Abstract
Background & Aims
Systemic inhibition of DNA methylation causes cancers in animals, in part by inducing genetic instability. Epidemiologic evidence linking low genomic methylation in systemic blood DNA to carcinogenesis is limited, however, specifically of the colorectum, where genetic instability is a primary etiologic factor. We examined genomic methylation of leukocyte DNA in relation to colorectal adenoma (CRA) among asymptomatic women (40-79 years) participating in a multi-center colonoscopy screening study (CONCeRN Study, 2000-2002).
Methods
Of all participants who completed self-administered risk factor and food frequency questionnaires, peripheral blood donation, and colonoscopy, 115 pairs of CRA cases and controls with matching age and month of blood draw were studied. Genomic methylation of leukocyte DNA was determined by liquid chromatography-mass spectrometry. Conditional logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI).
Results
Compared with women in the lowest tertile of genomic methylation, women in the second (OR, 0.72; 95% CI, 0.34-1.52) and third tertiles (OR, 0.17; 95% CI, 0.06-0.49) had lower risk of CRA (P trend = 0.002). The inverse relationship was stronger for non-advanced than for advanced adenoma and, less notably, for proximal than for distal adenoma. The association was also moderately more protective with low rather than high total folate intake, but did not differ by other nutrients involved in one-carbon metabolism or colorectal cancer risk factors.
Conclusions
Our findings regarding asymptomatic CRA implicate systemic genomic methylation as a potential etiologic factor for an early stage of colorectal adenoma.
Keywords: case-control study, colonoscopy, colorectal adenoma, folate, genomic DNA methylation
The proportion of methylated cytosine in genomic DNA is typically reduced in a wide range of tumors compared to that in normal tissue 1. This genomic hypomethylation may occur independently of 2 or in conjunction with 3 another cancer-associated aberrant methylation, CpG island hypermethylation in mostly promoters, that often silences tumor suppressor genes 4, 5. As in CpG island hypermethylation 6, genomic hypomethylation presents field effects, indicating that the alterations may occur in early stages of tumorigenesis. Genomic DNA methylation is lower in the colorectal tumor, for example, compared to normal mucosa within a patient with colorectal neoplasia 7 and is also lower in adjacent or distant normal-appearing mucosa of patients with either colorectal cancer or its precursor adenoma than in the mucosa of healthy controls 8, 9.
In addition to the genomic hypomethylation in tumor tissues, animal experiments support the notion that systemically low genomic methylation may be causally involved in tumorigenesis: transgenic mice with little DNA methyltransferase activity 10, 11 and rodents either treated with methyl inhibitors 12 or fed methyl-deficient diets (diets low in methionine, choline, folate, and vitamin B12) 13 all developed malignant tumors. The carcinogenic mechanism is proposed to be through chromosomal instability and activation of proto-oncogenes 14. Similarly, in limited human data from Britain, genomic methylation in systemic blood DNA was lower in symptomatic colorectal adenoma or cancer patients than in controls 8. This finding supports the causal evidence from animal experiments but might have been the result of advanced tumors. Also, it is of interest whether such an association exits in populations fortified with folic acid, such as the U.S., considering that the nutrient and other factors of one-carbon metabolism, which supplies methyl units, are one of few known determinants of genomic methylation, including age and gender 3, 15, 16.
Detection of altered genomic methylation in systemic blood DNA in the early stage of adenoma would offer stronger implications for its etiologic role and may render utility for easily obtainable diagnostics and prognostics 1. We examined whether genomic methylation of leukocyte DNA is associated with colorectal adenoma (CRA) among asymptomatic women participating in a U.S. colonoscopy screening study. We further investigated whether the association between genomic methylation and CRA differs by the range of genomic methylation, the stage and site of adenoma, nutritional factors of one-carbon metabolism, or various lifestyle risk factors of colorectal cancer.
Materials and Methods
The CONCeRN Study
The CONCeRN (COlorectal Neoplasia screening with Colonoscopy in asymptomatic women at Regional Navy/army medical centers) Study is a colonoscopy screening study with a primary objective to determine the efficacy of colonoscopy versus sigmoidoscopy as a screening tool for colorectal neoplasia, as described in detail elsewhere 17. Subjects were recruited between July 1999 and December 2002 among consecutive female patients at four medical centers (National Naval Medical Center, Bethesda, MD; Walter Reed Army Medical Center, Washington, DC; and Naval Medical Centers in San Diego, CA and Portsmouth, VA). Average-risk asymptomatic women aged 50-79 years or asymptomatic women aged 40-79 years with a family history of colorectal cancer among first-degree relatives were approached for the screening study. To ensure recruitment of asymptomatic women at average-risk, women were excluded if they had a screening in recent years or had a personal history of adenomas, colorectal cancer, inflammatory bowel disease, hereditary non-polyposis colorectal cancer syndrome, or familial adenomatous polyposis 17. The study was approved by institutional review boards at the National Cancer Institute and participating medical centers. All participants provided written informed consent.
Case Definition and Ascertainment
After standard bowel preparation, participants underwent colonoscopic examination by a gastroenterologist or colorectal surgeon. The location of the polyp was defined based on the depth of insertion of the colonoscope and anatomical landmarks (hepatic flexure, splenic flexure, and junction of the descending and sigmoid colon), and the diameter was estimated by a guidewire (Olympus Colonoscopy Measuring Guidewire). Final diagnosis of each polyp was made by an expert gastrointestinal pathologist who reviewed histologic specimens without knowledge of the initial diagnosis during colonoscopy. Based on the most advanced lesion found in the entire colon and rectum, cases of CRA were defined as persons with a pathologically verified adenoma of tubular, villous, or mixed type and of any size in the proximal colon (ascending and transverse) or the distal colon (descending and sigmoid colon and rectum), excluding hyperplastic polyps or benign lesions. Advanced adenomas were defined as cancer, high-grade dysplasia, villous adenoma, or adenomas ≥ 1cm.
Biospecimen and Questionnaire Data Collection
For the current study involving biospecimen and questionnaire data collection, participants recruited from January 2000 through 2002 were invited. Upon arrival at the medical centers for the colonoscopy, participants submitted two complete questionnaires that they had received in mail. The risk factor questionnaire queried information on demographic and known risk factors for colorectal cancer, including family history, menopausal status and use of hormone replacement therapy, smoking, current and past physical activity, and use of aspirin and other non-steroid anti-inflammatory drugs. Body mass index (kg/m2) was calculated from direct measures of height and weight.
The consumption frequency (10 possible categories) and portion size (3 possible ranges) for 124 food items over the past year was assessed using the diet history questionnaire (DHQ) 18, a self-administered food frequency questionnaire (FFQ) that was validated against data from four 24-hour dietary recalls and observed comparable with other FFQs 19. From the responses, nutrient intake was estimated using the Diet*Calc Analysis Program (version 1.4.3, 2005), based on the nutrient content information provided by the U.S. Department of Agriculture Survey Nutrient Database and the Nutrition Data Systems for Research (NDS-R) from the University of Minnesota. Nutrients that are commonly provided in multivitamins or single vitamin supplements were estimated separately by the source: from foods only, from supplements only, and in the total amount. Folate intake from food was estimated based on either pre-fortification content before year 1998 to account for long-term intake or post-fortification content. Post-fortification folate was further classified as natural folate (reduced polyglutamates) or synthetic folate (folic acid in grain products, breakfast cereals, supplements) and was estimated either in μg or in dietary folate equivalent (DFE: natural folate + synthetic folate × 1.7, to reflect higher absorption of the latter).
Case-Control Selection
Out of 990 women approached with questionnaires, 910 women (92% participation; 181 cases and 729 non-cases) provided the information and had undergone colonoscopy, and 865 women provided fasting venous blood samples. DNA was successfully extracted from 827 buffy coat samples (155 cases, 672 non-cases) using non-organic DNAQuikTM reagents at BioServe (Laurel, MD) and was stored at −70° C until assay. We selected all 115 adenoma cases with sufficient DNA available and matched each case to a control based on age at colonoscopy (± 2.5 years) and time of blood collection (± 2 months).
Genomic DNA Methylation Assay
DNA (1μg) was hydrolyzed by sequential enzyme digestion 20, 21. DNA bases as well as methylcytosine were separated by reversed-phase high-performance liquid chromatography (HPLC) and were quantified by electrospray ionization mass spectrometry (ESI/MS): exogenous isotopomers were used as internal standards 20, 21. Compared with radiolabelled methyl acceptance assays of genomic methylation that rely on methyl-sensitive endonuclease and methyltransferase, the LC/ESI/MS method is direct and has less variability 21.
Genomic DNA methylation was expressed as the relative amount of methylcytosine to total cytosine residues: percent methylation = [methylcytosine / (methylcytosine + unmethylated cytosine)] × 100. We inserted quality control samples from three volunteers randomly and blindly among study samples to estimate assay reproducibility. Mean coefficient of variation within subject was 8.6%.
Statistical Analysis
Characteristics of matched cases and controls were compared using a paired t-test or the Wilcoxon nonparametric test for continuous variables and the chi-square test for categorical variables. Food items and nutrients, except alcohol, were adjusted for energy intake by the nutrient density method 22. Percent genomic methylation and other continuous variables were categorized as binary or tertiles based on the distribution among controls. To account for potential non-linearity in the dose-response relationship of genomic methylation with CRA, a smoothing spline of the matching-adjusted association between the two was examined using the R package survival with the function pspline and 3 degrees of freedom 23. Conditional logistic regression was performed to estimate odds ratios (OR) and 95% confidence intervals (CI) for the association between genomic methylation and CRA. We examined known risk factors for colorectal cancer and dietary factors, such as fiber, fat, calcium, vitamin D, and fruits and vegetables, as potential confounders. Variables that changed the methylation-adenoma association by 10% or more were kept in the model. The final model included smoking history (never, smoked < 20 years or ≥ 20 years) and intake of selected red meats (energy-adjusted tertiles of the sum of daily gram intake of hamburger, steak, pork, bacon, and sausage; this 5-item variable consists of red meats commonly cooked by high temperature cooking methods and represents a proxy for heterocyclic amine intake 24). Linear trends for the categorical analyses were assessed using the score variable that contained median values of binary or tertile categories. Effect modification was evaluated using cross-product terms (P < 0.10 considered significant) and likelihood ratio test statistics comparing models with and without the product term.
Results
Compared with controls, cases were more likely to have never used postmenopausal hormone replacement therapy, more likely to have smoked for greater than 20 years, and consumed more of the selected red meats and processed meat (Table 1). Otherwise, cases did not significantly differ from controls with respect to demographics, study centers, and colorectal cancer risk factors, and known determinants of one-carbon metabolism.
Table 1.
Characteristics of cases and controls, the CONCeRN Study (2000-2002)
| Case | Control | ||||
|---|---|---|---|---|---|
| N | % or median | N | % or median | ||
| Percent genomic methylation | 115 | 5.32* | 115 | 5.36 | |
| Non-advanced adenoma | 90 | 5.29* | 90 | 5.36 | |
| Advanced adenoma | 25 | 5.36 | 25 | 5.39 | |
| Age | 115 | 61 | 115 | 61 | |
| Race | White | 88 | 77% | 92 | 80% |
| Black | 14 | 12% | 14 | 12% | |
| Others | 13 | 11% | 9 | 8% | |
| Study Center | Bethesa | 83 | 72% | 87 | 76% |
| Walter Reed | 12 | 10% | 9 | 8% | |
| San Diego | 13 | 11% | 13 | 11% | |
| Portsmouth | 7 | 6% | 6 | 5% | |
| Postmenopausal | Yes | 102 | 89% | 101 | 88% |
| Hormone replacement | Current | 35 | 30% | 53 | 46% |
| Former | 19 | 17% | 20 | 17% | |
| Never | 48 | 42%* | 28 | 24% | |
| Family history of colorectal cancer | Yes | 28 | 24% | 34 | 30% |
| Body Mass Index, kg/m2 | 113 | 26.6 | 113 | 25.5 | |
| Smoking history | Former | 45 | 40% | 43 | 38% |
| Current | 6 | 5% | 5 | 4% | |
| Smoking duration | > 20 years | 38 | 34%* | 23 | 20% |
| Smoking dose | > 1 pack/d | 13 | 11% | 10 | 9% |
| Exercise | Regular currently | 96 | 86% | 86 | 77% |
| Aspirin use | Regular | 35 | 30% | 41 | 36% |
| Alcohol, drinks/week | 83 | 2.7 | 89 | 1.7 | |
| Multivitamin supplement use, % yes | 115 | 46% | 115 | 46% | |
| Calories | 112 | 1370 | 112 | 1342 | |
| Daily dietary intake (per 1000kcal) | 112 | 112 | |||
| Pre-fortification | |||||
| Folate from food, μg | 172 | 179 | |||
| Total folate, μg | 369 | 319 | |||
| Post-fortification | |||||
| Folate from food, μg | 216 | 218 | |||
| Total folate, μg / dietary folate equivalent or DFE | 399 / 460 | 347 / 414 | |||
| Vitamin B12 from food, μg | 2.0 | 2.1 | |||
| Vitamin B6 from food, mg | 1.1 | 1.1 | |||
| Riboflavin from food, mg | 0.9 | 1.0 | |||
| Methionine, g | 0.8 | 0.8 | |||
| Selected red meat (hamburger, steak, pork, bacon, sausage), g | 10* | 8 | |||
| Processed meat, g | 7.1* | 5.7 | |||
P < 0.05 for comparison of cases versus controls using Wilcoxon non-parametric test for continuous variables and using chi-square test for categorical variables
Percent genomic methylation of leukocyte DNA was significantly lower among cases, specifically of non-advanced adenoma, than controls (Table 1). Women in the highest tertile of genomic methylation had 70% lower risk of adenoma overall (Table 2). The inverse association was stronger after adjustment for smoking history and red meat consumption but was not affected by other factors. A linear dose-response model fit to the entire data was significantly influenced by outliers: 7 of the 8 observations under 4% and 12 of the 18 observations under 4.75% genomic methylation were cases, yielding a substantially higher risk estimate associated with the low levels than the risk estimate for the rest of the data (Figure 1A). A smoothing spline to the bulk of the observations without the influential outliers (i.e., 7.5th-95th percentile or 4.75%-5.75%) showed halving of the CRA risk going from about 5% to 5.75% (Figure 1B). Cases with low (< 4.75%) or high (>5.75%) methylation values were not systematically different in histology from the others: most were non-advanced tubular adenomas.
Table 2.
Association* between percent genomic methylation of leukocyte DNA and colorectal adenoma, the CONCeRN Study (2000-2002)
| % genomic methylation of leukocyte DNA
|
||||
|---|---|---|---|---|
| Tertile 1
(2.76 – 5.29) |
Tertile 2
(5.29 – 5.43) |
Tertile 3
(5.43 – 6.42) |
P trend | |
| All adenoma | ||||
| N, case / control | 51 / 38 | 42 / 39 | 22 / 38 | |
| Unadjusted OR (95% CI) | 1.0 (reference) | 0.76 (0.41, 1.42) | 0.29 (0.12, 0.69) | 0.009 |
| Adjusted OR (95% CI) | 1.0 (reference) | 0.72 (0.34, 1.52) | 0.17 (0.06, 0.49) | 0.002 |
|
| ||||
| Below median
(2.76 – 5.36) |
Above median
(5.37 -6.42) |
P trend | ||
|
| ||||
| Non-advanced adenoma | ||||
| N, case / control | 61 / 46 | 29 / 44 | ||
| Unadjusted OR (95% CI) | 1.0 (reference) | 0.35 (0.16, 0.78) | 0.01 | |
| Adjusted OR (95% CI) | 1.0 (reference) | 0.27 (0.11, 0.66) | 0.004 | |
| Advanced adenoma | ||||
| N, case / control | 12 / 11 | 13 / 14 | ||
| Unadjusted OR (95% CI) | 1.0 (reference) | 0.80 (0.22, 2.98) | 0.74 | |
| Adjusted OR (95% CI) | 1.0 (reference) | 0.84 (0.15, 4.80) | 0.84 | |
|
| ||||
| Proximal adenoma† | ||||
| N, case / control | 47 / 37 | 30 / 40 | ||
| Unadjusted OR (95% CI) | 1.0 (reference) | 0.47 (0.21, 1.05) | 0.06 | |
| Adjusted OR (95% CI) | 1.0 (reference) | 0.31 (0.12, 0.82) | 0.02 | |
| Distal adenoma† | ||||
| N, case / control | 33 / 28 | 17 / 22 | ||
| Unadjusted OR (95% CI) | 1.0 (reference) | 0.55 (0.20, 1.48) | 0.23 | |
| Adjusted OR (95% CI) | 1.0 (reference) | 0.58 (0.20, 1.72) | 0.32 | |
OR, odds ratio; CI, confidence interval.
Unadjusted odds ratios were obtained from conditional logistic regression accounting for the matching factors (age and month of blood draw). Stratified analyses by stage or anatomic location were conducted on binary level of genomic methylation due to small numbers, comparing above versus below median level. Multivariable adjusted odds ratios were obtained from a conditional logistic model that included matching factors, smoking history (never, < 20 years, ≥ 20 years), and red meat intake (energy-adjusted tertiles of daily sum of hamburger, steak, pork, bacon, and sausage consumed in grams).
Proximal and distal adenoma subgroups include cases (and their matched controls; 12 pairs) that had both proximal and distal adenomas.
Figure 1.
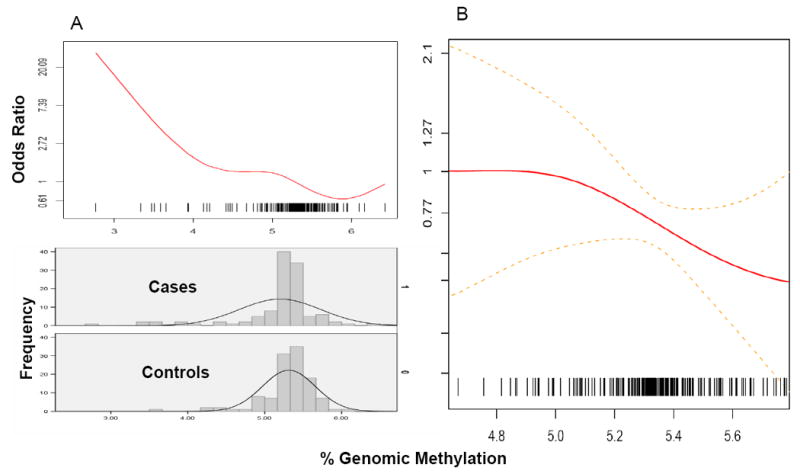
Smoothing spline of the odds ratio for leukocyte genomic methylation and colorectal adenoma for the entire range of observations (A), with frequencies among cases and controls indicated below, and for the range of 4.75% - 5.75% with 4.75% as the reference and with point-wise standard errors as dotted lines (B).
In analyses by CRA stage, the inverse association between genomic methylation and neoplasia was stronger when the outcome was restricted to non-advanced adenomas (Table 2), in part due to higher genomic methylation values in advanced adenoma patients compared to non-advanced adenoma patients (Table 1; interquartile range, 5.28-5.39 and 5.15-5.40, respectively). When considering anatomic location, the association was slightly stronger for adenomas in the proximal colon than in the distal colon. The small number of people (N = 12) who carried adenomas in both proximal and distal colon showed no significant association (data not shown). In case-case analyses that adjusted for age, season of blood draw, smoking history, and red meat intake, neither the contrast between non-advanced and advanced cases (P = 0.36) nor that between proximal and distal cases (P = 0.67) were statistically significant for the ORs comparing above versus below median methylation. Use of unconditional instead of conditional logistic regression or applying polytomous in place of case-case analyses yielded similar or slightly attenuated estimates (data not shown).
The methylation-CRA association was similarly inverse in women younger (OR for above versus below median methylation, 0.30; 95% CI, 0.12, 0.81) or older than 60 (OR, 0.42; 95% CI, 0.11, 1.65) and also did not significantly differ by family history of colorectal cancer, hormone replacement therapy, body mass index (normal-weight or overweight and obese combined), smoking history (never, former, or current), or red meat intake.
Among various indicators of folate intake examined, only total folate at pre-fortification levels and total DFE at post-fortification levels showed significant interactions with genomic methylation (Table 3). Genomic methylation showed a stronger inverse association with adenoma among low as compared to high folate consumers by these indicators. On the other hand, the folate-adenoma association was inverse among those with low genomic methylation but positive among those with high methylation. No significant effect modification was observed by other factors of one-carbon metabolism or combinations of factors (selected data in Table 3).
Table 3.
Association between percent genomic methylation of leukocyte DNA and colorectal adenoma stratified by levels of dietary factors of one-carbon metabolism, the CONCeRN Study (2000-2002)
| % genomic methylation of leukocyte DNA | ||||||||
|---|---|---|---|---|---|---|---|---|
| Below median (2.76 – 5.36) | Above median (5.37 -6.42) | |||||||
| N case / control | Odds ratio | 95% Confidence interval | N case / control | Odds ratio | 95% Confidence interval | P interaction | ||
| Folate (μg/1,000kcal) | ||||||||
| Pre-fortification intake | ||||||||
| Food-derived | ||||||||
| Low (< 178) | 37 / 30 | 1.0 | Referent | 23 / 26 | 0.38 | 0.14, 1.03 | ||
| High (≥ 178) | 35 / 26 | 0.96 | 0.45, 2.05 | 17 / 31 | 0.31 | 0.11, 0.85 | 0.79 | |
| Total | ||||||||
| Low (< 317) | 33 / 21 | 1.0 | Referent | 15 / 35 | 0.19 | 0.07, 0.51 | ||
| High (≥ 317) | 39 / 35 | 0.82 | 0.40, 1.68 | 25 / 22 | 0.51 | 0.20, 1.33 | 0.05 | |
| Post-fortification intake | ||||||||
| Total dietary folate equivalents | ||||||||
| Low (< 413) | 32 / 20 | 1.0 | Referent | 15 / 36 | 0.16 | 0.06, 0.48 | ||
| High (≥ 413) | 40 / 36 | 0.83 | 0.39, 1.78 | 25 / 21 | 0.52 | 0.20, 1.37 | 0.03 | |
| Vitamin B12 (μg/1,000kcal) | ||||||||
| Food-derived | ||||||||
| Low (< 2.1) | 44 / 31 | 1.0 | Referent | 17 / 25 | 0.35 | 0.13, 0.89 | ||
| High (≥ 2.1) | 28 / 25 | 0.76 | 0.34, 1.69 | 23 / 32 | 0.29 | 0.11, 0.76 | 0.82 | |
| Total | ||||||||
| Low (< 4.1) | 33 / 24 | 1.0 | Referent | 15 / 32 | 0.19 | 0.06, 0.55 | ||
| High (≥ 4.1) | 39 / 32 | 0.92 | 0.45, 1.88 | 25 / 25 | 0.49 | 0.19, 1.23 | 0.10 | |
| Methionine (mg/1,000kcal) | ||||||||
| Low (< 821) | 36 / 27 | 1.0 | Referent | 18 / 29 | 0.31 | 0.11, 0.87 | ||
| High (≥ 821) | 36 / 29 | 0.80 | 0.33, 1.92 | 22 / 28 | 0.30 | 0.10, 0.86 | 0.63 | |
| Alcohol | ||||||||
| Non-drinker | 28 / 17 | 1.0 | Referent | 11 / 16 | 0.22 | 0.06, 0.76 | ||
| Drinker | 44 / 39 | 0.52 | 0.24, 1.14 | 29 / 41 | 0.22 | 0.08, 0.60 | 0.39 | |
| Combined methyl-diet | ||||||||
| Low (low total DFE, drinker) | 24 / 13 | 1.0 | Referent | 13 / 28 | 0.18 | 0.06, 0.59 | ||
| Middle | 29 / 34 | 0.44 | 0.16, 1.20 | 20 / 22 | 0.28 | 0.09, 0.88 | ||
| High (high total DFE, non-drinker) | 20 / 10 | 1.89 | 0.62, 5.78 | 9 / 8 | 0.73 | 0.15, 3.59 | 0.31 | |
Discussion
Our finding of an inverse relationship between systemic genomic methylation and CRA is consistent with a previous case-control study 8 and demonstrates a dose-response. Compared to the highest tertile of genomic methylation, our finding is equivalent to ORs of 4.2 (95% CI, 1.5-11.8) for the middle tertile and 5.8 (95% CI, 2.0-16.6) for the lowest tertile, whereas the ORs were 6.68 (95% CI, 0.99, 45.12) and 10.27 (95% CI, 2.05, 51.46), respectively, in the previous study of 35 adenoma patients and 76 controls 8. Our study, which was based on colonoscopic examination of asymptomatic average-risk women in the folate-fortified U.S., tends to replicate the previous observation of symptomatic adenoma patients from the unfortified British population. Although neither genomic methylation levels nor dietary folate intake amounts in the previous study are directly comparable to ours due to different assessment methodologies applied, both studies essentially support the hypothesis that low genomic methylation in circulating leukocyte DNA may be associated with early colorectal tumorigenesis.
Reverse causation is theoretically possible in a cross-sectional study like ours, that is, presence of CRA might have lowered genomic methylation. However, it is unlikely that small, histologically non-advanced adenomas could reduce DNA methylation in leukocytes. Therefore, low levels of systemic genomic methylation may be a marker for some other etiologic factors that coincide systemically with aberrant methylation, such as inflammation 25. Alternatively, they may be a tumorigenic factor, reflecting limited systemic availability of methyl units to organs that are susceptible to hypomethylation-related chromosomal instability 26. Adequate genomic methylation in the target tissue is hypothesized to be protective against tumorigenesis because methylation stabilizes the genome 27, in part by reducing the loss of heterozygosity at the pericentromeric chromosomal regions 11, and thereby, preventing mutations and deletions that follow chromosomal instability.
It has been suggested that leukocyte DNA may be examined as a proxy for susceptible tissues regarding genomic methylation status 28, 29. To the best of our knowledge, however, conclusive human data are not available on the correlation between genomic methylation status in circulation and in colorectal mucosa. One previous study reported similarly and significantly lower systemic and local genomic methylation in adenoma patients compared to healthy controls, although they did not report the correlation between the two sources 8. Also, plasma homocysteine was significantly inversely correlated with colonic methylation, and with leukocyte methylation to a lesser degree 8, which points to homocysteine as the potential mechanism to link the systemic and local methylation: it is known that circulating homocysteine is in equilibrium with intracellular homocysteine 30 and that intracellular homocysteine and its substrate in a reversible state, S-adenosylhomocysteine, are potent inhibitors to methylation reactions 29.
Genomic DNA methylation is reportedly associated with and modified by folate status 29 and has been considered an important underlying mechanism for epidemiologic observations of a protective association between folate intake or status and colorectal adenoma 31, 32 or cancer 33. However, some inconsistencies existed among studies for such protective effects of folate 33, and a recent randomized trial detected an elevated risk of advanced and multiple adenoma recurrence with folic acid supplementation 34, possibly due to the high dose, synthetic form, or advanced disease stage at time of folate delivery 35-37. In our study, folate intake was not correlated with genomic methylation, and a non-significant inverse association with adenoma was detected for food-derived folate, especially of pre-fortification levels, but not for total intake, as in the previous meta-analysis 33. This suggests that increasing levels of folate, once a replete state is reached through fortification and supplementation, may not contribute to genomic methylation or prevention of colorectal tumorigenesis 20, 36. Also, while we a priori considered folate intake as an antecedent determinant of genomic methylation and did not hypothesize an interaction between the two, we found that the methylation-adenoma association was stronger with low rather than high total folate intake and that folate intake had moderately opposing associations with adenoma depending on methylation levels. This could be a chance finding, especially given the inconsistency among different indicators of folate intake. Alternatively, high total folate may increase promoter methylation 38 and thus, lessen the protective effects. At the least, our study suggests that genomic methylation may predict lower risk of CRA independently of folate or combined nutritional status for one-carbon metabolism among average-risk individuals on food supply fortified with folic acid. Larger and prospective studies are needed to determine the role of folate and one-carbon metabolism in the levels of systemic genomic methylation and its association with colorectal neoplasia.
We hypothesized that genomic methylation, like some other risk factors 39, may have different associations with the well-established stages of colorectal carcinogenesis. In support for an early involvement, local or systemic genomic methylation was reported to be similarly lower in patients with colon polyps or adenocarcinoma than in healthy controls 7, 8. Our findings of the higher level of genomic methylation in the few cases of advanced adenoma, and thus, the suggested less protective association for advanced than non-advanced adenoma, should be interpreted with caution considering that they are from cross-sectional data. Time series studies are needed before we can draw an inference that, for example, increases in genomic methylation may occur in later stages secondary to expanding promoter-associated CpG hypermethylation, especially with high folate intake. Similarly, our observation of a slightly more protective association for proximal compared to distal adenoma may reflect inadequate power rather than a potentially true anatomic subsite difference 40.
In addition to its relevance to colorectal carcinogenesis, genomic methylation status of leukocyte DNA has been linked to the risk of other cancers. Lower methylation in serum DNA of LINE1 transposons, commonly repeated elements in human genome 14, was found among gastric carcinoma patients as compared to matched controls 41 and was positively associated with the risk of head and neck squamous cell carcinoma (adjusted OR comparing the lowest to highest tertile of methylation, 1.6; 95% CI, 1.1-2.4) 42, possibly due to increased detrimental recombinations between repeated elements in the unmethylated regions of the target tissue as well systemic blood DNA 3. These independent findings from previous studies 8, 41, 42 along with our observation suggest low systemic genomic DNA methylation as a common etiology for tumors in multiple different sites that rely on loss of heterozygosity or chromosomal instability for tumorigenesis 26. This has implications for further studies of genomic DNA methylation in leukocytes in conjunction with that in target tissues, both for the underlying mechanism and for its potential to serve as a non-specific systemic marker of methyl-imbalance, for example, at levels lower than 4% among average-risk individuals, for some but not all cancers 43.
Our findings warrant replication in large prospective studies. Future studies also need to address the effect of nutritional status and potential gene-nutrient interactions using biomarkers and relevant genetic polymorphisms, respectively, for one-carbon metabolism 20.
Acknowledgments
We gratefully acknowledge the participation of the following investigators in the CONCeRN Study: Drs. J. Butler, P. Perdue, and P.J. Chandler, Bethesda, MD; Dr. C. Furlong, Portsmouth, VA; and Drs. J. Shad and R. Schindler, San Diego, CA. We also thank the contributions of Mary K. Keyes at Tufts University for assistance with assays; Shelley Niwa and Mark Stewart at Westat and Jane Curtin at the Information Management Services, Inc. for computer support; Janis Koci at Frederick repository for biospecimen handling; Dr. Amy F. Subar for advice on dietary assessment; Dr. Tom Fears for assistance in quality control analysis; Drs. Bill Kopp and Wen Shao at SAIC-Frederick for processing quality control samples; and Tawanda Roy at Nutritional Epidemiology Branch for research assistance.
Grant support: This research was supported by the Intramural Research Program of the National Institutes of Health at the National Cancer Institute (UL), by K07-CA108910-01A1 from the National Cancer Institute (AF), and by research grants from the American College of Gastroenterology and the American Society for Gastrointestinal Endoscopy.
Footnotes
** No Conflicts of interest exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 3.McCabe DC, Caudill MA. DNA methylation, genomic silencing, and links to nutrition and cancer. Nutr Rev. 2005;63:183–195. doi: 10.1301/nr.2005.jun.183-195. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 5.Toyota M, Issa JP. Epigenetic changes in solid and hematopoietic tumors. Semin Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Giovannucci E, Ogino S. DNA methylation, field effects, and colorectal cancer. J Natl Cancer Inst. 2005;97:1317–1319. doi: 10.1093/jnci/dji305. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 8.Pufulete M, Al Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, Emery PW, Sanders TA. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–1248. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 9.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 11.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 12.Thomas GA, Williams ED. Production of thyroid tumours in mice by demethylating agents. Carcinogenesis. 1992;13:1039–1042. doi: 10.1093/carcin/13.6.1039. [DOI] [PubMed] [Google Scholar]
- 13.Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26:185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 15.Davis CD, Uthus EO. DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 16.Jang H, Mason JB, Choi SW. Genetic and epigenetic interactions between folate and aging in carcinogenesis. J Nutr. 2005;135:2967S–2971S. doi: 10.1093/jn/135.12.2967S. [DOI] [PubMed] [Google Scholar]
- 17.Schoenfeld P, Cash B, Flood A, Dobhan R, Eastone J, Coyle W, Kikendall JW, Kim HM, Weiss DG, Emory T, Schatzkin A, Lieberman D. Colonoscopic screening of average-risk women for colorectal neoplasia. N Engl J Med. 2005;352:2061–2068. doi: 10.1056/NEJMoa042990. [DOI] [PubMed] [Google Scholar]
- 18.Diet History Questionnaire Version 1.0. Applied Research Program National Cancer Institute National Institutes of Health; 2000. [Google Scholar]
- 19.Subar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, McNutt S, McIntosh A, Rosenfeld S. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires : the Eating at America's Table Study. Am J Epidemiol. 2001;154:1089–1099. doi: 10.1093/aje/154.12.1089. [DOI] [PubMed] [Google Scholar]
- 20.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 22.Willett WC. Nutritional epidemiology. New York: Oxford University Press; 1998. [Google Scholar]
- 23.Therneau TM, Grambsch PM. Modeling Survival Data: extending the Cox model. Philadelphia: Springer-Verlag Inc.; 2000. [Google Scholar]
- 24.Sinha R, Kulldorff M, Chow WH, DeNobile J, Rothman N. Dietary intake of heterocyclic amines, meat-derived mutagenic activity, and risk of colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2001;10:559–562. [PubMed] [Google Scholar]
- 25.Risques RA, Rabinovitch PS, Brentnall TA. Cancer surveillance in inflammatory bowel disease: new molecular approaches. Curr Opin Gastroenterol. 2006;22:382–390. doi: 10.1097/01.mog.0000231812.95525.a7. [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130:129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 28.Piyathilake CJ, Johanning GL. Cellular vitamins, DNA methylation and cancer risk. J Nutr. 2002;132:2340S–2344S. doi: 10.1093/jn/132.8.2340S. [DOI] [PubMed] [Google Scholar]
- 29.Kim YI. Nutritional epigenetics: impact of folate deficiency on DNA methylation and colon cancer susceptibility. J Nutr. 2005;135:2703–2709. doi: 10.1093/jn/135.11.2703. [DOI] [PubMed] [Google Scholar]
- 30.Blom HJ. Consequences of homocysteine export and oxidation in the vascular system. Semin Thromb Hemost. 2000;26:227–232. doi: 10.1055/s-2000-8467. [DOI] [PubMed] [Google Scholar]
- 31.Giovannucci E, Stampfer MJ, Colditz GA, Rimm EB, Trichopoulos D, Rosner BA, Speizer FE, Willett WC. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J Natl Cancer Inst. 1993;85:875–884. doi: 10.1093/jnci/85.11.875. [DOI] [PubMed] [Google Scholar]
- 32.Baron JA, Sandler RS, Haile RW, Mandel JS, Mott LA, Greenberg ER. Folate intake, alcohol consumption, cigarette smoking, and risk of colorectal adenomas. J Natl Cancer Inst. 1998;90:57–62. doi: 10.1093/jnci/90.1.57. [DOI] [PubMed] [Google Scholar]
- 33.Sanjoaquin MA, Allen N, Couto E, Roddam AW, Key TJ. Folate intake and colorectal cancer risk: a meta-analytical approach. Int J Cancer. 2005;113:825–828. doi: 10.1002/ijc.20648. [DOI] [PubMed] [Google Scholar]
- 34.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, Keown-Eyssen G, Summers RW, Rothstein RI, Burke CA, Snover DC, Church TR, Allen JI, Robertson DJ, Beck GJ, Bond JH, Byers T, Mandel JS, Mott LA, Pearson LH, Barry EL, Rees JR, Marcon N, Saibil F, Ueland PM, Greenberg ER. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 35.Kim YI. Role of folate in colon cancer development and progression. J Nutr. 2003;133:3731S–3739S. doi: 10.1093/jn/133.11.3731S. [DOI] [PubMed] [Google Scholar]
- 36.Mason JB, Dickstein A, Jacques PF, Haggarty P, Selhub J, Dallal G, Rosenberg IH. A Temporal Association between Folic Acid Fortification and an Increase in Colorectal Cancer Rates May Be Illuminating Important Biological Principles: A Hypothesis. Cancer Epidemiol Biomarkers Prev. 2007;16:1325–1329. doi: 10.1158/1055-9965.EPI-07-0329. [DOI] [PubMed] [Google Scholar]
- 37.Ulrich CM, Potter JD. Folate and cancer--timing is everything. JAMA. 2007;297:2408–2409. doi: 10.1001/jama.297.21.2408. [DOI] [PubMed] [Google Scholar]
- 38.van den DM, Pellis L, Crott JW, van EM, Friederich P, Nagengast FM, van Bergeijk JD, de Boer SY, Mason JB, Kok FJ, Keijer J, Kampman E. Folic Acid and Vitamin B-12 Supplementation Does Not Favorably Influence Uracil Incorporation and Promoter Methylation in Rectal Mucosa DNA of Subjects with Previous Colorectal Adenomas. J Nutr. 2007;137:2114–2120. doi: 10.1093/jn/137.9.2114. [DOI] [PubMed] [Google Scholar]
- 39.Slattery ML, Edwards SL, Samowitz W. Stage of colon cancer at diagnosis: implications for risk factor associations? Int J Epidemiol. 1998;27:382–387. doi: 10.1093/ije/27.3.382. [DOI] [PubMed] [Google Scholar]
- 40.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–408. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 41.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 42.Ting HD, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 43.Woodson K, Mason J, Choi SW, Hartman T, Tangrea J, Virtamo J, Taylor PR, Albanes D. Hypomethylation of p53 in peripheral blood DNA is associated with the development of lung cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:69–74. [PubMed] [Google Scholar]


