Abstract
The infusion of TIL586 along with interleukin-2 into the autologous patient with metastatic melanoma resulted in the objective regression of tumor. A gene encoding a tumor antigen recognized by TIL586 was previously isolated and shown to encode gp75 or TRP-1. Here we report that TRP-2 was identified as a second tumor antigen recognized by a HLA-A31– restricted CTL clone derived from the TIL586 cell line. The peptide LLPGGRPYR epitope was subsequently identified from the coding region of TRP-2 based on studies of the recognition of truncated TRP-2 cDNAs and the HLA-A31 binding motif. This epitope peptide was capable of sensitizing target cells for lysis by a CTL clone at 1 nM peptide concentration. Although some modified peptides could be recognized by the CTL clone, none were found to be better recognized by T cells than the parental peptide. Like other melamona differentiation antigens, TRP-2 was only expressed in melanoma, melanocytes, and retina, but not in other human tissues tested.
The adoptive transfer of tumor infiltrating lymphocytes (TIL)1 plus IL-2 into the autologous patient with metastatic melanoma can result in the objective regression of tumor (1, 2), suggesting that T cells play an important role in tumor rejection in vivo. To understand the molecular basis of T cell–mediated antitumor responses, effort has been directed toward the identification of tumor antigens recognized by T cells and has led to the identification of a number of genes encoding tumor antigens in human melanoma (3–8). These antigens can be divided into several classes based on their expression pattern. One class of tumor antigens such as MAGE1, MAGE3, BAGE, and GAGE are encoded by genes that are expressed only in tumor, testis, and placenta, but not other normal human tissues (9–12). The second class of antigens such as MART-1/Melan-A (13, 14), gp100 (15), tyrosinase (16, 17), and gp75/TRP-1 (18) are differentiation antigens encoded by genes that are expressed only in melanocytes, melanomas, and normal retinal tissue. These latter antigens are nonmutated self proteins. However, several mutated antigens were also identified to be recognized by T cells, including CDK4 (19), β-catenin (20), and MUM-1 (21).
Recently, we cloned the gp75/TRP-1 gene encoding a tumor antigen recognized by the HLA-A31–restricted TIL586 (18), which was previously shown to have in vivo antitumor activity when infused along with IL-2 into the autologous patient with melanoma. Surprisingly, the peptide recognized by TIL586 was found to be derived from the gene product translated from an alternative open reading frame of the TRP-1 gene (22). Site-directed mutagenesis experiments indicated that the ATG start codon in the alternative open reading frame was essential for generating the epitope recognized by TIL586. 6 of 15 T cell clones established from the TIL586 cell line were capable of recognizing 586mel tumor cells as well as 586EBV B cells pulsed with the peptide ORF3P derived from the alternative open reading frame of the TRP-1 gene (22). However, some T cell clones isolated from the same TIL586 line did not recognize TRP-1– and HLA-A31–transfected COS-7 cells and its ORF3P (MSLQRQFLR) peptide-pulsed 586EBV B cells, although they were capable of recognizing 586mel and HLAA31+ melanocytes.
In this report, we describe the identification of TRP-2 as a new tumor antigen recognized by CTL clones derived from tumor reactive TIL line. TRP-2 is a member of the tyrosinase-related gene family. Further experiments have allowed us to identify the epitope peptide from the TRP-2 coding sequence recognized by T cell clones, suggesting that T cells recognized a nonmutated self peptide.
Materials and Methods
Chemicals and Reagents.
The following chemicals and reagents were purchased from the sources indicated: RPMI 1640, AIM-V media, Lipofectamine, G418 (GIBCO BRL, Gaithersburg, MD); the eukaryotic expression vector pCR3 (Invitrogen, San Diego, CA); anti–HLA-A31 monoclonal antibody (One lambda; Canoga Park, CA); anti–immunoglobulin M antibody conjugated with fluorescein isothiocyanate (Vector Labs., Inc., Burlingame, CA).
T Cell Clones and Lines.
TIL586 were isolated from the tumor specimen of a patient with metastatic melanoma and grown in medium containing IL-2 (6,000 IU/ml) (Chiron Corp., Emeryville, CA) for 32–60 d as previously described (23). TIL586 were predominantly CD8+ T cells. The T cell clones were generated by limiting dilution methods (at 1 cell/well) from the TIL586 cell line, and used allogeneic PBL (1 × 103 cells/well) as feeder cells in RPMI1640 containing 10% human AB sera and 500 IU IL-2. After 12 d, T cell clones were then expanded in AIM-V medium containing 6,000 IU/ml IL-2. To obtain a optimal expansion, we used the OKT3 expansion method described by Walter et al. (24). Briefly, on day 0, 5 × 104–5 × 105 T cells were cocultured with HLA-A31+ PBL (500:1, PBL/T cell) and 586EBV B cells (100:1, EBV/T cell) in 25 ml RPMI 1640 containing 11% human sera, 30 ng/ ml OKT3 antibody, and antibiotics. On day 1, IL-2 was added at final concentration of 180 IU/ml. The medium was changed with fresh medium containing 11% human sera, and 180 IU/ml of IL-2 on day 5. The medium was then changed every 3 d. On days 12–14, T cells were harvested, counted, and cryopreserved.
Melanoma cell lines 397mel, 397mel/A31, 586mel, 624mel, 624mel/A31, and EBV transformed B cell lines 586EBV and 1510EBV were established in our laboratory and cultured in RPMI 1640 medium containing 10% FCS. Normal cultured melanocytes derived from infant foreskin (NHEM680; Clonetics Corp., San Diego, CA) were cultured in melanocyte growth medium (MGM; Clonetics). The COS-7 cell line was provided by Dr. W. Leonard (National Institutes of Health).
GM-CSF Secretion Assay.
DNA transfection and GM-CSF assays were performed as previously described (18). Briefly, 200 ng of DNA encoding antigens and 50 ng of the HLA-A31 DNA were mixed with 2 μl of lipofectamine in 100 μl of DMEM for 15–45 min. The DNA/lipofectamine mixture was then added to the COS-7 (5 × 104) cells and incubated overnight. The following day, cells were washed twice with DMEM medium. TIL586 was added at a concentration of 1 × 105 cells/well in AIM-V medium containing 120 IU/ml of IL-2. For T cell clones, only 1–2 × 104 cells/well were added. After 18–24 h incubation, 100 μl of supernatant was collected and GM-CSF was measured in a standard ELISA assay (R & D Sys., Inc., Minneapolis, MN). For testing peptide recognition, 586EBV or T2 cells were incubated with peptides at 37°C for 90 min, and then washed three times with AIM-V medium containing 120 IU/ml of IL-2. T cells were added and incubated for an additional 18–24 h; 100 μl of supernatant was collected for GM-CSF assay.
ExoIII/S1 Deletion Constructions and Subcloning.
TRP-2 cDNA was a gift of Dr. Shibahara (25), and subcloned into the pCR3 vector with a CMV promoter for expression. To make a series of deletions, the plasmid pCR3 containing TRP-2 cDNA was digested with XbaI and filled in with α-phosphorothioate deoxyribonucleotide triphosphates to block ExoIII nuclease digestion. The linearized DNA was subjected to the second restriction enzyme digestion to generate one end sensitive to ExoIII. ExoIII nuclease/Mung bean nuclease deletion was performed according to the manufacturer's instructions (Stratagene Corp., La Jolla, CA). All deletion constructs were sequenced to determine the location of DNA sequence being removed. pTA plasmid was a derivative of pCR3-TRP2, in which an ApaI DNA fragment was deleted from the 3′ end of TRP-2 gene. pTK was created after removal of a KpnI DNA fragment from the 3′ end of the TRP-2 gene. pTP was generated by deleting an internal PstI fragment and religation.
Northern Blot Analysis.
Total RNA was isolated by the guanidine isothiocyanate/cesium chloride centrifugation method. Total RNA from human normal tissue was purchased from Clontech (Palo Alto, CA). 20 μg of total RNA was subjected to electrophoresis in a 1.2% agarose formaldehyde gel and transferred to a nylon membrane. A 2.0-kb DNA fragment of the TRP-2 gene was labeled with 32P-α-CTP by the random priming method. Prehybridization and hybridization were performed according to the QuickHyb protocol (Stratagene Corp.). Membranes were washed twice with 2× SSC/0.1% SDS at room temperature for 15 min and twice with 0.1× SSC/0.1% SDS at 60°C for 30 min. The autoradiography was performed at −70°C.
Cytotoxicity Assays.
Cytolysis was determined by use of calcein AM ( Molecular Probes Inc., Eugene, OR). Briefly, T2 or 586EBV B cells were pulsed with peptides in RPMI1640/ 5%FCS for 90 min. Tumor cells and the peptide pulsed EBV B cells were labeled with calcein AM (15 μl of 1 mg/ml calcein AM for every 1 × 106 cells) for 30 min at 37°C. After incubation, cells were washed three times with AIM V/120 IU IL-2. 1 × 103 target cells were mixed with T cells at various E/T (effector/target ratio). After 4 h incubation at 37°C, 5 μl of bovine hemoglobin quench solution containing ethidium bromide was added. The plate was read by lambda scan. The percentage of lysis was calculated from the following equation: [1 − (A − B)/(C − B)] × 100 where A is the reading of nonlysed cells in the presence of T cells, B is background signal value, and C is the maximum signal value from target cells.
The peptides were synthesized by a solid-phase method using a peptide synthesizer (AMS 422; Gilson Co., Inc., Worthington, OH). Some peptides were purified by HPLC and became >98% in purity. The peptide mass of some peptides was confirmed by mass spectrometry analysis.
Results
Recognition of New Antigens on Tumor Cells by CTL Clones.
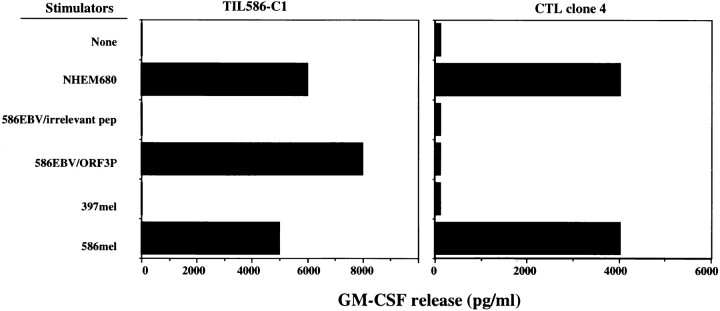
In previous studies, we isolated a number of T cell clones from the TIL586 cell line by the limiting dilution method (22). Among them, six clones recognized 586EBV B cells pulsed with the ORF3P peptide derived from a gene product translated from an alternative open reading of the TRP1/gp75 gene, and the autologous 586mel tumor cells, but did not recognize 586EBV B cells pulsed with an irrelevant peptide. TIL586-C1 was a representative of these T cell clones as shown in Fig. 1. However, several T cell clones isolated from the same TIL586 cell line recognized neither 586EBV B cells pulsed with the TRP-1 peptide ORF3P nor COS cells transfected with TRP-1 and HLA-A31 cDNAs, but were capable of recognizing 586mel as well as HLA-A31+ NHEM680 melanocyte line (Fig. 1). These results suggested that these T cell clones recognized additional tumor antigens on the 586mel tumor cells. These T cell clones were then expanded to obtain enough cells for screening cDNA libraries or testing other cDNAs for recognition by methods described in the Materials and Methods section. One of the clones, CTL clone 4, was successfully expanded and used for further studies as described below.
Figure 1.
Recognition of various target cells and the antigenic peptide by CTL clones derived from TIL586. T cell clones were generated by limiting dilution (1 cell/well) from the TIL586 cell line and were further expanded in AIM-V medium containing 6,000 IU/ml IL-2. GM-CSF secretion by CTL clone 586TILC1 (left) and clone 4 (right) was measured after coculturing with a normal melanocyte cell line NHEM680 (HLA-A31+), 586EBV B cells pulsed with the ORF3P peptide or irrelevant peptide, 397mel or 586mel cells.
Identification of a cDNA Encoding a Tumor Antigen Recognized by T Cell Clones.
To determine the HLA molecule responsible for presenting antigen to CTL clone 4, we transfected HLA-A31 cDNA into HLA-A31–negative tumor lines such as 397mel and 624mel and tested for recognition by the CTL clone. Transfectants of 397mel and 624mel expressing HLA-A31 were significantly recognized by CTL clone 4 (Table 1). Furthermore, these T cells were also capable of recognizing the HLA-A31–positive allogeneic tumor line 1353mel, indicating that recognition of the tumor antigen by CTL clone 4 was HLA-A31 restricted.
Table 1.
Specific Secretion of GM-CSF by CTL Clone 4 is HLA-A31–Restricted
| Stimulators | GM-CSF secretion | |||||
|---|---|---|---|---|---|---|
| Cell lines | Transfected gene | HLA-31 expression | ||||
| pg/ml | ||||||
| None | None | − | <10 | |||
| 397mel (TRP1−/TRP2+) | None | − | 23 | |||
| 397mel (TRP1−/TRP2+) | HLA-A31 | + | 2,840 | |||
| 624mel (TRP1+/TRP2+) | None | − | 39 | |||
| 624mel (TRP1+/TRP2+) | HLA-A31 | + | 670 | |||
| 1353mel (TRP1+/TRP2+) | None | + | 879 | |||
| 586mel (TRP1+/TRP2+) | None | + | >4,000 | |||
| 586EBVB | None | + | 29 | |||
| COS-7 | None | − | 35 | |||
| COS-7 | HLA-A31 | + | 30 | |||
GM-CSF in the supernatant was measured after 24 h incubation of 2 × 104 CTL clone 4 cells with either melanoma cell lines or COS-7 transfected with the HLA-A31 cDNA.
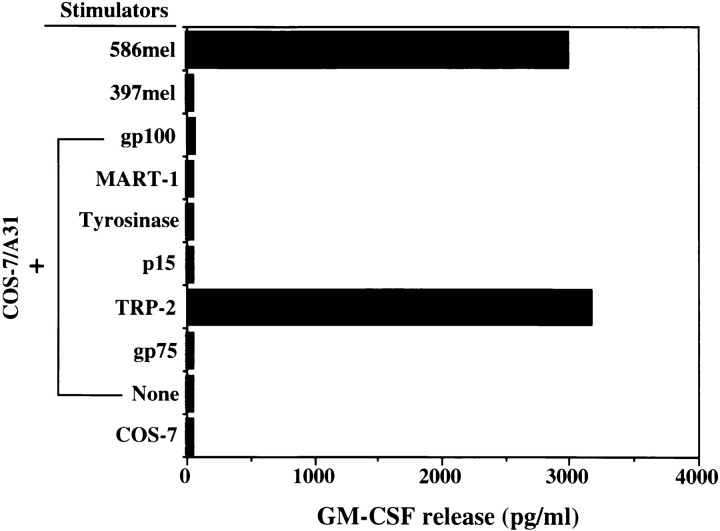
Since only a limited number of T cells were available, we first tested whether or not these T cells recognized previously identified tumor antigens or melanocyte-lineage differentiation proteins. Recognition of COS-7 cells transfected with HLA-A31 cDNA and genes encoding the known tumor antigens or putative antigens including MART-1 (13), gp75 (18), gp100 (15), tyrosinase (17), p15 (26), and TRP-2 (25, 27) by CTL clone 4 was tested. COS cells transfected with HLA-A31 alone or TRP-2 alone did not confer recognition by the T cell clones. However, COS cells transfected with HLA-A31 and TRP-2 cDNA stimulated GMCSF release from T cells, whereas COS cells transfected with HLA-A31 and other genes did not, indicating that the T cell clone 4 recognized TRP-2 as a tumor antigen in an HLA-A31 restricted manner (Table 1 and Fig. 2). TRP-2 is located on the human chromosome 13 and has been shown to be a member of the tyrosinase-related gene family and shares a 40–45% amino acid sequence identity with tyrosinase and gp75/TRP-1 (25, 27). TRP-2 encodes a protein with 519 amino acids and has been demonstrated to have DOPAchrome tautomerase activity involved in melanin synthesis (27).
Figure 2.
Identification of TRP-2 as a tumor antigen recognized by CTL clone 4. GM-CSF release by CTL clone 4 was measured after coculture with COS-7 cotransfected with the HLA-A31 cDNA along with genes encoding MART-1, gp75/TRP-1, gp100, tyrosinase, p15, and TRP-2. Control stimulator cells included 586mel, 397mel, COS-7 alone, and COS-7 transfected with the HLA-A31 cDNA.
Expression of the TRP-2 Gene.
Northern blot analyses were performed using TRP-2 cDNA as a probe to evaluate the expression pattern of TRP-2 in different tissues. Normal retinal tissue was shown to be the only positive expression of TRP-2 among the normal human tissues tested. The expression pattern of TRP-2 in melanoma cell lines and other cell lines is listed in Table 2. 22 of 27 melanoma cell lines were found to express TRP-2. The Burkitt's B cell line Daudi and the breast cancer cell line MDA231 were negative, in agreement with previous results (27). Thus, like tyrosinase, TRP-1, gp100, and MART-1, the expression pattern of this gene appeared to be restricted to melanomas, normal melanocyte cell lines, and retina.
Table 2.
Expression of TRP-2 in Different Cell Lines and Human Tissues Tested
| Cell lines | Expression of TRP-2 | |
|---|---|---|
| 397mel | + | |
| 526mel | + | |
| 501mel | + | |
| 537mel | − | |
| 553Bmel | + | |
| 586mel | + | |
| 624mel | + | |
| 677mel | + | |
| 679mel | − | |
| 697mel | + | |
| 729mel | − | |
| 894mel | + | |
| 836mel | − | |
| 888mel | + | |
| 928mel | + | |
| 1290mel | + | |
| 1300mel | + | |
| 952mel | + | |
| HT144 | + | |
| 1011mel | + | |
| 1088mel | + | |
| SK23 | + | |
| SK28 | + | |
| Maisel | + | |
| Groves | + | |
| WN266 | + | |
| A375 | − | |
| 586EBVB | − | |
| Melanocytes | ||
| FM906 | + | |
| FM680 | + | |
| Other tumor lines | ||
| Daudi | − | |
| MDA231 | − | |
| Normal tissues | ||
| Retina | + | |
| Testis | − | |
| Brain | − | |
| Spleen | − | |
| Liver | − | |
| Fetal liver | − | |
| Thymus | − | |
| Lung | − |
Expression of TRP-2 was tested by Northern blot analysis with 10–20 μg of total RNA and probed with the TRP-2 cDNA fragment. Daudi is a Burkitt's B cell line and MDA231 is a breast cancer cell line.
The Peptide Epitopes Recognized by T Cells.
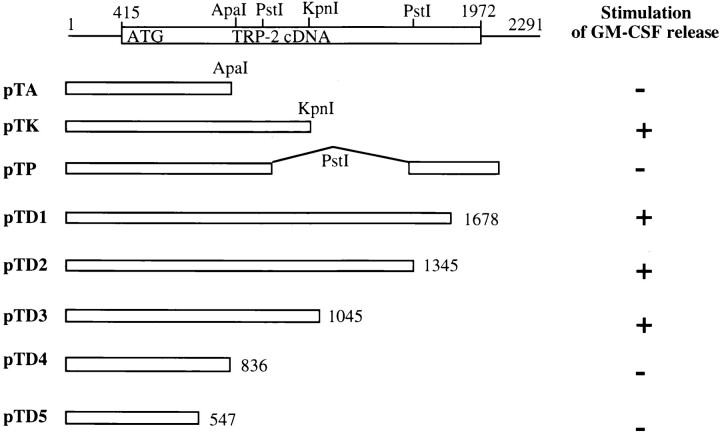
To determine the antigenic epitopes from TRP-2, a series of nested deletions of the TRP-2 gene from the 3′ end using ExoIII/S1 nuclease, as well as DNA fragments encoding the truncated form of TRP-2, were generated. These deletions and subcloned constructs were transfected into COS-7 cells along with the pBK-CMV plasmid containing the HLA-A31 cDNA. Recognition of the transfected COS cells was tested with the CTL clone 4 by measuring GM-CSF cytokine release from the CTL clone. Fig. 3 indicates that pTD1, 2, and 3 constructs retained the ability to stimulate cytokine release from the CTL clone 4, but pTD4 and pTD5 lost the stimulating activity to the CTL clone 4, indicating that the epitope(s) recognized by the CTL clone 4 was located in the region of nucleotides 836–1045. This was consistent with results obtained by the subcloning experiments. Although pTA and pTP lost the ability to stimulate cytokine release from CTL clone 4, pTK still remained positive in the cytokine release assay. Therefore, the epitopes resided in a DNA fragment flanked by the first PstI and KpnI sites as shown in Fig. 4.
Figure 3.
Construction of deletions and subclones of the TRP-2 gene and T cell recognition. The full-length cDNA of TRP-2 which comprises the 1,557-bp open reading frame is shown. Nucleotides are numbered from the first nucleotide from the 5′ untranslated region of TRP-2 cDNA. A series of deletion constructs and subcloning of DNA fragments were made. T cell recognition of each construct was determined after coculturing CTL clone 4 with COS-7 cotransfected with the DNA fragments shown above and the HLA-A31 gene.
Figure 4.
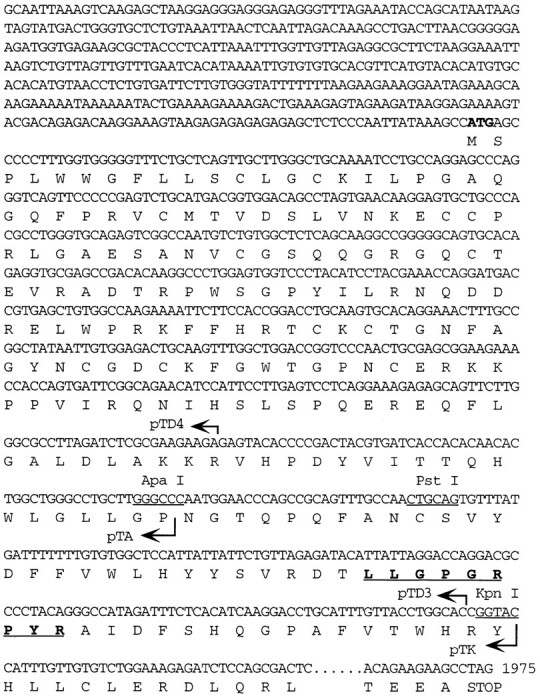
Antigenic peptide and partial coding sequence of TRP-2. The partial nucleotide and amino acid sequences of the TRP-2 gene are shown. The length and 3′ terminus of the DNA fragments in pTD4, pTA, pTD3, and pTK are indicated by arrows and the restriction sites for ApaI, PstI, and KpnI, are marked. The antigenic peptide sequence recognized by CTL clone 4 is in bold and underlined.
To identify the epitopes from the coding region of this small DNA fragment, we made five synthetic peptides based on the peptide binding motif for HLA-A31 (hydrophobic residues at position 2 and positively charged residues at position 9) (28). These peptides were pulsed onto 586EBV B cells and tested for their ability to stimulate cytokine release by CTL clone 4. As shown in Table 3, peptide TRP197–205 was strongly recognized by CTL clone 4 when pulsed on 586EBV B cells. The recognition of this peptide by CTL clone 4 was observed only when the peptide was pulsed onto HLA-A31+ EBV B cells such as 586EBV and 1510EBV, but not onto HLA-A31–negative T2 cells. CTL clone 4 did not recognize the ORF3P peptide derived from the alternative open reading frame of the TRP-1 gene. These results demonstrated that TIL586-C1 specifically recognized the ORF3P peptide derived from TRP-1 and CTL clone 4 specifically recognized the peptide derived from TRP2. No cross reactivity was observed, although both ORF3P and TRP197−205 were presented to T cells by HLA-A31 molecules.
Table 3.
Identification of Synthetic Peptides with Reactivity to T Cell Clones
| Target cells pulsed with peptide | GM-CSF release | |||||
|---|---|---|---|---|---|---|
| TIL586-C1 (TRP-1) | CTL clone 4 (TRP-2) | |||||
| pg/ml | ||||||
| 586EBV + TRP186–194 | VWLHYYSVR (TRP-2) | <50 | <50 | |||
| 586EBV + TRP185–194 | FVWLHYYSVR (TRP-2) | <50 | <50 | |||
| 586EBV + TRP194–202 | RDTLLGPGR (TRP-2) | <50 | <50 | |||
| 586EBV + TRP197–205 | LLGPGRPYR (TRP-2) | <50 | >4,000 | |||
| 586EBV + TRP213–221 | GPAFVTWHR (TRP-2) | <50 | <50 | |||
| 586EBV + ORF3P | MSLQRQFLR (TRP-1) | >8,000 | <50 | |||
| 1510EBV + TRP197–205 | LLGPGRPYR (TRP-2) | <50 | >4,000 | |||
| 1510EBV + ORF3P | MSLQRQFLR (TRP-1) | >6,000 | <50 | |||
| T2 + TRP197–205 | LLGPGRPYR (TRP-2) | <50 | <50 | |||
| T2 + ORF3P | MSLQRQFLR (TRP-1) | <50 | <50 | |||
| 586EBV + none | <50 | <50 | ||||
| 586mel + none | >5,000 | >3,000 | ||||
586EBV cells were incubated with individual peptides at a concentration of 1 μg/ml for 90 min. GM-CSF release was measured after coincubation of peptide-loaded 586EBV cells with T cell clones recognizing either TRP-1 or -2. GM-CSF secretion by T cells alone without stimulators was subtracted. 586EBV and 1510EBV were EBV transformed B cell lines expressing HLA-A31.
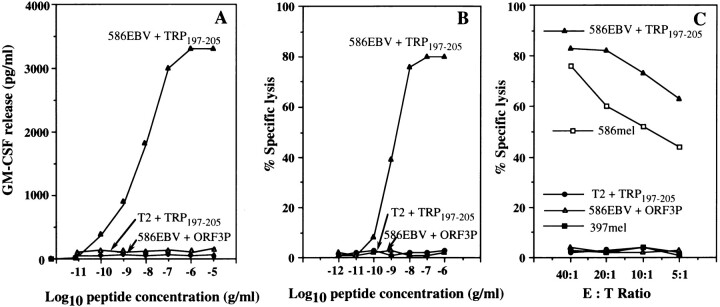
Characterization of TRP197–205 Peptide.
Titration experiments demonstrated that 1 nM of peptide was sufficient to stimulate GM-CSF release from the T cell clone 4 and the stimulation reached a plateau at 500 nM (Fig. 4 A). Lysis of 586EBV B cells pulsed with TRP197−205 by CTL clone 4 was also determined at various peptide concentrations (Fig. 4 B). Similar to cytokine release assays, lysis of target cells by the CTL clone 4 was detected at 1 nM peptide concentration. Maximum lysis was seen at 100 nM of peptide concentration. CTL clone 4 was capable of lysing 586EBV pulsed with TRP197-205 and 586mel tumor cells even at low E/T, but failed to lyse 586EBV B cells alone, or 586 EBV B cells pulsed with the control peptide ORF3P, or the HLA-A31 negative 397mel line (Fig. 4).
The majority of human melanoma antigens identified to date are nonmutated self-antigens, and the T cell recognition and binding affinity of these self-peptides to the corresponding MHC molecules have in some instances been improved by substitution of amino acids at anchor residues (29). A number of synthetic peptides including 8- or 10-mer, and modified peptides as indicated in Table 4 were made and tested for recognition by CTL clone 4 when pulsed onto 586EBV B cells. The 10-mer TLLGPGRPYR, in which one amino acid was extended at the NH2 terminus of TRP197−205, was still recognized by CTL clone 4 when pulsed on 586EBV B cells, but the reactivity was ∼60% of the overlapping 9-mer LLGPGRPYR. In contrast, peptides LLGPGRPYRA or LGPGRPYR had little or no reactivity compared with the overlapping 9-mer LLGPGRPYR. These results showed that the 9-mer LLGPGRPYR was better recognized than its derivatives, the 8- or 10-mers. Based on the binding motif of HLA-A31, we generated a few modified peptides with substitution of amino acids at anchor residues positions 2 and 9 as well as at other positions. Although the Arg residue at position 9 in the COOH terminus could be substituted with the Lys residue, the modified peptide retained only 60% of the activity of the parental peptide. No activity was detected by other amino acid substitutions at this position (Table 4). Substitution of a Leu residue at position 2 with either Ser, Ile, or Val residues retained the same activity or reduced the activity to 60% of the parental peptide while substitution with Ala or Phe at this position dramatically reduced the ability to stimulate cytokine release from T cells (Table 4). Other modifications at positions 1, 3, and 6 of the peptide either completely abrogated or dramatically reduced T cell recognition.
Table 4.
Comparison of T Cell Reactivity of Modified Peptides
| Target cells pulsed with peptides | CTL clone 4 | |
|---|---|---|
| GM-CSF release | ||
| pg/ml | ||
| 586EBV + LLGPGRPYR | 3,450 | |
| 586EBV + TLLGPGRPYR | 2,100 | |
| 586EBV + LLGPGRPYRA | 545 | |
| 586EBV + LGPGRPYR | <50 | |
| 586EBV + LIGPGRPYR | 2,545 | |
| 586EBV + LVGPGRPYR | 2,100 | |
| 586EBV + LSGPGRPYR | 3,300 | |
| 586EBV + LAGPGRPYR | 550 | |
| 586EBV + LFGPGRPYR | <50 | |
| 586EBV + LLGPGRPYK | 2,000 | |
| 586EBV + LLGPGRPYH | <50 | |
| 586EBV + ALGPGRPYR | <50 | |
| 586EBV + RLGPGRPYR | <50 | |
| 586EBV + KLGPGRPYR | 420 | |
| 586EBV + LLLPGRPYR | <50 | |
| 586EBV + LLFPGRPYR | <50 | |
| 586EBV + LLAPGRPYR | <50 | |
| 586EBV + LLGPGFPYR | 738 | |
| 586EBV + LLGPGAPYR | <50 | |
| 586EBV + LLGPGIPYR | <50 | |
| 586EBV + LLGPGVPYR | <50 | |
| 586EBV + LLGPGKPYR | 321 | |
| 586EBV | <50 | |
| 586mel | >3,000 |
586EBV cells were incubated with individual peptides at a concentration of 0.5 μg/ml for 90 min. GM-CSF release was measured after coincubation of peptide-loaded 586EBV cells with the CTL clone 4 cells. GM-CSF secretion by T cells alone without stimulators was subtracted. 586EBV was a EBV transformed B cell line expressing HLA-A31.
Discussion
In the last few years, tyrosinase, MART-1, gp100, and TRP-1/gp75 have been identified as human melanoma antigens recognized by T cells derived from human PBLs or TILs. Here, we provide evidence that TRP-2, another melanoma/melanocyte differentiation antigen of the tyrosinase protein family, is indeed a new tumor antigen recognized by HLA-A31–restricted T cells. TIL586 cell line recognized both COS-7 cells transfected with HLA-A31 plus TRP-2 cDNA, and HLA-A31–positive EBV B cells pulsed with the TRP197−205 peptide derived from TRP-2. However, recognition of TRP-2 by TIL586 cell line was much lower than that of TRP-1 or the ORF3P peptide (data not shown). This is probably due to a difference in the percentage of subsets of the T cell population. Isolation of these T cell clones from TIL586 allowed us to demonstrate that the TRP-2 gene product is the second tumor antigen recognized by this CTL. Of interest, two additional CTL clones isolated from TIL586 were found to recognize tumor 586mel, but failed to recognize TRP-1 and -2 in the context of HLA-A31 (Wang and Rosenberg, unpublished data), indicating that CTLs derived from melanoma patient 586 recognized additional tumor antigens. T cell recognition of multiple tumor antigens in a single tumor has been previously observed. TIL lines derived from many HLA-A2 patients recognized both MART-1 and gp100 (30, 31). Several TIL lines obtained from patient 888 at different stages of disease recognized tyrosinase (17), p15 (26), and β-catenin (20); several CTL clones derived from the patient MZ2 recognized MAGE (9), BAGE (12), and GAGE (11), respectively. Recognition of multiple tumor antigens by TIL may explain the effective in vivo antitumor activity of these T cells when infused into the autologous patients. It has been demonstrated that tumor reactive CTL can be readily generated from PBL of healthy individuals as well as melanoma patients by stimulation with peptides (32–34). The effectiveness of the adoptive transfer of peptide-specific CTL depends on the expression of the targeted tumor antigens on tumor cells. Antigen-loss variants may overgrow, as suggested by the observation that antigen-loss variants occurred in vivo (35). Therefore, use of multiple antigen- or multiple epitope-specific CTLs (consisting of subsets of T cell populations) may be more effective in the treatment of cancer patients and have advantages over CTL that recognize a single antigen or epitope.
TRP-2 belongs to a class of shared, differentiation antigens including tyrosinase, MART-1/Melan-A, gp100, and TRP-1 or gp75. TRP-2 is one of the most highly expressed glycoproteins in human pigmented melanocytic cells and melanoma. While its biological function is not completely known, TRP-2 has been shown to have DOPAchrome tautomerase activity which is involved in melanin production (27). Mutation of this gene in black-haired mice may be responsible for hair color change (36). A direct correlation was observed between melanin content and the expression of the four melanogenic proteins: tyrosinase, gp100, TRP-1/gp75, and TRP-2 (37, 38). The TRP-2 protein contains two putative copper-binding sites, cysteinerich regions, and a transmembrane domain. Human TRP-2 has been mapped to chromosome 13 while the mouse counterpart has been mapped to chromosome 14 in the region of the coat color mutation slaty. There is about a 40% amino acid sequence identity between TRP-2 and tyrosinase or TRP-1/gp75, but no CTL line or clone has been found that recognizes a common peptide epitope among the tyrosinase protein family.
The 9-mer TRP197−205 peptide recognized by CTL clone 4 is located at one of the copper binding sites in the coding region of TRP-2. This peptide most efficiently stimulated cytokine release from T cells compared with other peptides including modified peptides tested in this study. This was in agreement with the predicted HLA-A31–binding motif, which indicates that Leu at position 2 and Arg at position 9 are the favorable residues. Although Leu at position 2 and Arg at position 9 could be replaced with Ile and Ser at position 2 and Lys at position 9, respectively, with little or minor loss of T cell recognition, substitutions of amino acids at positions 1, 3, or 6 led to dramatic or complete loss of reactivity. It remained to be tested whether this is due to the change of binding affinity of the modified peptides to HLA-A31, or to abrogation of T cell receptor interacting with the peptide/MHC complex.
Analysis of the structural similarities in HLA-A3, A11, A31, A33, and A68 and their peptide binding motif has suggested the existence of the A3-like supermotif (39). A single epitope peptide could cross-react with HLA-A3, A11, A31, A33, and A68 molecules which are cumulatively expressed in ∼40–50% of the general population. It has been reported that the same peptide epitope derived from Hepatitis B virus nucleocapsid protein could be presented by HLA-A31 and -A68 molecules and recognized by the corresponding HLA-A31– or -A68–restricted CTL (40). We tested whether HLA-A31–restricted T cells recognized TRP-1 and TRP-2 epitopes when pulsed onto HLA-A3–positive EBV B cells. Interestingly, weak recognition was detected based on GM-CSF release from T cells (data not shown). However, no recognition of HLA-A3–positive tumor cells was detected. We are now studying the binding of these HLA-A31–epitope peptides to HLA-A3 and A11 to determine whether CTL restricted by HLA-A3, -A11, -A33, and -A68 could be generated from PBL using HLAA31–binding peptides.
Since TRP-2 is a nonmutated self antigen, this raises questions about the nature and mechanism of immune responses to self antigens on growing tumor cells. Although shared differentiation antigens like those encoded by tyrosinase, MART-1/Melan-A, gp100, TRP-1, and -2 may serve as useful targets for the immunotherapy of patients with melanoma, anti-tumor activity might lead to self-tissue destruction or autoimmune disease as well. Local depigmentation occurring in melanoma patients has been reported to correlate with prolonged survival (41, 42) and clinical response to chemoimmunotherapy (43). The depigmentation resulting from the destruction of melanocytes as a consequence of an immune response directed against these differentiation antigens may be observed in patients with melanoma responding to IL-2–based immunotherapy (44). Patient 586 experienced partial regression (>50% reduction) of all measurable lesions after receiving the infusion of TIL586 cells plus IL-2. However, no adverse effects related to melanocyte destruction were observed. These results suggest that CTLs directed against shared differentiation antigens may mediate tumor destruction without damage to normal tissues. Hara et al. have recently demonstrated that passive immunization with a mouse monoclonal antibody against TRP-1/gp75 TA99 induced protection against and rejection of the TRP-1+ B16F10 melanoma in syngeneic mice (45). There was no evidence of decrease in pigmentation, inflammation, or changes in cellular morphology or tissue architecture in the eyes of mice treated with antibody. Although coat color changes were observed in regenerating hair on the trunk, the threshold dose required for coat color changes was fivefold greater than that required for tumor regression (45).
The use of a mouse B16 melanoma model may be helpful in evaluating the potential autoimmune responses and maximum antitumor activity induced by self-antigens. However, mouse melanoma antigens with properties similar to the MART-1, gp100, tyrosinase, TRP-1, and -2 antigens found in human melanoma have not been identified. Recently, mouse TRP-2 has been identified as a tumor antigen recognized by a CTL line against B16 melanoma (Bloom, M.B., and J.C. Yang, personal communication). Like human TRP-2, mouse TRP-2 was frequently expressed in the samples of melanoma of Tyr-SV40E transgenic mice (46). These results suggested that TRP-2 is a tumor antigen recognized by CTL in both mouse and human melanoma. Mouse TRP-2 may represent an ideal tumor antigen to be tested in mouse models and to evaluate questions important for the development of effective immunotherapies in humans.
Figure 5.
Characterization of the antigenic peptide recognized by CTL clone 4. (A) GM-CSF release by T cells at different peptide concentrations. 586EBV (A31+) were pulsed with the TRP197–205 peptide ( ) and T2 (non-A31) cells were pulsed with the TRP197–205 (
) and T2 (non-A31) cells were pulsed with the TRP197–205 ( ) at various peptide concentrations for 90 min. ORF3P as a control peptide was pulsed onto 586EBV B cells (--▵--). GM-CSF release by CTL clone 4 was determined after coincubation with 586EBV B cells pulsed with TRP197–205 and ORF3P, and T2 cells pulsed with TRP197–205. (B) Sensitization of the target cells for lysis by CTL clone 4 at different peptide concentrations. 586EBV B cells were incubated with TRP197–205 (
) at various peptide concentrations for 90 min. ORF3P as a control peptide was pulsed onto 586EBV B cells (--▵--). GM-CSF release by CTL clone 4 was determined after coincubation with 586EBV B cells pulsed with TRP197–205 and ORF3P, and T2 cells pulsed with TRP197–205. (B) Sensitization of the target cells for lysis by CTL clone 4 at different peptide concentrations. 586EBV B cells were incubated with TRP197–205 ( ), an irrelevant peptide ORF3P (--▵--), and T2 cells pulsed with TRP197–205 (
), an irrelevant peptide ORF3P (--▵--), and T2 cells pulsed with TRP197–205 ( ) at various peptide concentrations. After peptide incubation, target cells were labeled for 30 min. Following washes, cytolytic activity of CTL clone 4 at an E/T of 40:1 was measured after a 4 h incubation of T cells with target cells. (C) Lysis of the target cells by CTL clone 4 at different E/T. Target 586EBV cells were separately incubated with TRP197–205 (
) at various peptide concentrations. After peptide incubation, target cells were labeled for 30 min. Following washes, cytolytic activity of CTL clone 4 at an E/T of 40:1 was measured after a 4 h incubation of T cells with target cells. (C) Lysis of the target cells by CTL clone 4 at different E/T. Target 586EBV cells were separately incubated with TRP197–205 ( ) or the irrelevant peptides ORF3P (--▵--), and target T2 cells were incubated with the TRP197–205 peptide (
) or the irrelevant peptides ORF3P (--▵--), and target T2 cells were incubated with the TRP197–205 peptide ( ) for 90 min. 586mel (
) for 90 min. 586mel ( ) and 397mel (
) and 397mel ( ) were used as positive and negative controls, respectively.
) were used as positive and negative controls, respectively.
Acknowledgments
We would like to thank Drs. Robert Bright, S. Topalian, and John Wunderlich for providing cell lines, Shigeki Shibahara for providing a TRP-2 cDNA clone, and P. Robbins and M. El-Gamil for helpful discussions on T cell expansion. We also thank A. Mixon and E.B. Fitzgerald for performing FACS® analysis.
Footnotes
1 Abbreviations used in this paper: E/T, effector/target ratio; TIL, tumor infiltrating lymphocyte.
References
- 1.Rosenberg SA, Packard BS, Aebersold PM, Solomon D, Topalian SL, Toy ST, Simon P, Lotze MT, Yang JC, Seipp CA, et al. Use of tumor infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. N Engl J Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yannelli JY, Yang JC. Treatment of patients with metastatic melanoma using autologous tumor–infiltrating lymphocytes and interleukin-2. J Natl Cancer Inst. 1994;86:1159–1166. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 3.Boon T, Cerottini J-C, Van Den Eynde B, Van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 4.Houghton AN. Cancer antigens: immune recognition of self and alterted self. J Exp Med. 1994;180:1–4. doi: 10.1084/jem.180.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsomides TJ, Eisen HN. T-cell antigens in cancer. Proc Natl Acad Sci USA. 1994;91:3487–3489. doi: 10.1073/pnas.91.9.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardoll DM. News and views: a new look for the 1990s. Nature (Lond) 1994;369:357–358. doi: 10.1038/369357a0. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg SA. The development of new cancer therapies based on the molecular identification of cancer regression antigens. Cancer J Sci Am. 1995;1:90–100. [PubMed] [Google Scholar]
- 8.Wang RF, Rosenberg SA. Human tumor antigens recognized by T lymphocytes: implications for cancer therapy. J Leukocyte Biol. 1996;60:296–309. doi: 10.1002/jlb.60.3.296. [DOI] [PubMed] [Google Scholar]
- 9.Van der Bruggen P, Traversari C, Chomez P, Lurquin C, DePlaen E, Van Den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science (Wash DC) 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 10.Gaugler B, Van Den Eynde B, Van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethe B, Brasseur F, Boon T. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böel P, Wildmann C, Sensi ML, Brasseur R, Renauld JC, Coulie P, Boon T, Van der Bruggen P. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami Y, Eliyahu S, Delgaldo CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen ED, Lurquin C, Szikora J-P, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Identification of a human melanoma antigen recognized by tumor infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brichard V, Van Pel A, Wölfel T, Wölfel C, De Plaen E, Lethë B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins PF, El-Gamil M, Kawakami Y, Stevens E, Yannelli J, Rosenberg SA. Recognition of tyrosinase by tumor infiltrating lymphocytes from a patient responding to immunotherapy. Cancer Res. 1994;54:3124–3126. [PubMed] [Google Scholar]
- 18.Wang RF, Robbins PF, Kawakami Y, Kang XQ, Rosenberg SA. Identification of a gene encoding a melanoma tumor antigen recognized by HLA-A31–restricted tumor-infiltrating lymphocytes. J Exp Med. 1995;181:799–804. doi: 10.1084/jem.181.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer K-H Zum Buschenfelde, and D. Beach. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science (Wash DC) 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 20.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulie PG, Lehmann F, Lethe B, Herman J, Lurquin C, Andrawiss M, Boon T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92:7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topalian S, Solomon D, Avis FP, Chang AE, Freeksen DL, Linehan WM, Lotze MT, Robertson CN, Seipp CA, Simon P, Simpson CG, Rosenberg SA. Immunotherapy of patients with advanced cancer using tumor infiltrating lymphocytes and recombinant interleukin-2: a pilot study. J Clin Oncol. 1988;6:839–853. doi: 10.1200/JCO.1988.6.5.839. [DOI] [PubMed] [Google Scholar]
- 24.Walter EA, Greenberg PD, Gibert MJ, Finch RJ, Watanabe KS, Thomas ED, Riddell SR. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama K, Suzuki H, Yasumoto K-I, Tomita Y, Shibahara S. Molecular cloning and functional analysis of a cDNA coding for human DOPAchrome tautomerase/ tyrosinase–related protein-2. Biochim Biophys Acta. 1994;1217:317–321. doi: 10.1016/0167-4781(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 26.Robbins PF, El-Gamil M, Li YF, Topalian SL, Rivoltini L, Sakaguchi K, Appella E, Kawakami Y, Rosenberg SA. Cloning of a new gene encoding an antigen recognized by melanoma-specific HLA-A24 restricted tumor-infiltrating lymphocytes. J Immunol. 1995;154:5944–5950. [PubMed] [Google Scholar]
- 27.Bouchard B, Del Marmol V, Jackson IJ, Cherif D, Dubertret L. Molecular characterization of a human tyrosinase–related–protein-2 cDNA. Eur J Biochem. 1994;219:127–134. doi: 10.1111/j.1432-1033.1994.tb19922.x. [DOI] [PubMed] [Google Scholar]
- 28.Rammensee H-G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 29.Parkhurst MR, Salgaller M, Southwood S, Robbins P, Sette A, Rosenberg SA, Kawakami Y. Improved induction of melanoma reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A0201 binding residues. J Immunol. 1996;157:2537–2548. [PubMed] [Google Scholar]
- 30.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JB, Appella E, Rosenberg SA. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2 restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins PF, Sette A, Appella E, Rosenberg SA. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 32.Rivoltini L, Kawakami Y, Sakaguchi K, Southwood S, Sette A, Robbins PF, Marincola FM, Salgaller ML, Yannelli JR, Appella E. Induction of tumor-reactive CTL from peripheral blood and tumor-infiltrating lymphocytes of melanoma patients by in vitro stimulation with an immunodominant peptide of the human melanoma antigen MART-1. J Immunol. 1995;154:2257–2265. [PubMed] [Google Scholar]
- 33.Visseren MJ, Van Elsas A, van der Voort EI, Ressing ME, Kast WM, Schrier PI, Melief CJ. CTL specific for the tyrosinase autoantigen can be induced from healthy donor blood to lyse melanoma cells. J Immunol. 1995;154:3991–3998. [PubMed] [Google Scholar]
- 34.Wölfel T, Schneider J, Meyer K-H Zum Buschenfelde, H.-G. Rammensee, O. Rotzschke, and K. Falk. Isolation of naturally processed peptides recognized by cytolytic T lymphocytes (CTL) on human melanoma cells in association with HLA-A2.1. Int J Cancer. 1994;57:413–418. doi: 10.1002/ijc.2910570320. [DOI] [PubMed] [Google Scholar]
- 35.Jager E, Ringhoffer M, Karbach J, Arand M, Oesch F, Knuth A. Inverse relationship of melanocyte differentiation antigen expression in melanoma tissue and CD8+cytotoxic-T-cell responses: evidence for immunoselection of antigen-loss variants in vivo. Int J Cancer. 1996;66:470–476. doi: 10.1002/(SICI)1097-0215(19960516)66:4<470::AID-IJC10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Jackson I, Chambers DM, Tsukamoto K, Copeland NG, Gilbert DJ, Jenkins NA, Hearing V. A second tyrosinase-related protein, TRP-2, maps to and is mutated at the mouse slatylocus. EMBO (Eur Mol Biol Organ) J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halahan, R., and G. Moellmann. 1993. White mutants in mice shedding light on humans. J. Invest. Dermatol. 100(Suppl.): 176s–185s. [PubMed]
- 38.Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. TIG (Trends Genet) 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 39.Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo R, Chesnut RW, Sette A. Definition of an HLA-A3–like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 40.Missale G, Redeker A, Person J, Fowler P, Guilhot S, Schlicht HJ, Ferrari C, Chisari FV. HLA-A31– and HLA-Aw68–restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med. 1993;177:751–762. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordlund JJ, Kirkwood M, Forget BM, Milton G, Albert DM, Lerner AB. Vitiligo in patients with metastatic melanoma: a good prognostic sign. J Am Acad Dermatol. 1983;9:689–695. doi: 10.1016/s0190-9622(83)70182-9. [DOI] [PubMed] [Google Scholar]
- 42.Bystryn J-C, Rigel D, Friedman RJ, Kopf A. Prognostic significance of hypopigmentation in malignant melanoma. Arch Dermatol. 1987;123:1053–1055. [PubMed] [Google Scholar]
- 43.Richards JM, Mehta N, Ramming K, Skosey P. Sequential chemoimmunotherapy in the treatment of metastatic melanoma. J Clin Oncol. 1992;10:1338–1343. doi: 10.1200/JCO.1992.10.8.1338. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg SA, White DE. Vitiligo in patients with melanoma: normal tissue antigens can be targeted for cancer immunotherapy. J Immunother. 1996;19:81–84. [PubMed] [Google Scholar]
- 45.Hara I, Takechi Y, Houghton AN. Implicating a role for immune recognition of self in tumor rejection: passive immunization against the brownlocus protein. J Exp Med. 1995;182:1609–1614. doi: 10.1084/jem.182.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlow SJ, Hearing VJ, Sakai C, Urabe K, Zhou BK, Silvers WK. Changes in expression of putative antigens encoded by pigment genes in mouse melanomas at different stages of malignant progression. Proc Natl Acad Sci USA. 1995;92:10152–10156. doi: 10.1073/pnas.92.22.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]