major histocompatibility complex class I heterodimers in the endoplasmic reticulum of intact cells precedes the binding of peptide. J Exp Med. 1993;178:1971–1980. doi: 10.1084/jem.178.6.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neefjes JJ, Dierx J, Ploegh HL. Anchor residue modification and class I stability. Eur J Immunol. 1993;23:840–845. doi: 10.1002/eji.1830230411. [DOI] [PubMed] [Google Scholar]
- 10.Rock KL, Gamble S, Rothstein L, Gramm C, Benacerraf B. Dissociation of β2-microglobulin leads to the accumulation of a substantial pool of inactive class I MHC heavy chains on the cell surface. Cell. 1991;65:611–620. doi: 10.1016/0092-8674(91)90093-e. [DOI] [PubMed] [Google Scholar]
- 11.Smith MJ, Parker JMR, Hodges RS, Barber BH. The preparation and characterization of anti-peptide heteroantisera recognizing subregions of the intracytoplasmic domain of class I H-2 antigens. Mol Immunol. 1986;23:1077–1092. doi: 10.1016/0161-5890(86)90006-4. [DOI] [PubMed] [Google Scholar]
- 12.Hammerling GJ, Rusch E, Tada N, Kimura S, Hammerling U. Localization of allodeterminants on H-2Kbantigens determined with monoclonal antibodies and H-2 mutant mice. Proc Acad Natl Sci USA. 1982;79:4737–4741. doi: 10.1073/pnas.79.15.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machold RP, Andree S, Van Kaer L, Ljunggren H, Ploegh HL. Peptide influences the folding and intracellular transport of free major histocompatibility complex heavy chains. J Exp Med. 1995;181:1111–1122. doi: 10.1084/jem.181.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JD, Myers NB, Gorka J, Hansen TH. Model for the in vivo assembly of nascent Ld class I molecules and for the expression of unfolded Ldmolecules at the cell surface. J Exp Med. 1993;178:2035–2046. doi: 10.1084/jem.178.6.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carreno BM, Solheim JC, Harris M, Stroynowski I, Connolly JM, Hansen TH. TAP associates with a unique class I conformation, whereas calnexin associates with multiple class I forms in mouse and man. J Immunol. 1995;155:4726–4733. [PubMed] [Google Scholar]
- 16.Ljunggren H, Karre K. Host resistance directed selectively against H-2-deficient lymphoma variants: analysis of the mechanism. J Exp Med. 1985;162:1745–1759. doi: 10.1084/jem.162.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Ljunggren HG, Oudshoorn-Snoek M, Masucci MG, Ploegh HL. High resolution one-dimensional isoelectric focusing of murine MHC class I antigens. Immunogenetics. 1990;32:440–450. doi: 10.1007/BF00241639. [DOI] [PubMed] [Google Scholar]
- 19.Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 20.Flaherty, L., E. Elliott, J.A. Tine, A.C. Walsh, and J.B. Waters. 1990. Immunogenetics of the Q and TL regions in mouse. Crit. Rev. Immunol. 10, no. 2:131–171. [PubMed]
- 21.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb . Science (Wash DC) 1992;257:919–926. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 22.Bijlmakers MJE, Neefjes JJ, Wojcik-Jacobs EHM, Ploegh HL. The assembly of H-2Kb class I molecules translated in vitrorequires oxidized glutathione and peptide. Eur J Immunol. 1993;23:1305–1313. doi: 10.1002/eji.1830230618. [DOI] [PubMed] [Google Scholar]
- 23.Braakman I, Helenius J, Helenius A. Manipulating disulfide formation and protein folding in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan A, van den Broek L, Bolscher J, Vermaas DJ, Pastoors L, van Boeckel C, Ploegh H. Introduction of oxygen into the alkyl chain of N-decyl-dNM decreases lipophilicity and results in increased retention of glucose residues on N-linked oligosaccharides. Glycobiology. 1994;4:141–149. doi: 10.1093/glycob/4.2.141. [DOI] [PubMed] [Google Scholar]
- 25.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solheim JC, Johnson NA, Carreno BM, Lie WR, Hansen TH. Beta 2-microglobulin with an endoplasmic reticulum retention signal increases the surface expression of folded class I MHC molecules. Eur J Immunol. 1995;25:3011–3016. doi: 10.1002/eji.1830251104. [DOI] [PubMed] [Google Scholar]
- 27.Galvin K, Krishna S, Ponchel F, Frohlich M, Cummings DE, Carlson R, Wands JR, Isselbacher KJ, Pillai S, Ozturk M. The major histocompatibility complex class I antigen binding protein p88 is the product of the calnexin gene. Proc Natl Acad Sci USA. 1992;89:8452–8456. doi: 10.1073/pnas.89.18.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ljunggren HG, Stam NS, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TNM, Townsend A, Karre K, Ploegh HL. Empty MHC class I molecules come out in the cold. Nature (Lond) 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 29.Yilla M, Tan A, Ito K, Miwa K, Ploegh HL. Involvement of the vacuolar H+-ATPases in the secretory pathway of HepG2 cells. J Biol Chem. 1993;268:19092–19100. [PubMed] [Google Scholar]
- 30.Degen E, Williams DB. Participation of a novel 88-kD protein in the biogenesis of murine class I histocompatibility molecules. J Cell Biol. 1991;112:1099–1115. doi: 10.1083/jcb.112.6.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tector M, Salter RD. Calnexin influences folding of human class I histocompatibility proteins but not their assembly with β2-microglobulin. J Biol Chem. 1995;270:19638–19642. doi: 10.1074/jbc.270.33.19638. [DOI] [PubMed] [Google Scholar]
- 32.Townsend A, Ohlen C, Bastin J, Ljunggren HG, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature (Lond) 1989;340:443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- 33.Rock KL, Gramm C, Benacerraf B. Low temperatures and peptides favor the formation of class I heterodimers on RMA/S cells at the cell surface. Proc Natl Acad Sci USA. 1991;88:4200–4204. doi: 10.1073/pnas.88.10.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rock KL. A new foreign policy: MHC class I molecules monitor the outside world. Immunol Today. 1996;17:131–137. doi: 10.1016/0167-5699(96)80605-0. [DOI] [PubMed] [Google Scholar]