Abstract
The Escherichia coli Cpx envelope stress system is comprised of three proteins; the periplasmic regulatory CpxP, the inner membrane sensor kinase CpxA, and the cytoplasmic transcriptional activator CpxR. Although misfolded envelope proteins activate the Cpx system, the molecular mechanism by which this signal is sensed remains largely unknown. In an attempt to reconstitute the Cpx system from purified proteins, we failed to produce the small CpxP protein in its natural periplasmic compartment, but a high protein level was achieved when it was produced in the cytoplasm. Silent base mutations in the first codons of the cpxP gene encoding the signal sequence or substitution by two well-characterized signal sequences, those of MalE and DsbA, resulted in a large increase of the CpxP level in the periplasm. Our results support the hypothesis that periplasmic expression could be inhibited by sequence elements in the early coding signal sequence region of cpxP.
Keywords: periplasm, signal sequence, synonymous codon, stress response, protein production
In Escherichia coli, most envelope proteins are transported in an unfolded state through the SecYEG membrane-embedded translocon. After translocation, mature envelope proteins are subsequently sorted out in the periplasm, where resident chaperones, folding catalysts, and proteases prevent these nascent proteins from misfolding and aggregating (Mogensen and Otzen 2005). Because the periplasm is only separated from the extracellular milieu by the porous outer membrane, it is more susceptible to changes in the external environment that can damage or unfold periplasmic proteins (Missiakas and Raina 1997). Also, cells sense and respond to the accumulation of incorrectly folded proteins in the periplasm by two signaling pathways, the σE and Cpx pathways (Raivio and Silhavy 1999). Although both envelope stress responses activate the expression of genes encoding periplasmic protein folding factors, the mechanisms by which they achieve this regulation are quite different (Duguay and Silhavy 2004).
The σE response is mainly activated by unfolded or unassembled outer membrane proteins in the periplasm displaying a C-terminal tripeptide that is normally buried in native membrane-embedded proteins (Walsh et al. 2003). This specific peptide activates a series of proteolytic cleavages that destroy the anti-σ factor RseA and release σE to direct transcription of genes encoding periplasmic folding factors (Alba and Gross 2004).
The Cpx system is a typical two-component signal transduction system consisting of the inner membrane kinase CpxA (Weber and Silverman 1988) and the cytoplasmic response regulator CpxR (Dong et al. 1993). When an envelope stress signal is sensed, CpxA autophosphorylates on a conserved histidine in the cytoplasmic kinase domain and then transfers phosphate to a conserved aspartate in the N-terminal domain of CpxR. Phosphorylated CpxR activates the transcription of genes encoding the periplasmic DsbA, PpiA, and PpiD folding catalysts (Danese and Silhavy 1997; Pogliano et al. 1997; Dartigalongue and Raina 1998), the DegP protease (Danese et al. 1995), as well as genes involved in lipid and lipopolysaccharide metabolism (De Wulf et al. 2002). Together, these proteins serve to ensure proper biogenesis of the bacterial envelope by preventing any perturbation in periplasmic protein folding (Raivio and Silhavy 2001). The Cpx pathway is induced by elevated pH (Nakayama and Watanabe 1995), altered inner membrane composition (Danese et al. 1998), and overproduction of envelope proteins like the membrane lipoprotein NlpE (Snyder et al. 1995) or misfolded pili subunits (Hung et al. 2001) and variants of the maltose-binding protein (Hunke and Betton 2003). The Cpx regulon is inhibited by CpxP (Danese and Silhavy 1998), a small protein that could bind the periplasmic sensor domain of CpxA (Fleischer et al. 2007) and is degraded in the presence of Cpx inducers (Buelow and Raivio 2005; Isaac et al. 2005). Indeed, it has been recently shown that CpxP is required for suppressing the toxicity associated with misfolded P pilus subunits and that it is efficiently degraded by the periplasmic DegP protease with these misfolded substrates (Isaac et al. 2005). In contrast to the σE response, the precise molecular mechanism by which the Cpx system senses the presence of incorrectly folded proteins in the periplasm is not known. Although CpxP modulates the activity of CpxA, the protein does not appear to be required for signaling all Cpx stresses (DiGiuseppe and Silhavy 2003). To try to understand the function of CpxP, we decided to carry out biochemical studies with purified components. Because we were interested only in native proteins, we produced the wild-type mature CpxP in the periplasm without any additional tag or fusion sequences. Here, we show that the signal sequence of CpxP exerted a strong negative effect on its periplasmic production. Silent mutations in the gene region encoding the signal sequence increase the production of CpxP in the periplasm, suggesting that cpxP expression could be inhibited by the formation of mRNA secondary structures. When other well-characterized signal sequences were fused to the mature CpxP, efficient production of functionally active CpxP was found. Finally, this study enabled us to produce large amounts of CpxP in the bacterial periplasm.
Results
Cytoplasmic versus periplasmic expression
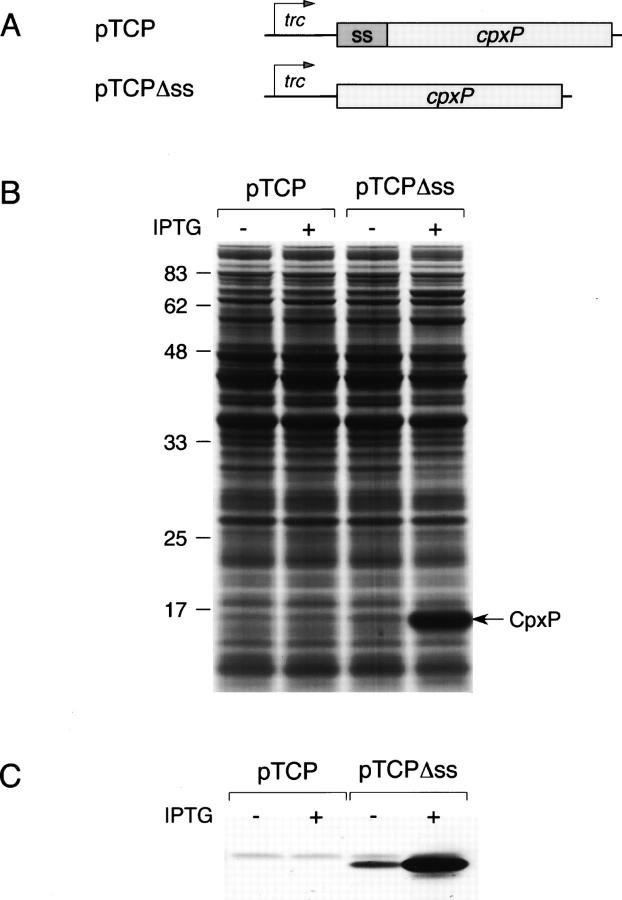
The cpxP gene, either with or without its own signal sequence, was cloned under the control of the trc promoter on the pTrc99A expression vector, and a ΔcpxP strain was transformed with the resulting pTCP and pTCPΔss plasmids. Following induction of the trc promoter by IPTG, no CpxP protein was detectable from cells carrying pTCP, by Coomassie Blue staining, or immunoblotting (Fig. 1). In contrast, the cytoplasmic production of CpxP in cells carrying pTCPΔss resulted in high protein levels. This observation led us to consider that CpxP could be rapidly degraded by proteases when produced with its signal sequence and more likely by periplasmic DegP, as was previously shown in the presence of P pilus subunits (Isaac et al. 2005). However, the expression from pTCP in a ΔdegP strain did not improve the steady-state level of CpxP in the periplasm (data not shown). Therefore, we hypothesized that the dramatic difference of the CpxP level observed between cytoplasmic and periplasmic expression might be due to effects exerted at the translational level. Since both cpxP mRNAs transcribed from pTCP and pTCPΔss contained the same 5′ untranslated region but differed in the translational initiation region, we performed two sets of experiments to investigate whether this critical sequence downstream of the start codon prevents the efficient production of CpxP in the periplasm.
Figure 1.
Cytoplasmic versus periplasmic production of CpxP. (A) Schematic representation of genetic constructs used for the overexpression of cpxP under trc promoter control. (B) MM2 strain harboring pTCP or pTCPΔss was grown at 37°C either with or without IPTG induction. Whole-cell extracts normalized to the same amount of cells were separated by SDS-PAGE and stained by Coomassie Blue. The position of CpxP is indicated by an arrow. (C) Immunoblots of whole-cell extracts from cells carrying pTCP or pTCPΔss using anti-CpxP antibodies.
Optimization of the signal sequence of CpxP
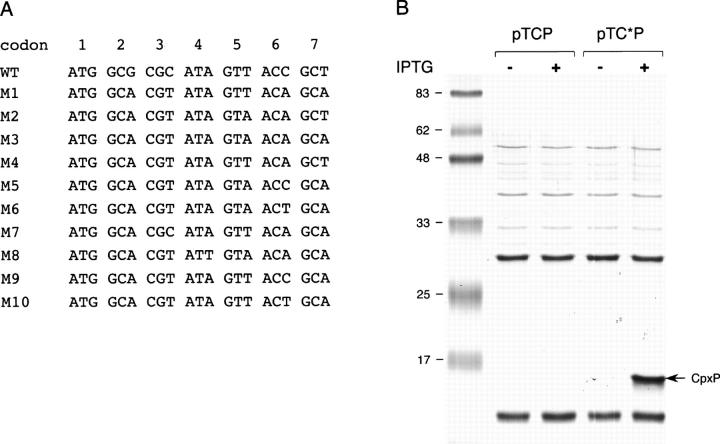
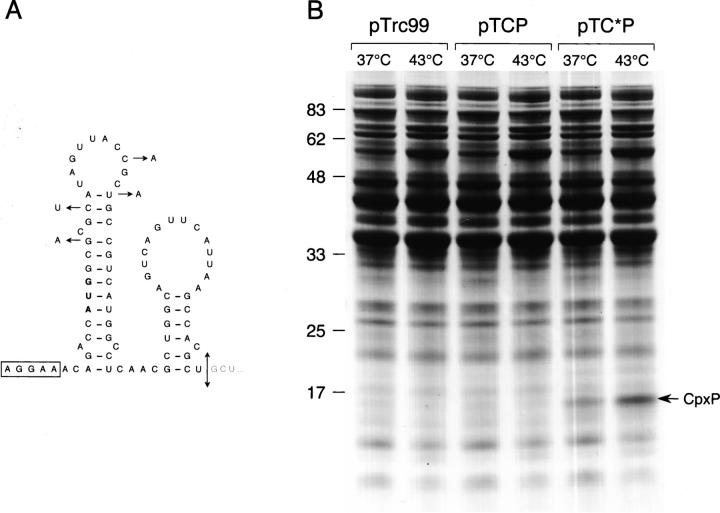
It is generally accepted that gene expression at the translational level can be determined by the efficiency of the initiation process (de Smit and van Duin 1990). In an attempt to mutagenize the cpxP mRNA sequence in the region downstream of the start codon, silent base substitutions were designed to minimize both GC content and base-pair probability by using the knowledge-based software ProteoExpert (Fig. 2A). By this methodology, recently developed for improving the translational efficiency of mRNAs in cell-free expression (Voges et al. 2004), high protein levels from an initially poorly expressed gene have been achieved in E. coli cells (Betton 2004). The first set M1 of silent base mutations was introduced into the pTC*P plasmid, creating a CpxP signal sequence derivative by maintaining the same amino acid sequence as that encoded by pTCP. Following induction, spheroplasts of cells carrying pTCP or pTC*P were fractionated and periplasmic fractions were analyzed by SDS-PAGE (Fig. 2B). Remarkably, the expression of cpxP with the mutated signal sequence resulted in the presence of large amounts of CpxP in the periplasm. Although the quantification of this effect could not be realized because of the absence of CpxP in the wild-type condition, the four base substitutions dramatically increased the production of the protein. However, the precise contribution of specific sequence motifs or features in the mRNA cannot be estimated by the software. Nevertheless, we investigated whether base-pair probability in the mutated signal sequence could help to explain this positive effect on cpxP expression. A putative mRNA secondary structure was predicted for the cpxP gene encoding the signal sequence by the Mfold program (Fig. 3A). Among the four base substitutions that increased the production of CpxP, three may affect base pairing of this hypothetical stem–loop structure. To test whether this putative mRNA secondary structure would be disrupted by an increase of temperature, and thus enhancing the translation efficiency, we compared the level of CpxP in cells grown at 37°C and at 43°C (Fig. 3B). While the steady-state level of CpxP produced with the wild-type signal sequence remained undetectable even at high temperature, a twofold increase between 37°C and 43°C was found for the CpxP with the optimized signal sequence. This result is consistent with a temperature destabilization of a mRNA structure that limits the synthesis of CpxP.
Figure 2.
Silent mutations enhancing the periplasmic production of CpxP. (A) The 10 sets of silent base substitutions within codons 2–7 of cpxP proposed by ProteoExpert are ranked as the indicated order with M1 having the highest predicted expression score. (B) MM2 strain harboring pTCP or pTC*P was grown at 37°C either with or without IPTG induction, then fractionated by spheroplast preparation. Periplasmic fractions normalized to the same amount of cells were analyzed by SDS-PAGE and stained by Coomassie Blue. The position of CpxP is indicated by an arrow.
Figure 3.
Heat-induced production of CpxP. (A) Putative mRNA secondary structure predicted by the Mfold program for the 5′ region encoding the CpxP signal sequence. The Shine–Dalgarno sequence is boxed and the initiation codon is shown in boldface letters. The four silent base substitutions introduced in pTC*P and the corresponding cleavage site of the CpxP signal sequence are indicated by arrows. (B) MM2 strain harboring pTrc99, pTCP, or pTC*P was grown with IPTG induction at 37°C or 43°C. Whole-cell extracts normalized to the same amount of cells were separated by SDS-PAGE and stained by Coomassie Blue. The position of CpxP is indicated by an arrow.
Signal sequence of MalE and DsbA promote high-level of CpxP
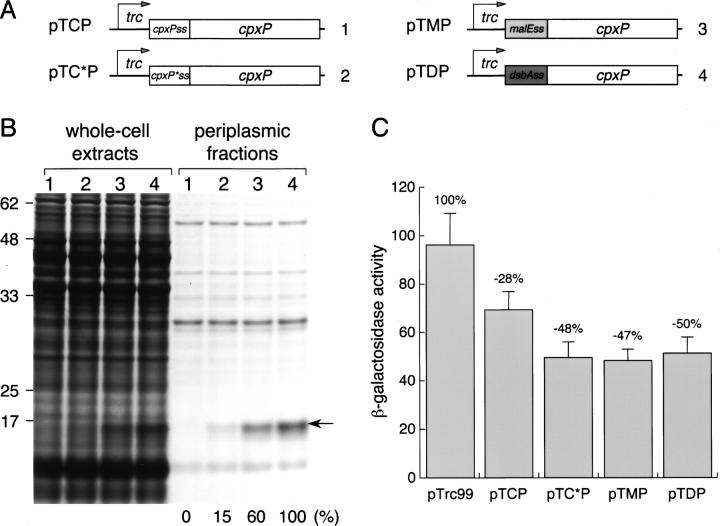
To analyze if the absence of periplasmic CpxP was determined uniquely by its signal sequence or by features related to its mature sequence, we tested whether other signal sequences were able to efficiently produce periplasmic CpxP. MalE and DsbA are two well-characterized E. coli periplasmic proteins, and it was recently shown that their signal sequences can promote post- and cotranslational precursor translocation, respectively (Schierle et al. 2003). Thus, the mature sequence of CpxP was fused in-frame with the signal sequence of MalE or DsbA (Fig. 4A). After fractionation, large amounts of periplasmic CpxP were detected in cells producing the protein fused to both signal sequences (Fig. 4B). The MalE and DsbA signal sequences showed an increased protein yield of fourfold and sevenfold, compared to the mutated CpxPss, respectively. The N-terminal amino acid sequence obtained for periplasmic CpxP was AEVG, indicating that the three corresponding precursors were correctly processed during translocation. Thus, CpxP fused to these different signal sequences revealed that large amounts of protein could be achieved in the periplasm, confirming that its signal sequence has a negative effect on CpxP synthesis.
Figure 4.
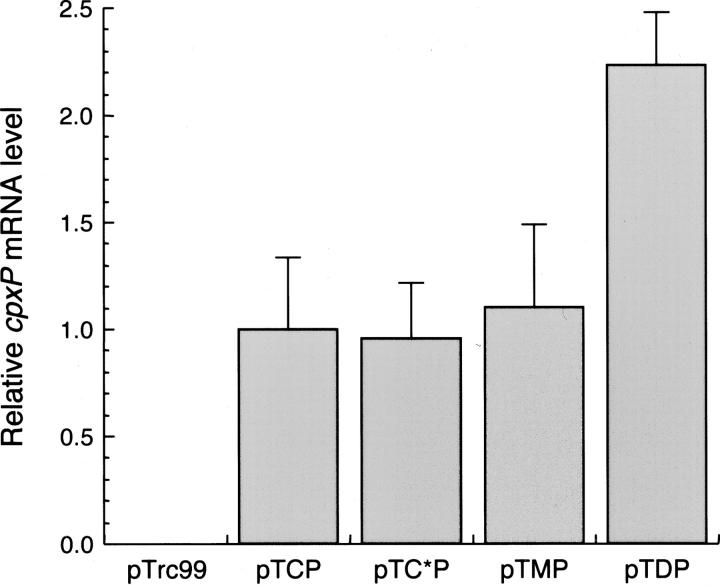
Signal sequences that further increase the periplasmic level of CpxP. (A) Schematic representation of genetic constructs using the CpxP signal sequence (pTCP), the mutated CpxP signal sequence (pTC*P), the MalE signal sequence (pTMP), and the DsbA signal sequence (pTDP) fused to the mature coding sequence of CpxP. (B) MM2 strain harboring pTCP (1), pTC*P (2), pTMP (3), and pTDP (4) was grown at 37°C with IPTG induction, then fractionated by spheroplast preparation. Whole-cell extracts and periplasmic fractions, normalized to the same amount of cells, were separated by SDS-PAGE and stained by Coomassie Blue. The position of CpxP is indicated by an arrow and the relative amount of this protein is indicated under the corresponding lane. (C) Effect of cpxP overexpression on the activity of its own promoter. SR4046 strain, carrying a chromosomal cpxP-lacZ fusion and transformed by the indicated plasmids, was grown at 37°C with IPTG induction. Miller units of lacZ encoded β-galactosidase were calculated using the average of four independent experiments. Inhibition of the cpxP promoter activity is indicated at the top of the bars.
Although CpxP is not required for Cpx signaling, it can negatively regulate this response, probably via a direct binding to the periplasmic sensing domain of CpxA (Fleischer et al. 2007). To see what effect increased Cpx levels could have on the Cpx regulon, we quantified the corresponding inhibition using a cpxP-lacZ transcriptional gene fusion (Fig. 4C). Interestingly, the inhibition of cpxP expression seemed to be saturated by these large amounts of periplasmic CpxP. About 50% of the maximal inhibitory signal was observed with the undetectable protein level from pTCP, and this signal was saturated by the amounts of CpxP from pTC*P. Although we did not determine the cellular level of CpxA, this saturation would seem to reflect a direct interaction between periplasmic CpxP and the membrane-bound CpxA.
Quantification of steady-state mRNA levels
An important parameter that might influence protein synthesis in E. coli is mRNA stability. To test whether differences in mRNA levels could explain the variation in protein levels, we measured steady-state cpxP mRNA levels from cells carrying the various plasmids encoding CpxP by quantitative real-time RT-PCR (Fig. 5). The relative abundance of the endogenous 16S rRNA was used as an internal control for data normalization. Except for pTDP, the quantification of cpxP mRNA expressed from all pTrc99 derivative plasmids gave a similar value. Interestingly, the twofold increase of cpxP mRNA level found for pTDP could be correlated with the twofold increase of CpxP when produced with the DsbAss. Since all genes were expressed in identical conditions from the same trc promoter, this increase is almost certainly not derived from a transcriptional effect. However, these comparative data clearly indicate that the steady-state level of cpxP mRNA from pTCP was not a limiting factor for producing the corresponding protein.
Figure 5.
Quantification of cpxP mRNA levels. The steady-state level of cpxP mRNA in MM2 strain transformed by the indicated plasmids encoding CpxP was quantified using real-time RT-PCR. The relative change in cpxP mRNA transcripts, normalized to 16S RNA transcripts, was measured using the average of four independent determinations with their standard deviations.
The signal sequence of CpxP exerts a negative effect on malE expression
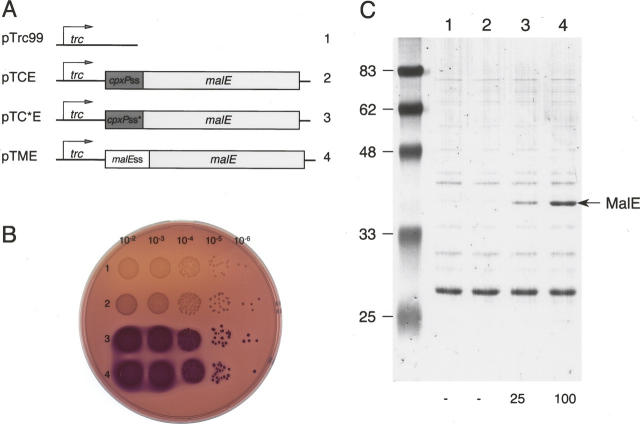
To further test whether the signal sequence of CpxP limits the production of another protein, we fused the mature sequence of MalE to either the wild-type or optimized CpxPss (Fig. 6A), and compared the resulting maltose phenotypes and periplasmic protein levels. While no protein could be detected when fused to the CpxPss, this periplasmic amount of MalE was able to complement to some extent the chromosomal deletion ΔmalE444 for maltose uptake, as indicated by the light red color of the corresponding colonies on the maltose MacConkey plate (Fig. 6B). Like CpxP, the periplasmic level of MalE varied from undetectable to detectable amounts when using the wild-type and mutated CpxPss, yet substantially lower than the MalEss. The doubling time (DT) in liquid M63B1-maltose minimal medium provides a more objective quantification of periplasmically active MalE (Table 2). The decrease of DT values determined for cells carrying the various plasmids was well correlated with the increase of periplasmic MalE levels found in the corresponding cells (Fig. 6C). In agreement with the previous results, the optimized CpxPss improved the production of MalE in the periplasm.
Figure 6.
Signal sequence of CpxP limits the production of MalE. (A) Schematic representation of genetic constructs using the CpxP signal sequence (pTCE), the mutated CpxP signal sequence (pTC*E), and the MalE signal sequence (pTME) fused to the mature coding sequence of MalE. (B) MacConkey agar plate showing the maltose phenotype of PM9 strain carrying pTrc99 (1), pTCE (2), pTC*E (3), and pTME (4). Overnight cultures were spotted after serial dilutions as indicated, and the plate was incubated for 24 h at 37°C. (C) PM9 strain harboring pTrc99 (1), pTCE (2), pTC*E (3), and pTME (4) was grown at 37°C with IPTG induction, then fractionated by spheroplast preparation. Periplasmic fractions normalized to the same amount of cells were separated by SDS-PAGE and stained by Coomassie Blue. The position of MalE is indicated by an arrow and the relative amount of this protein is indicated under the corresponding lane.
Table 2.
Maltose phenotype
Discussion
We have attempted to understand why CpxP was not detectably produced in cells containing a cpxP-overexpressing plasmid. Taken together, our experiments indicated that the presence of the signal sequence strongly limits the quantity of CpxP in the periplasm. The substitution of the CpxP signal sequence by that of MalE or DsbA dramatically shifts the level of CpxP protein from no detectable to high periplasmic amounts. We hypothesized that the low cpxP expression level might be due to inefficient translation initiation. Indeed, the finding that silent base substitutions in the first codons of cpxP, which would disrupt or abolish a hypothetical mRNA secondary structure and result in a large increase of CpxP levels, suggests that the predicted structure may be directly or indirectly related to translation efficiency. Additional support for this hypothesis can be found in the further increase of CpxP levels in cells grown at 43°C. When the CpxP signal sequence was fused to the mature MalE sequence, the same effects were also observed on the periplasmic levels of MalE.
A current model for the role of CpxP suggests that in unstressed cells the protein should be bound to the periplasmic domain of the CpxA kinase, keeping its activity at a ground level (Isaac et al. 2005). It was recently shown that such a protein–protein interaction occurs in vitro with both purified proteins (Fleischer et al. 2007). In the presence of misfolded proteins, CpxP dissociates from CpxA and binds these competing proteins. Then, the resulting complexes are delivered to DegP, which degrades both the misfolded substrates and CpxP. Thus, it was proposed that CpxP functions as a periplasmic adaptor protein to activate or modulate the activity or the specificity of DegP (Isaac et al. 2005). This model predicts that in the absence of DegP detectable levels of CpxP could be achieved. Although we were limited by the sensitivity of the detection technique that we used, no CpxP could be detected in a degP minus strain carrying pTCP. It appears that extensive proteolysis of CpxP could not entirely explain our failure to steadily produce this protein in the periplasm. Therefore, we searched for other explanations and explored the possibility that the translational initiation region of cpxP in this context was critical for its expression. Other possibilities like extensive degradation of the CpxP precursor in the cytoplasm or the inability of the CpxP signal sequence to promote efficient precursor export have also been envisaged to explain this failure. However, under the conditions we tested, no precursor was ever detected in the production of CpxP, and the large difference in CpxP levels between the two identical amino acid precursor sequences produced either from pTCP or from pTC*P cannot be explained by its rapid proteolysis. In addition, our studies show that CpxP was stable enough at 43°C to be steadily detected by Coomassie Blue staining. This result was not expected because heat shock conditions might result in an increased level of envelope-stressed proteins that activate the synthesis of DegP (Alba and Gross 2004) and consequently degrade CpxP. Thus, high temperatures, per se, are unlikely to represent an inducing signal for the Cpx response, even if a functional Cpx system is required to alleviate the toxicity of misfolded periplasmic proteins under heat shock conditions (Hunke and Betton 2003). It has been predicted that a correlation should exist between the ability of a stressed periplasmic protein to activate the Cpx response and the CpxP-mediated proteolysis of that specific protein by DegP.
While other more complex mechanisms may be involved, we could not exclude that formation of the predicted mRNA secondary structure may negatively affect cpxP expression in our experiments. Beside the translational control proposed for rpoH (Morita et al. 1999), a similar mechanism involving the translational initiation sequence encoding the signal sequence of a lytic transglycosylase was suggested to regulate its gene expression (Serruto and Galeotti 2004). Our experiments, obtained with recombinant cpxP overexpression, did not attempt to address whether such a translational control is physiologically relevant for the chromosomal expression of cpxP. Indeed, it is difficult to compare plasmid and chromosomal expression since the 5′-untranslated region (5′-UTR) of transcripts from both cpxP genes are different and a significant part of this region is included in the predicted stem–loop structure of the mRNA. However, when cpxP was cloned into a different expression vector (T7 based) with a different 5′-UTR, we observed a similar inefficient protein production in the periplasm, suggesting that the 3′-translated region of cpxP mRNA encoding the signal sequence has an intrinsic tendency to form a stable secondary structure.
Although the level of CpxP when fused to the signal sequence of MalE was lower than when fused to that of DsbA, export of CpxP may not have a strong requirement for the cotranslational mode or SRP-dependent pathway. The observed difference between these constructs can be easily explained at the transcriptional level by their mRNA abundance. It was hypothesized that nonoptimal codons present in the signal sequence of exported proteins via the Sec-dependent pathway may play an important role in the coupling of translation to export, by slowing down the translation rate (Burns and Beacham 1985). This interesting hypothesis has recently received experimental support using the signal sequence of MalE as an exported-protein reporter (Zalucki and Jennings 2007). In this case, the six codons that have been changed to their most optimal synonymous codons resulted in a 20-fold decrease of the periplasmic MalE level. The values of the codon adaptation index or CAI (an index measuring the codon usage bias in a gene [Sharp and Li 1987] that is proportional to the translation rate of individual codons) for the wild-type and optimized signal sequences were 0.175 and 0.366, respectively. In our study, the difference between CAI values calculated for the wild-type and optimized signal sequences is only 0.025 (0.171 vs. 0.145). Although it would be surprising that even a small change in codon usage dramatically influences gene expression, the increased production of CpxP when fused to the optimized signal sequence is compatible with a decreased rate of translation.
Finally, by applying a knowledge-based software developed previously for enhancing the efficiency of translational initiation for in vitro expression, we demonstrated that this technology works in vivo as well for homologous expression and that it may be useful for engineering the production of difficult to export proteins with biotechnical interest. In addition, these results can be extended to modulate the production of periplasmic proteins at desired levels. Here, these optimizations enabled us to produce enough CpxP in the bacterial periplasm for future biochemical studies of the extracytoplasmic Cpx stress response.
Materials and Methods
Bacterial strains and growth conditions
E. coli strains pop6590 (ΔmalE444 recA1 srl∷Tn10), SR4640 (cpxP-lacZ), and MM2 (pop6590 ΔcpxP) are derivatives of MC4100. MM2 was constructed by introducing the cpxP∷aphA (KmR) allele by standard P1 transduction. Luria broth (LB) and M63B1 growth media and MacConkey agar (Difco) were as described by Miller (1992). Cells were generally grown at 37°C in LB medium supplemented with 0.1 mg/mL ampicillin. To induce expression from the trc promoter, IPTG was used at 0.5 mM.
Plasmids constructions
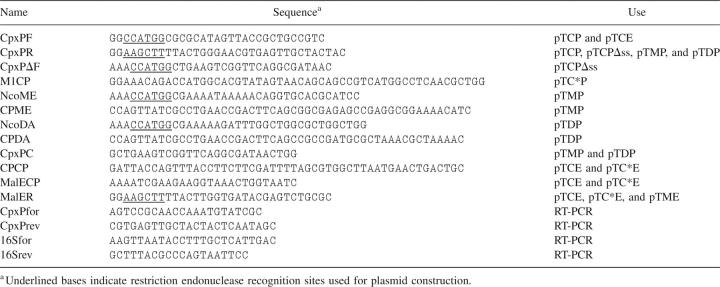
All plasmids generated in this study are derivatives of the pTrc99 expression vector (Amersham Pharmacia). pTCP and pTCPΔss were constructed by cloning DNA fragments containing the wild-type and signal sequence deleted cpxP amplified by PCR from E. coli MC4100 chromosomal DNA using the primer pairs CpxPF/CpxPR and CpxPΔF/CpxPR (Table 1), respectively. Silent mutations into the first seven codons of cpxP were introduced by the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene) using the mutagenic primer M1CP and pTCP as DNA template. The substitution of the signal sequence was generated in two steps by overlap extension PCR. First, the region of cpxP, malE, and dsbA encoding the signal sequence was amplified using the primer pairs CpxPF/CPCP, NcoME/CPME, and NcoDA/CPDA and cpxP and malE encoding the mature sequence using CpxPC/CpxPR and CPCP/MalECP, respectively. The DNA fragments containing malE were amplified from a plasmid that carries a single base change altering its internal NcoI site. Then, these PCR products were assembled using appropriate outer primers (Table 1) in a second PCR step, and the final DNA fragments were digested with NcoI and HindIII and inserted into pTrc99.
Table 1.
Oligonucleotides used in this study
Cell fractionation
SR4640 cells carrying the various pTrc99 derivatives were grown at 37°C in LB supplemented with 0.1 mg/mL ampicillin to mid-log phase and induced with IPTG. After 1 h (A600 ≈ 1.5), cells were normalized to the same amount and harvested by centrifugation. The pellets were resuspended in 25 mM Tris-HCl (pH 7.5) buffer containing 0.5 M sucrose. Spheroplasts were prepared by adding lysozyme (0.2 mg/mL) and EDTA (10 mM, pH 7.5), and suspensions were incubated for 30 min at 4°C before centrifugation at 14,000g for 5 min. Supernatants representing the periplasmic fractions were withdrawn, mixed with XT sample buffer (Bio-Rad), and boiled. Cell extracts were analyzed on 12% Bis-Tris polyacrylamide gels (Criterion, Bio-Rad). Proteins were either visualized by Coomassie Blue staining or electrotransferred onto nitrocellulose membranes prior to immunodetection with rabbit polyclonal antibodies against purified CpxP, generated by Eurogentec, and with alkaline phosphatase-coupled antiserum against rabbit immunoglobulins. The immunoblots were developed with nitroblue tetrazolium and 5-bromo 4-chloro 3-indolyl phosphate. For quantitative analysis, gels were scanned with an Image master VDS camera (Amersham Biotech).
Quantitative real-time RT-PCR
MM2 cells carrying the various pTrc99 derivatives were grown at 37°C in LB supplemented with 0.1 mg/mL ampicillin and 0.5 mM IPTG. After 2 h (A600 ≈ 0.6), triplicate samples were collected and total RNA from each culture was isolated by a method using Macaloid clay as described (Kuipers et al. 1993). DNA contamination was removed using the TURBO DNA-free kit (Ambion), and cDNA synthesis was performed using the reverse transcriptase as described previously. The primer pairs CpxPfor/CpxPrev and 16Sfor/16Srev (Table 1) were used to amplify a 149-bp fragment of the cpxP gene-specific cDNA and a 117-bp fragment of the 16S rRNA-specific cDNA, respectively. The later was used as an internal standard. A quantitative PCR was performed on the MyiQ PCR detection system (Bio-Rad) in a total volume of 25 μL using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions. Each specific amplicon was verified by the presence of a single melting-temperature peak and the relative abundance of cpxP mRNAs normalized to 16S rRNA were calculated as described previously (Livak and Schmittgen 2001).
β-galactosidase assays
Cells (SR4640 strain) carrying pTrc99, pTCP, pTC*P, pTMP, and pTDP were grown at 37°C in LB medium and induced by IPTG. The β-galactosidase activity of permeabilized cells was assayed as previously described (Betton et al. 1996), and the specific activity in Miller units was calculated using the average of four independent determinations with their standard deviations.
Computer analysis
Silent base substitutions in cpxP encoding the signal sequence and cloned into pTrc99 were designed by the commercial software ProteoExpert (https://ssl.biomax.de/ProteoExpert/html/index.jsp). From all silent mutations in the first seven codons this software calculates the top 10 gene sequences with the highest expression score (Voges et al. 2004). The prediction of base pairing in cpxP mRNA sequence restricted to 100 bases that encode the signal sequence were obtained by the Mfold program (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi), which uses the method of Zuker (1989) to determine optimal and suboptimal secondary structures for nucleic acids.
Acknowledgments
We thank Olivier Poupel for his invaluable assistance with RT-PCR analysis and Emmett Johnson for critical reading of the manuscript. This work was supported by grants from the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), and the Agence Nationale de la Recherche (06-BLAN-023904). M.M. is a recipient of Fondation de la Recherche Médicale (FRM).
Footnotes
Reprint requests to: Jean-Michel Betton, Unité Biochimie Structurale, Institut Pasteur, URA CNRS 2185, 25 rue du Docteur Roux, 75724 Paris cedex 15, France; e-mail: jmbetton@pasteur.fr; fax: 331-4568-8604.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073047807.
References
- Alba B.M. and Gross, C.A. 2004. Regulation of the Escherichia coli σ-dependent envelope stress response. Mol. Microbiol. 52: 613–619. [DOI] [PubMed] [Google Scholar]
- Betton J.M. 2004. High-throughput cloning and expression strategies for protein production. Biochimie 86: 601–605. [DOI] [PubMed] [Google Scholar]
- Betton J.M., Boscus, D., Missiakas, D., Raina, S., and Hofnung, M. 1996. Probing the structural role of an αβ loop of maltose-binding protein by mutagenesis: Heat-shock induction by loop variants of the maltose-binding protein that form periplasmic inclusion bodies. J. Mol. Biol. 262: 140–150. [DOI] [PubMed] [Google Scholar]
- Buelow D.R. and Raivio, T.L. 2005. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J. Bacteriol. 187: 6622–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D.M. and Beacham, I.R. 1985. Rare codons in E. coli and S. typhimurium signal sequences. FEBS Lett. 189: 318–324. [DOI] [PubMed] [Google Scholar]
- Danese P.N. and Silhavy, T.J. 1997. The σ(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli . Genes & Dev. 11: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Danese P.N. and Silhavy, T.J. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180: 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese P.N., Snyder, W.B., Cosma, C.L., Davis, L.J., and Silhavy, T.J. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes & Dev. 9: 387–398. [DOI] [PubMed] [Google Scholar]
- Danese P.N., Oliver, G.R., Barr, K., Bowman, G.D., Rick, P.D., and Silhavy, T.J. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli . J. Bacteriol. 180: 5875–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartigalongue C. and Raina, S. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli . EMBO J. 17: 3968–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M.H. and van Duin, J. 1990. Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acid Res. Mol. Biol. 38: 1–35. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McGuire, A.M., Liu, X., and Lin, E.C. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli . J. Biol. Chem. 277: 26652–26661. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe P.A. and Silhavy, T.J. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185: 2432–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Iuchi, S., Kwan, H.S., Lu, Z., and Lin, E.C. 1993. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli . Gene 136: 227–230. [DOI] [PubMed] [Google Scholar]
- Duguay A.R. and Silhavy, T.J. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694: 121–134. [DOI] [PubMed] [Google Scholar]
- Fleischer R., Heermann, R., Jung, K., and Hunke, S. 2007. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli . J. Biol. Chem. 282: 8583–8593. [DOI] [PubMed] [Google Scholar]
- Hung D.L., Raivio, T.L., Jones, C.H., Silhavy, T.J., and Hultgren, S.J. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20: 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunke S. and Betton, J.M. 2003. Temperature effect on inclusion body formation and stress response in the periplasm of Escherichia coli . Mol. Microbiol. 50: 1579–1589. [DOI] [PubMed] [Google Scholar]
- Isaac D.D., Pinkner, J.S., Hultgren, S.J., and Silhavy, T.J. 2005. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc. Natl. Acad. Sci. 102: 17775–17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers O.P., Beerthuyzen, M.M., Siezen, R.J., and De Vos, W.M. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216: 281–291. [DOI] [PubMed] [Google Scholar]
- Livak K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Miller J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Missiakas D. and Raina, S. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179: 2465–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen J.E. and Otzen, D.E. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57: 326–346. [DOI] [PubMed] [Google Scholar]
- Morita M.T., Tanaka, Y., Kodama, T.S., Kyogoku, Y., Yanagi, H., and Yura, T. 1999. Translational induction of heat shock transcription factor σ32: Evidence for a built-in RNA thermosensor. Genes & Dev. 13: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S. and Watanabe, H. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177: 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J., Lynch, A.S., Belin, D., Lin, E.C., and Beckwith, J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes & Dev. 11: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Raivio T.L. and Silhavy, T.J. 1999. The σE and Cpx regulatory pathways: Overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2: 159–165. [DOI] [PubMed] [Google Scholar]
- Raivio T.L. and Silhavy, T.J. 2001. Periplasmic stress and ECF σ factors. Annu. Rev. Microbiol. 55: 591–624. [DOI] [PubMed] [Google Scholar]
- Schierle C.F., Berkmen, M., Huber, D., Kumamoto, C., Boyd, D., and Beckwith, J. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185: 5706–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D. and Galeotti, C.L. 2004. The signal peptide sequence of a lytic transglycosylase of Neisseria meningitidis is involved in regulation of gene expression. Microbiol. 150: 1427–1437. [DOI] [PubMed] [Google Scholar]
- Sharp P.M. and Li, W.H. 1987. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder W.B., Davis, L.J., Danese, P.N., Cosma, C.L., and Silhavy, T.J. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177: 4216–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Watzele, M., Nemetz, C., Wizemann, S., and Buchberger, B. 2004. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem. Biophys. Res. Commun. 318: 601–614. [DOI] [PubMed] [Google Scholar]
- Walsh N.P., Alba, B.M., Bose, B., Gross, C.A., and Sauer, R.T. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113: 61–71. [DOI] [PubMed] [Google Scholar]
- Weber R.F. and Silverman, P.M. 1988. The cpx proteins of Escherichia coli K12. Structure of the cpxA polypeptide as an inner membrane component. J. Mol. Biol. 203: 467–478. [DOI] [PubMed] [Google Scholar]
- Zalucki Y.M. and Jennings, M.P. 2007. Experimental confirmation of a key role for nonoptimal codons in protein export. Biochem. Biophys. Res. Commun. 355: 143–148. [DOI] [PubMed] [Google Scholar]
- Zuker M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]