Abstract
FYVE domain proteins play key roles in regulating membrane traffic in eukaryotic cells. The FYVE domain displays a remarkable specificity for the head group of the target lipid, phosphatidylinositol 3-phosphate (PtdIns[3]P). We have identified five putative FYVE domain proteins in the genome of the protozoan parasite Leishmania major, three of which are predicted to contain a functional PtdIns(3)P-binding site. The FYVE domain of one of these proteins, LmFYVE-1, bound PtdIns(3)P in liposome-binding assays and targeted GFP to acidified late endosomes/lysosomes in mammalian cells. The high-resolution solution structure of its N-terminal FYVE domain (LmFYVE-1[1–79]) was solved by nuclear magnetic resonance. Functionally significant clusters of residues of the LmFYVE-1 domain involved in PtdIns(3)P binding and dependence on low pH for tight binding were identified. This structure is the first trypanosomatid membrane trafficking protein to be determined and has been refined to high precision and accuracy using residual dipolar couplings.
Keywords: NMR structure, FYVE, membrane trafficking, residual dipolar couplings
A large number of eukaryotic proteins contain a FYVE domain that is responsible for mediating the recruitment of these proteins to a range of organelles such as endosomes, multivesiscular bodies, and phagosomes (for review, see Kutateladze 2006). Membrane recruitment follows the binding of the FYVE domain to membrane-embedded phosphatidylinositol-3-phosphate (PtdIns[3]P). Recruitment is also facilitated by insertion of part of the FYVE domain into the target membrane (Kutateladze et al. 2004) and, depending on the specific characteristics of the FYVE domain structure, is enhanced by dimerization (Hayakawa et al. 2004), electrostatic interactions with other phospholipids, and acidification (Lee et al. 2005). The FYVE domain is defined by three conserved elements: an N-terminal WxxD, a central polybasic RrHHCR, and the C-terminal RVC motif. Each of these elements form part of the PtdIns(3)P-binding site as observed in solution and crystal structures of the Saccharomyces cerevisiae Vps27, Drosophila Hrs, and human EEA1 FYVE domains (Misra and Hurley 1999; Mao et al. 2000; Dumas et al. 2001; Kutateladze and Overduin 2001). Complementary mutagenesis studies confirm that these motifs are essential for PtdIns(3)P binding and also indicate that other neighboring residues can dramatically affect the avidity and binding properties of the full-length FYVE protein for endogenous pools of PtdIns(3)P and the subcellular distribution of these proteins (Hayakawa et al. 2004). Further studies on the three-dimensional structure should provide further insights into the functional significance of sequence heterogeneity in FYVE domains.
Parasitic protozoa are highly divergent unicellular eukaryotes that can cause a number of important diseases in humans. Many of these parasites depend on endocytosis and lysosomal degradation for nutrient uptake, differentiation, and removal of opsonic host proteins (McConville et al. 2002). Endocytic trafficking is particularly important for the trypanosomatid parasites that are the cause of African sleeping sickness (Trypanosoma brucei), Chagas disease (T. cruzi), and human leishmaniasis (Leishmania spp). In all cases, endocytosis and secretion occur at a specialized invagination of the plasma membrane, termed the flagellar pocket, which surrounds the single flagellum (McConville et al. 2002; Field et al. 2007). Organelles involved in secretion (i.e., the Golgi apparatus) and endocytosis (endosomes, multivesicular bodies) are also polarized and generally localized in close proximity to the flagellar pocket. While the ultrastructure of the secretory and endocytic apparatus in these parasites is reasonably well characterized, much less is known about the molecular machinery involved and its diversity in other eukaryotes. It has been shown recently that the single PtdIns(3)P kinase of T. brucei is essential for secretory and endocytic trafficking as well as the partitioning of the Golgi apparatus, indicating that PtdIns(3)P plays a key role in these events (Hall et al. 2006). A number of FYVE domain-containing proteins have also been annotated in the genomes of the trypanosomatid parasites (Berriman et al. 2005). Interestingly, the FYVE domain is the only one of these trypanosomatid proteins to be characterized that was found to be nonfunctional (Kunz et al. 2005). We have identified five putative FYVE domain proteins (LmFYVE) in the L. major genome. Two of these proteins are predicted to be nonbinding to PtdIns(3)P, while the other three contain the three motifs required for PtdIns(3)P binding. In this study, we show that one of these proteins, LmFYVE-1, binds PtdIns(3)P in vitro and is targeted to late endosomes/lysosomes in animal cells. The complete solution structure of this FYVE domain was then determined by NMR and its dynamic properties upon interaction with the putative ligand PtdIns(3)P investigated.
Results and Discussion
Bioinformatic analysis of the L. major genome identified five genes that are predicted to encode FYVE-domain proteins (Supplemental Fig. S1). All of these proteins are predicted to contain the WxxD and RrHHCR motifs. However, two of these proteins (LmFYVE-4 and LmFYVE-5) lack the RVC motif and the eighth cysteine residue that is conserved in all functional FYVE domains, and are not predicted to bind PtdIns(3)P. LmFYVE-5 shares domain and sequence similarity with yeast FAB1 and mammalian PIKfyve, a well-conserved PtdIns(3)P 5-kinase that is required for the formation of multivesicular bodies in the late endosome/lysosome network. In all other species, this protein is thought to be recruited to the limiting membrane of late endosomes via interactions with its substrate, PtdIns(3)P. While the FYVE domain is still present in the L. major FAB1 paralog, the absence of the RVC motif raises the prospect that L. major FAB1 might be recruited to the abundant multivesicular bodies of promastigote stages (Mullin et al. 2001) by alternative mechanisms.
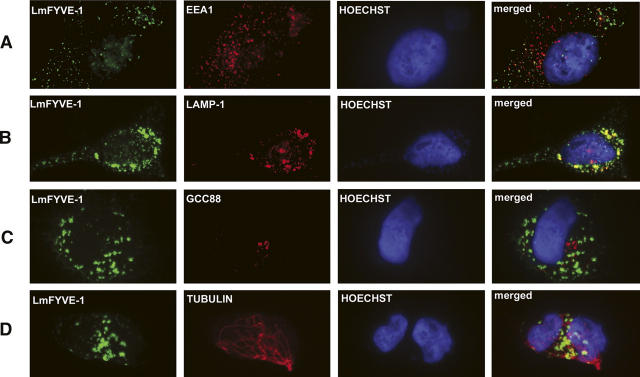
To test whether the other L. major FYVE domain proteins bind PtdIns(3)P, a GST fusion containing the LmFYVE-1 FYVE domain (LmFYVE-1[1–79]) was immobilized on agarose beads, and binding to PtdIns(3)P-containing liposomes was measured (Supplemental Fig. S2). While GST alone did not bind PtdIns(3)P liposomes, significant binding was observed when GST was fused to a single copy of the LmFYVE-1 domain. Increased binding was observed when GST was fused with a tandem repeat of the FYVE domain of the mammalian endosomal protein Hrs, reflecting the greater avidity of tandem or dimerized FYVE domains for membrane-embedded PtdIns(3)P. When a GFP-LmFYVE-1(1–79) fusion protein was expressed in animal cells, fluorescence was detected in punctate structures throughout the cytoplasm. When transfected cells were fixed and co-labeled with antibodies to the endosomal marker EEA1, only weak partial overlap was observed (Fig. 1A). However, strong overlap was observed with the late-endosome/lysosome marker LAMP-1. No overlap was observed with markers for the Golgi apparatus or tubulin. These data suggest that Leishmania LmFYVE-1(1–79) is selectively recruited to the more acidified compartments of the endocytic pathways. As indicated below, the affinity of LmFYVE-1(1–79) for PtdIns(3)P increases for acidic pH, suggesting that the polybasic motif of this domain may make it more sensitive to variation of the intracellular pH, resulting in a tropism for late endocytic/lysosomal compartments. Collectively, these data strongly suggest that LmFYVE-1 contains a functional PtdIns(3)P-binding FYVE domain.
Figure 1.
Intracellular localization of LmFYVE-1 in HeLa cells. HeLa cells were transiently transfected with GFP-LmFYVE-1, and, after 24 h of transfection, cells were fixed in 4% paraformaldehyde, permeabilized with 1% Triton-X-100, and co-stained for antibodies to EEA1 (A), Lamp-1 (B), GCC88 (C), and Tubulin (D), followed by anti-Ig Alexa594. The coverslips were mounted in Mowiol 4–88 containing Hoechst dye. Cells were viewed with a Zeiss Axioplan2 microscope equipped with a AxioCam Mrm digital camera.
Solution structure of the L. major LmFYVE-1(1–79) domain
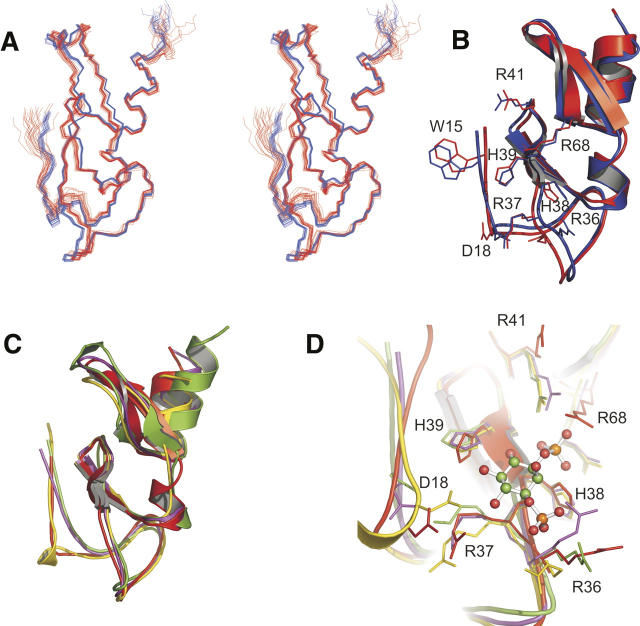
The high-resolution NMR structure of LmFYVE-1(1–79) was determined using three-dimensional heteronuclear triple-resonance and 15N/13C-edited NOESY experiments, and refined using multiple sets of residual dipolar couplings (RDCs) collected in a liquid crystalline PF1 bacteriophage medium (Table 1; Fig. 2A,B). The solution structure reveals the topology expected for a FYVE domain: two short double-stranded antiparallel β-sheets and a C-terminal α-helix, with the elements of secondary structure anchored by two tetrahedral coordinated Zn2+ ions. The coordination of two Zn2+ ions is supported by the downfield shifted resonances of the cysteine Cβs for Cys 24, Cys 27, Cys 48, Cys 51 (ligating Zn1), and Cys 40, Cys 43, Cys 70, Cys 73 (ligating Zn2).
Table 1.
Summary of statistics for the final ensemble of 20 conformersa
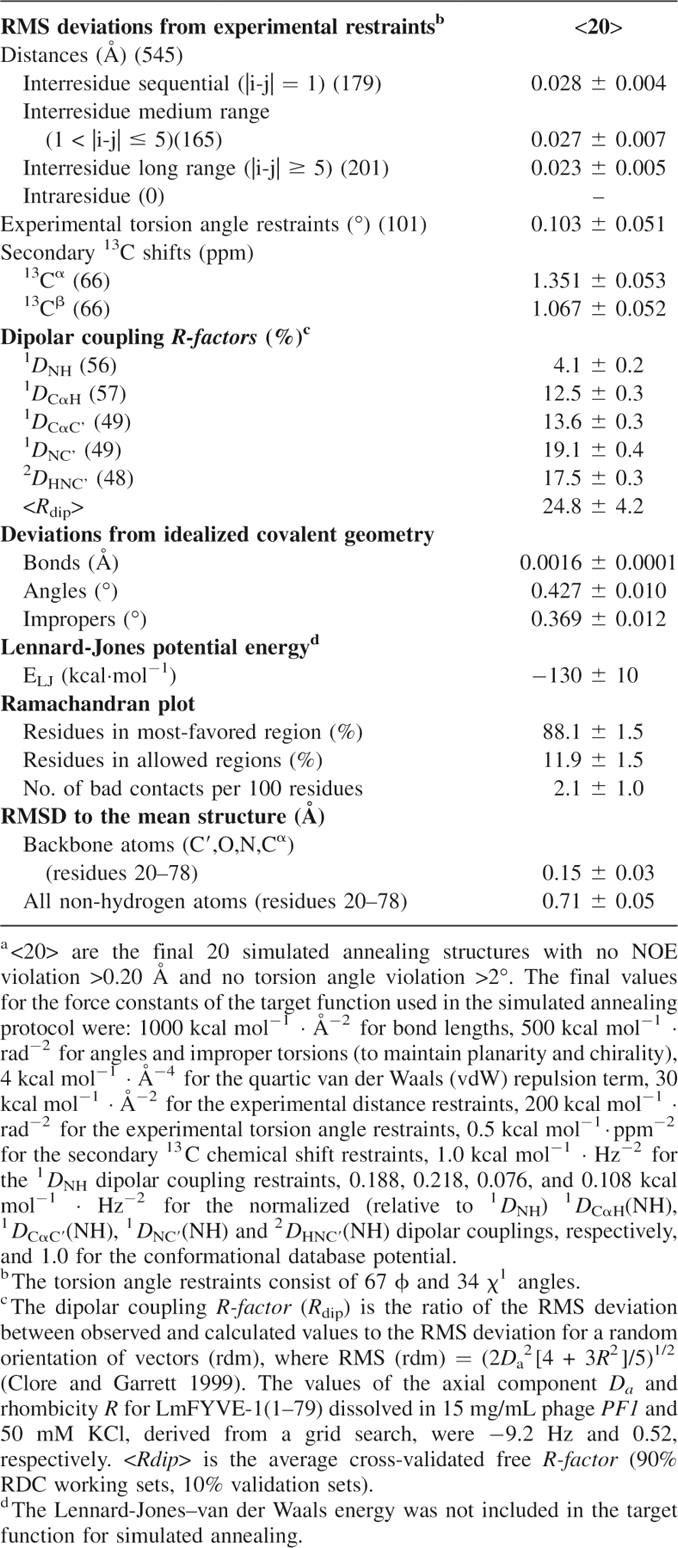
Figure 2.
FYVE domain of LmFYVE-1(1–79). (A) Stereoview of LmFYVE-1(1–79). (Blue) Structure refined against RDCs, (red) unrefined structure. (B) Cartoon representation of LmFYVE-1(1–79) showing selected side chains. (C) Overlay of FYVE domain structures. (Red) LmFYVE-1(1–79), (yellow) EEA1(1347–1410), (purple) HRS(156–218), (green) Vps27 (168–230). (D) Overlay of FYVE domain active site showing positioning of functional residues. The EEA1-bound ITP ligand as observed in the EEA1 crystal structure is shown as a ball and stick model.
The three characteristic motifs shown to be important for substrate recognition by FYVE domain proteins are present: the N-terminal WxxD motif (located in a region of irregular secondary structure prior to the β1 strand, including Trp 15 and Asp 18), the polybasic motif (located along the β1 strand, from Arg 36 to Arg 41), and the RVC motif (located along the β4 strand, including Arg 68). The putative membrane insertion region termed the turret loop or membrane insertion loop (MIL) is also present as a loop of essentially hydrophobic residues (Phe 31 to Val 35) prior to the β1 strand. Refinement of the solution structure using RDCs led to small adjustments of the position of the protein backbone (Fig. 2A,B). While improving the fit of this FYVE domain to other similar proteins during structural alignment (Supplemental Table 2), the major impact of the RDCs is observed as an increase in precision and as a measure of backbone accuracy (equivalent to a 2.5 Å resolution crystal structure).
LmFYVE-1(1–79) binds PtdIns(3)P in vitro
LmFYVE-1(1–79) was titrated with diC8-PtdIns(3)P, and significant chemical shift perturbations were observed in the signature PtdIns(3)P-binding motifs (Supplemental Fig. S3). The PtdIns(3)P-binding site is localized along β1, incorporating the polybasic motif, WxxD motif, and adjacent acidic residues. The side chains of both His-38 and His-39 are also positioned in the primary binding site, the amide resonances of which disappear upon PtdIns(3)P addition due to exchange broadening. The average Kd determined at pH 7.5 was 59 ± 13 μM, considerably lower than the previously determined Kd of 1300 ± 400 μM for diC4-PtdIns(3)P binding to the EEA1 FYVE domain at neutral pH (Lee et al. 2005). To determine whether protonation of His-38 and His-39 at acidic pH activates a histidine switch, enhancing the affinity of LmFYVE-1 for PtdIns(3)P as it does for EEA1 (Lee et al. 2005), we titrated LmFYVE-1(1–79) with diC8-PtdIns(3)P at pH 6.1. An average Kd of 29 ± 9 μM was determined, indicating higher affinity binding to PtdIns(3)P at low pH. This compared with a Kd of 70 ± 2 μM determined for diC4-PtdIns(3)P binding to the EEA1 FYVE domain at pH 6.0 (Lee et al. 2005). As late endosomes and lysosomes may be enriched in PtdIns(3,5)P2, we conducted similar titrations of LmFYVE-1(1–79) with diC8-PtdIns(3,5)P at pH 7.5 and found no significant binding. Collectively, these data strongly suggest that LmFYVE-1(1–79) binds preferentially to PtdIns(3)P on acidified late endosomal compartments.
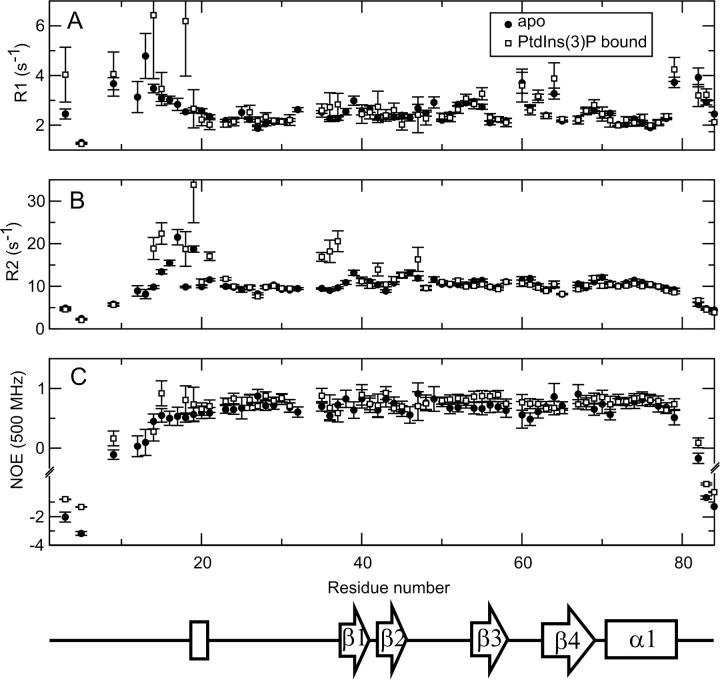
Heteronuclear relaxation data were acquired for LmFYVE-1(1–79), and a change in dynamics between the free and diC4-PtdIns(3)P bound forms was observed (Fig. 3). In the bound state, the flexibility of the N-terminal residues from Tyr 14 to Gly 26 decreases. In particular, the heteronuclear NOE for the conserved WxxD motif residue Trp 15 increases from 0.55 to 0.72, suggesting that upon PtdIns(3)P head-group binding, residues of the WxxD motif become less flexible. Residues Val 35 and Arg 36, prior to the first β-strand, also show a decrease in flexibility on the nanosecond timescale when LmFYVE-1(1–79) binds PtdIns(3)P, with the heteronuclear NOE increasing from 0.70 to 0.82 and 0.54 to 0.79, respectively. The average longitudinal (R1) and transverse (R2) relaxation rates measured for the free and PtdIns(3)P-bound FYVE domain of LmFYVE-1 are 2.66 ± 0.68 s−1 and 9.92 ± 2.88 s−1, and 2.70 ± 0.89 s−1 and 10.98 ± 4.83 s−1, respectively. Within experimental error, no significant difference in these average rates across the entire domain is evident. However, on close inspection, two regions of the protein sequence, from Tyr 14 to Ala 23 and from Val 35 to Arg 37, show a general increase in R2, which is generally taken as an indicator of mobility on the millisecond timescale resulting from chemical exchange. The N-terminal region (from Leu 3 to Glu 19) also shows some differences in R1 between the free and bound state with an increase observed for Tyr 14, Trp 15, and Glu 18 upon PtdIns(3)P binding.
Figure 3.
Heteronuclear NMR relaxation data. The longitudinal, R1 (A), and transverse, R2 (B), autorelaxation rates, and 1H-15N NOE (C) for LmFYVE-1(1–79) acquired at 500 MHz. Data were collected in the absence (black circles) and presence (open circles) of 1.5 mM PtdIns(3)P at a protein concentration of 0.5 mM. The sequence position of secondary structure elements is indicated.
The importance of the unstructured N-terminal region in ligand binding is highlighted by the observed decrease in mobility for residues upon PtdIns(3)P ligation, as indicated by the increased heteronuclear NOEs measured across the WxxD motif. The implication is that a small conformational rearrangement accompanies binding of PtdIns(3)P by the FYVE domain. This was suggested by the investigators of the EEA1 NMR structure (Kutateladze and Overduin 2001) but was disregarded by the investigators of the EEA1 crystal structure (Dumas et al. 2001), who claim that no conformational change is required to accommodate the PtdIns(3)P head group in the binding pocket.
Comparison of LmFYVE-1(1–79) with other FYVE domain structures
The backbone pairwise RMSD values for the alignment of LmFYVE-1(1–79) to the solution structure of EEA1 and the X-ray crystal structures of EEA1, HRS, and Vps27 are summarized in Supplemental Table 2. It is clear that a high degree of structural conservation is present for this structural unit. Each of the crystal structures fits within 1.7 Å of the mean RDC-refined LmFYVE-1(1–79) structure, with the fit to the solution structure of EEA1 at almost 2 Å for all residues.
The FYVE domain scaffold is essentially identical for LmFYVE-1(1–79) from L. major, human EEA1, Drosophila HRS, and S. cerevisiae Vps27 (Fig. 2C); however, a significant deviation in the position of the backbone for the N-terminal irregularly structured region is observed between these structures. Functionally, this could impact on the substrate recognition and cellular compartment targeting properties of FYVE domain proteins. A helical loop structure involving residues of the WxxD motif is observed for the LmFYVE-1(1–79) and EEA1 FYVE domains but is absent for HRS and Vps27 due to a three-residue deletion in the sequence (creating a shortened Wx-D motif for the latter two proteins). The crystal structure of EEA1 in complex with the PtdIns(3)P substrate analog inositol 1,3-bisphosphate shows that the conserved aspartate of the WxxD motif forms important contacts with the 5- and 6-hydroxyl groups of the inositol ring (Fig. 2D). This interaction is considered to be crucial for stereoselectivity and the exclusion of other phosphoinositides and is supported by mutagenesis. Indeed, from the overlaid structures of the FYVE domain PtdIns(3)P-binding site (Fig. 2D), it is clear that the side-chain positions of the conserved aspartate are distinct for the Vps27 and HRS proteins compared with LmFYVE-1(1–79) and EEA1, suggesting that a functional difference in the binding mode for each group is likely.
The position of the backbone of the MIL for LmFYVE-1(1–79), EEA1, HRS, and Vps27 is essentially identical; however, this loop does vary in its degree of hydrophobicity between the four proteins (Supplemental Fig. S1). Residues at the tip of the MIL for EEA1 (Val 1367 and Thr 1368) have been shown to penetrate into monolayers and DPC micelles (Kutateladze and Overduin 2001; Kutateladze et al. 2004) while an 11-residue insertion of generally hydrophobic residues in the MIL of the FYVE domain of FENS-1 has been shown to enhance membrane affinity and endosomal localization (Blatner et al. 2004). Mutagenesis of the central MIL residues of EEA1 (ValThr 1368 to GlyGly and GluGlu), HRS (Phe 173 to Ala), and Vps27 (LeuLeu 186 to AlaAla) abrogates endosomal localization, decreases membrane penetration, and affinity for membrane-bound PtdIns(3)P (Kutateladze et al. 1999; Stahelin et al. 2002). The equivalent residues for LmFYVE-1(1–79), ThrThr 34, may not penetrate the membrane as deeply as those for EEA1 due to a general decrease in the hydrophobicity of the MIL.
Basic and polar residues adjacent to the MIL have also been shown to be critical for membrane insertion of FYVE domain proteins, and recent computational studies indicate that the PtdIns(3)P binding event itself facilitates membrane binding through a reduction of the positive potential of the FYVE domain surface (Diraviyam et al. 2003). LmFYVE-1(1–79) shares the same distribution of surface-exposed basic residues as the EEA1, HRS, and Vps27 FYVE domains (Supplemental Fig. S4), forming a large region of positive potential surrounding the PtdIns(3)P-binding site.
Dimerization has also been shown to increase affinity for PtdIns(3)P and enhance endosomal localization for several FYVE domain-containing proteins (Gillooly et al. 2000; Dumas et al. 2001; Blatner et al. 2004; Hayakawa et al. 2004). The EEA1 crystal structure shows a homodimer formed primarily from contacts in the coiled-coil region, but also from the FYVE domain involving the side chains of the conserved tryptophan of the WxxD motif and a proline residue of the β3-β4 hairpin loop (Dumas et al. 2001). The HRS crystal structure also shows a homodimer with contacts primarily formed between the adjacent helical VHS domains and also the β3-β4 loop of the FYVE domain (Mao et al. 2000). The functional significance of the β3-β4 loop region may be either to mediate a dimer interaction between FYVE domains (Mao et al. 2000) or to interact with the membrane surface (Dumas et al. 2001). The proposed model for membrane interactions for the β3-β4 loop of EEA1 relies upon positioning of the tandem lysine residues, Lys-1396 and Lys-1397, close to the head-group region of the lipid bilayer. For LmFYVE-1(1–79), these residues are replaced by Thr 64 and Glu 65, in HRS by Glu 204 and Lys 205, and in Vps27 by Tyr 216 and Glu 217. The nature of this interaction may be the requirement for at least one polar side chain; however, whether this loop is involved in dimer formation or membrane binding is unknown for LmFYVE-1(1–79) and Vps27. The EEA1 sequence contains a two-residue deletion in the β3-β4 loop, while LmFYVE-1(1–79), Vps27, and HRS share a common glycine residue, which may allow a higher degree of flexibility of this loop relative to that of EEA1.
Conclusion
Our results suggest that the structure of the FYVE domain evolved early in eukaryotic evolution and is highly conserved in both metazoan (human, Drosophila) and unicellular (yeast, L. major) eukaryotes. While the isolated FYVE domains of Vps27, HRS, and EEA1 have been shown to specifically bind membrane-embedded PtdIns(3)P with nanomolar affinity in vitro (Gaullier et al. 2000; Stahelin et al. 2002; Blatner et al. 2004), they do not localize in vivo to late endosomal structures, indicating that additional domains restrict their subcellular localization (Hayakawa et al. 2004). In contrast, LmFYVE-1(1–79) has the capacity to target reporter proteins to the late endosome–lysosome network structures despite exhibiting only micromolar affinity for a water-soluble PtdIns(3)P analog at neutral pH. As LmFYVE-1(1–79) shows increased affinity for PtdIns(3)P at lower pH in vitro, it is possible that the binding affinity of the FYVE domain of LmFYVE-1 increases in vivo, when its PtdIns(3)P ligand is associated with acidic organelles. Interestingly, mammalian-infective stages of Leishmania are exposed to low pH conditions when they invade and occupy the phagolysosome of host macrophages, conditions that are known to result in acidification of the parasite lysosome (Mullin et al. 2001). It will be important to determine whether stage-specific changes in parasite lysosome pH influence the recruitment of LmFYVE-1 to late endosomes/lysosomes of the parasite and membrane traffic to these compartments.
Only small structural variations are observed between the FYVE domain structures presently solved. Differences in the positions of residues in the WxxD motif are observed between Vps27/HRS and EEA1/LmFYVE-1 and could alter the specifics of substrate recognition and membrane targeting. However, both Vps27 and HRS can target PtdIns(3)P in vitro with nanomolar affinity although they contain a shortened WxxD motif (Wx-D), suggesting that this region is flexible and conformational change occurs on selection and binding of substrate PtdIns(3)P. Here, we show that interaction of PtdIns(3)P with LmFYVE-1(1–79) does lead to a decrease in the mobility of N-terminal residues, including those of the WxxD motif.
Materials and Methods
Cloning and expression of epitope-tagged LmFYVE-1
All cloning was prepared in Escherichia coli XL1-Blue-MRF′ (Stratagene). The following synthetic oligonucleotides were used to PCR amplify the various DNA constructs used in this study: primer 1, CGGGATCCATAGCCGACAAAGTCGTC, 5′ forward primer for pEGFPN3-LmFYVE-1 with a BamHI site; primer 2, CGGGATCCATAGCCGACAAAGTCGTC, 3′ reverse primer for pEGFPN3-LmFYVE-1 with a XhoI site; primer 3 CGGATCCATGGGAGAGAAGCAATCGAAG, 5′ forward primer for pGEX-6P-3 with a BamHI site; primer 4, CCTGCTCGAGCTAGCTCCCTGCCATGTTGCT, 3′ reverse primer for pGEX-6P-3 with a XhoI site.
Protein expression and purification
Bacterial expression of GST-LmFYVE-1 and GST-Hrs fusion protein was induced by addition of 0.3 mM isopropyl β-D-thiogalactoside (IPTG) to recombinant BL-21(DE3) cultures for 3 h at 37°C. Cells were pelleted at 8000g for 10 min at 4°C and stored at −20°C until required. Cell pellets were solubilized with 1% TX-100 and purified on a glutathione-Sepharose column (Amersham Biosciences) according to the manufacturer's instructions. Finally, purified fusion proteins were desalted on a PD10 column (Amersham Biosciences) and stored at −70°C.
Transfection
HeLa cells were transiently transfected with GFP-LmFYVE-1 using FuGene transfection reagent (Roche) according to the manufacturer's instructions. Immunofluorescence of transfected cells was performed 24 h after transfection.
Immunofluorescence
Cells grown on coverslips were fixed in 4% paraformaldehyde for 15 min, free aldehyde groups were quenched in 50 mM NH4Cl/PBS, and the cells were permeabilized with 1% Triton X-100 in PBS for 4 min. The fixed cells were probed with anti-EEA1 (BD Transduction Laboratories), anti-LAMP-1 (PharMingen International), anti-GCC88 (kindly provided by P. Gleeson, University of Melbourne), and anti-Tubulin (Molecular Probes) antibodies for 30 min at room temperature. After washing three times in PBS, the cells were incubated with AlexaFluor 594-conjugated secondary antibodies (Molecular Probes) for 30 min at room temperature. All antibodies were diluted in PBS/BSA. The coverslips were mounted in Mowiol 4–88 (Calbiochem) containing Hoechst dye. Cells were viewed with a Zeiss Axioplan2 microscope equipped with a AxioCam Mrm digital camera.
Liposome-binding assays
L-3-Phosphatidyl[N-methyl-3H]choline 1,2-dipalmitoyl (85 Ci/mmol) was obtained from GE Healthcare. Phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) were obtained from Sigma. Phosphatidylinositol 3-phosphate (PtdIns[3]P) was obtained from Echlon. The liposome-binding assay was performed as follows and as described (Gaullier et al. 2000). Liposomes were prepared at 0.35 mg/mL containing 63%–65% PC, 20% PS, 15% PE, 2% Phosphatidylinositol (PI), and a trace of [3H] PC, suspended in liposome buffer (20 mM Tris-HCl, pH 7.5, 60 mM NaCl), and sonicated on ice. GST fusion proteins were desalted into the liposome buffer and 20 μg of GST or molar equivalent of GST-LmFYVE-1 or GST-Hrs fusion proteins immobilized on 50 μL of 50% (v/v) glutathione–Sepharose beads by rotation for 30 min at room temperature. The beads were washed three times and resuspended in 100 μL of liposome buffer. One hundred microliters of sonicated 3H-labeled liposomes were added and the mixture incubated for 30 min with gentle rotation at room temperature. The beads were washed twice in liposome buffer, the supernatants collected, and radioactivity determined. The percentage of 3H-labeled liposomes bound to the Sepharose beads was calculated as follows: [c.p.m. in pellet/total c.p.m. (test beads)] − [c.p.m. in pellet/total c.p.m. (beads alone)]. All experiments were carried out in triplicate.
NMR spectroscopy
LmFYVE-1(1–79) from Leishmania major was expressed uniformly 15N and 13C labeled by published methods (Cai et al. 1998). Each 400-μL NMR sample contained 0.7 mM LmFYVE-1(1–79) in 50 mM KH2PO4/K2HPO4, pH 7.3, 50 mM KCl, 5 mM DTT, 2% (w/v) NaN3, 5% (v/v) D2O. PF1 filamentous phage was prepared in-house by standard procedures (Hansen et al. 2000) and was added to a final concentration of 10 mg/mL for the aligned samples. Alignment of PF1 in the magnetic field was tested by monitoring the observed quadrupolar splitting of the solvent 2H signal. NMR spectra of LmFYVE-1(1–79) were collected at 20°C on a Varian Inova 600 MHz spectrometer, using a 5-mm 1H, 13C, 15N single Z-axis gradient probe. Sequential assignments of backbone resonances were obtained using standard triple resonance experiments (Cavanagh et al. 2006), and assignment of side-chain 1H and 13C resonances were obtained from 3D HCCH-TOCSY and 3D HCCH-COSY spectra, and 3D H(CCO)NH and C(CO)NH spectra. Structural constraints were obtained from 3D [15N]-NOESY-HSQC, 3D [13C]-NOESY-HSQC and [13C]-HSQC-NOESY spectra.
N-HN, N-C′, HN-C′, Cα-C′, and Cα-Hα RDCs were obtained as described previously (Mertens and Gooley 2005). Spectra were processed using the program NMRPipe (Delaglio et al. 1995) and analyzed with XEASY (Bartels et al. 1995) or SPARKY 3.100 (T.D. Goddard and D.G. Kneller, SPARKY 3. University of California, San Francisco).
Structure determination
Initial structures were generated using the fast torsion angle dynamics program CYANA 1.0.7 (Guntert et al. 1997), making use of the CANDID module for automated NOE assignment (Herrmann et al. 2002). Final structure calculations using NOE-derived distance restraints and experimentally determined dihedral angles were then conducted in torsion angle space in the program Xplor-NIH (Schwieters et al. 2003) with additional refinement incorporating RDCs (Table 1), as described previously (Mertens and Gooley 2005). A database potential of mean force acting to exclude rarely observed conformations of the protein backbone during a structure calculation was also included (Kuszewski et al. 1996). One hundred trial structures of each protein were calculated, and an ensemble of the 20 best structures with lowest NOE and dihedral angle violations was chosen to represent the final structures (Table 1).
NMR titration of LmFYVE-1(1–79) with PtdIns(3)P
Changes in the average chemical shift observed in the 1HN and 15N dimensions of the 2D [15N,1H]-HSQC experiment, collected at 600 or 800 MHz at 20°C, were followed by titrating 30 or 65 μM LmFYVE-1(1–79) with 0–857 μM diC8-PtdIns(3)P (pH 7.5), 0–355 μM diC8-PtdIns(3)P (pH 6.1), and 0–613 μM diC8-PtdIns(3,5)P2 (pH 7.5). Average dissociation constants for the binding of dioctanoyl PtdIns(3)P (diC8-PtdIns[3]P) by the FYVE domain of LmFYVE-1 were calculated from NMR titration data with the program xcrvfit, (v3.0.6, R. Boyko and B. Sykes) using nonlinear least-squares fitting analysis assuming fast exchange regime and a one-site second-order binding model. The HN and 15N were fitted for the following residues: Gly 26, Gly 28, Ser 52, Arg 61, Val 46, His 54, Arg 68 (HN only), Cys 73, Asp 50, and Ala 23. Due to line broadening or resonance overlap, Val 46 and His 54 were not used at pH 7.5.
Relaxation and dynamics
15N T1 measurements at 500 MHz were recorded with relaxation delays of 20, 80 (two experiments), 140, 300, 500 (two experiments), 750, 900, and 1500 ms. 15N T2 measurements at 500 MHz were recorded with relaxation delays of 10, 30, 50, 70, 90, 110 (two experiments), 130, 150, 170, 190 (two experiments), and 250 ms. The recycle delay time was set to 1 s. Relaxation times were obtained from an analysis of the exponential decay of peak intensities using the program Curvefit (A.G. Palmer 1998). Peak intensities were measured in SPARKY and determined from peak heights. Heteronuclear [1H]-15N NOEs at both 500 MHz and 600 MHz were obtained using a gradient sensitivity-enhanced 1H-15N NOE experiment (Farrow et al. 1994), recorded with and without 1H saturation as two interleaved data matrices of 128 × 512 complex points acquired in the F1 (15N) and F2 (1H) dimensions, respectively. A relaxation delay of 2 s prior to 3 s of 1H saturation was used for the NOE spectra, and a 5-s relaxation delay was used for the reference spectra.
Data bank accession codes
The coordinates of the ensemble of 20 LmFYVE-1(1–79) structures refined using RDCs and a representative mean structure have been deposited in the Protein Data Bank with accession code 1Z2Q. The NMR chemical shifts and restraints have been deposited at the BMRB with accession number 6594.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NH&MRC) grants to P.R.G. and M.J.M. M.J.M. is a NH&MRC Principal Research Fellow.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Paul R. Gooley, Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, Victoria 3010, Australia; e-mail: prg@unimelb.edu.au; fax: 61-3-9348-1421; or Malcolm J. McConville, Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, Victoria 3010, Australia; e-mail: malcolmm@unimelb.edu.au; fax: (03) 9348-1421.
Abbreviations: LmFYVE-1, Leishmania major FYVE domain containing protein 1; PtdIns(3)P, phosphatidylinositol 3-phosphate; HSQC, heteronuclear single-quantum coherence; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; RMSD, root mean square deviation.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073009807.
References
- Bartels C., Xia, T.H., Billeter, M., Guntert, P., and Wuthrich, K. 1995. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR 6: 1–10. [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D.C., Lennard, N.J., Caler, E., Hamlin, N.E., Haas, B., et al. 2005. The genome of the African trypanosome Trypanosoma brucei . Science 309: 416–422. [DOI] [PubMed] [Google Scholar]
- Blatner N.R., Stahelin, R.V., Diraviyam, K., Hawkins, P.T., Hong, W., Murray, D., and Cho, W. 2004. The molecular basis of the differential subcellular localization of FYVE domains. J. Biol. Chem. 279: 53818–53827. [DOI] [PubMed] [Google Scholar]
- Cai M.L., Huang, Y., Sakaguchi, K., Clore, G.M., Gronenborn, A.M., and Craigie, R. 1998. An efficient and cost-effective isotope labeling protocol for proteins expressed in Escherichia coli . J. Biomol. NMR 11: 97–102. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother, W.J., Palmer, A.G., Skelton, N.J., and Rance, M. 2006. Protein NMR spectroscopy: Principles and practice, 2nd ed. Academic Press, New York.
- Clore G.M. and Garrett, D.S. 1999. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 121: 9008–9012. [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Diraviyam K., Stahelin, R.V., Cho, W., and Murray, D. 2003. Computer modeling of the membrane interaction of FYVE domains. J. Mol. Biol. 328: 721–736. [DOI] [PubMed] [Google Scholar]
- Dumas J.J., Merithew, E., Sudharshan, E., Rajamani, D., Hayes, S., Lawe, D., Corvera, S., and Lambright, D.G. 2001. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell 8: 947–958. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Field M.C., Natesan, S.K.A., Gabernet-Castello, C., and Koumandou, V.L. 2007. Intracellular trafficking in the trypanosomatids. Traffic 8: 629–639. [DOI] [PubMed] [Google Scholar]
- Gaullier J.M., Ronning, E., Gillooly, D.J., and Stenmark, H. 2000. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes—Role of conserved residues. J. Biol. Chem. 275: 24595–24600. [DOI] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow, I.C., Lindsay, M., Gould, R., Bryant, N.J., Gaullier, J.M., Parton, R.G., and Stenmark, H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19: 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntert P., Mumenthaler, C., and Wuthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Hall B.S., Gabernet-Castello, C., Voak, A., Goulding, D., Natesan, S.K., and Field, M.C. 2006. TbVps34, the trypanosome orthologue of Vps34, is required for Golgi complex segregation. J. Biol. Chem. 281: 27600–27612. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Hanson, P., and Pardi, A. 2000. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzymol. 317: 220–240. [DOI] [PubMed] [Google Scholar]
- Hayakawa A., Hayes, S.J., Lawe, D.C., Sudharshan, E., Tuft, R., Fogarty, K., Lambright, D., and Corvera, S. 2004. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 279: 5958–5966. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Guntert, P., and Wuthrich, K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319: 209–227. [DOI] [PubMed] [Google Scholar]
- Kunz S., Oberholzer, M., and Seebeck, T. 2005. A FYVE-containing unusual cyclic nucleotide phosphodiesterase from Trypanosoma cruzi . FEBS J. 272: 6412–6422. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn, A.M., and Clore, G.M. 1996. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Sci. 5: 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T.G. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta 1761: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T. and Overduin, M. 2001. Structural mechanism of endosome docking by the FYVE domain. Science 291: 1793–1796. [DOI] [PubMed] [Google Scholar]
- Kutateladze T.G., Ogburn, K.D., Watson, W.T., de Beer, T., Emr, S.D., Burd, C.G., and Overduin, M. 1999. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell 3: 805–811. [DOI] [PubMed] [Google Scholar]
- Kutateladze T.G., Capelluto, D.G., Ferguson, C.G., Cheever, M.L., Kutateladze, A.G., Prestwich, G.D., and Overduin, M. 2004. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279: 3050–3057. [DOI] [PubMed] [Google Scholar]
- Lee S.A., Eyeson, R., Cheever, M.L., Geng, J., Verkhusha, V.V., Burd, C., Overduin, M., and Kutateladze, T.G. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. 102: 13052–13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.X., Nickitenko, A., Duan, X.Q., Lloyd, T.E., Wu, M.N., Bellen, H., and Quiocho, F.A. 2000. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100: 447–456. [DOI] [PubMed] [Google Scholar]
- McConville M.J., Mullin, K.A., Ilgoutz, S.C., and Teasdale, R.D. 2002. Secretory pathway of trypanosomatid parasites. Microbiol. Mol. Biol. Rev. 66: 122–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens H.D.T. and Gooley, P.R. 2005. Validating the use of database potentials in protein structure determination by NMR. FEBS Lett. 579: 5542–5548. [DOI] [PubMed] [Google Scholar]
- Misra S. and Hurley, J.H. 1999. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97: 657–666. [DOI] [PubMed] [Google Scholar]
- Mullin K.A., Foth, B.J., Ilgoutz, S.C., Callaghan, J.M., Zawadzki, J.L., McFadden, G.I., and McConville, M.J. 2001. Regulated degradation of an endoplasmic reticulum membrane protein in a tubular lysosome in Leishmania mexicana . Mol. Biol. Cell 12: 2364–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160: 65–73. [DOI] [PubMed] [Google Scholar]
- Stahelin R.V., Long, F., Diraviyam, K., Bruzik, K.S., Murray, D., and Cho, W. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 277: 26379–26388. [DOI] [PubMed] [Google Scholar]