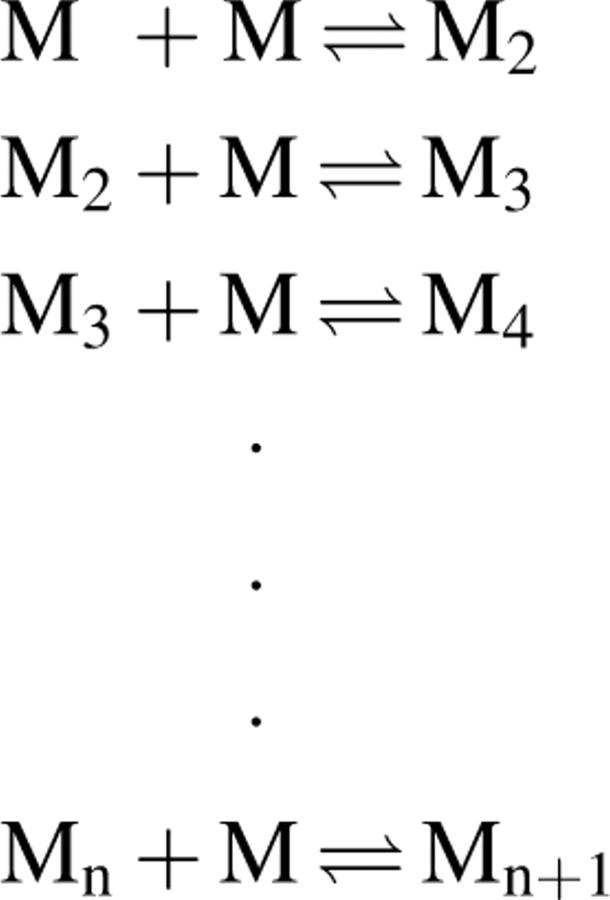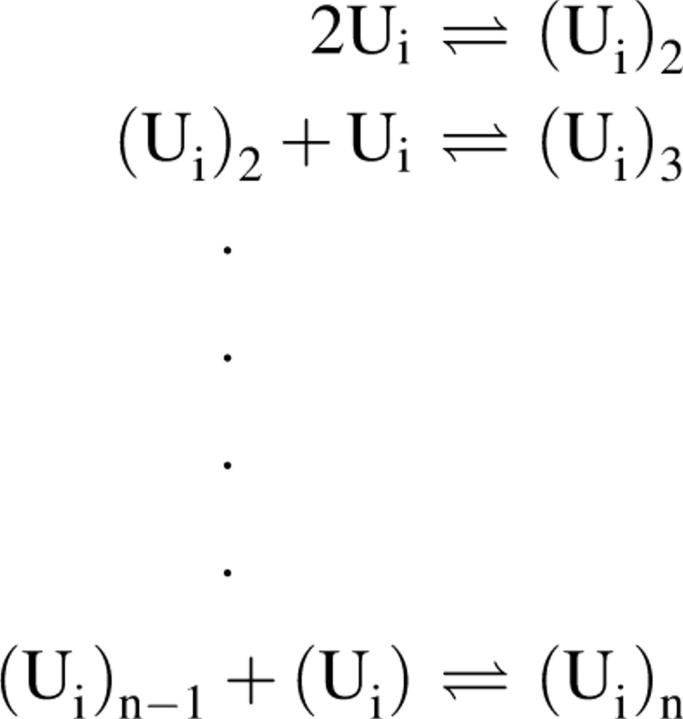Abstract
Amyloid formation typically follows a time course in which there is a long lag period followed by a rapid formation of fibrils. In this review, I show that the standard mechanisms of polymerization need to be expanded to consider that the monomeric proteins/peptides involved in amyloid formation are intrinsically disordered and exist as an ensemble of disordered-collapsed states. The review focuses primarily on events which occur in the long lag period defining these as protein folding issues, coupled with formation of oligomers. Experimental methods to explore folding and oligomerization issues over a wide range of protein concentrations using primarily fluorescence and 19F-NMR methods are discussed.
Keywords: intrinsically disordered proteins, nucleation, oligomers, polymerization, fluorescence spectroscopy, NMR
The formation of amyloid plaques in many neurodegenerative diseases is the consequence of aggregation of peptides/proteins that have been characterized as intrinsically disordered. Intrinsically disordered proteins (IDPs; also called natively disordered, natively unfolded, or intrinsically unstructured) may be defined as those proteins that appear to lack a defined stably structured state. They are ensembles of states in which side-chain and backbone positions may deviate significantly from some equilibrium position. In contrast to polymerization processes that start with structured monomers and may be driven by nucleotide binding or hydrolysis, amyloid formation involves interactions between ensembles of unstructured monomeric states with the final product, the fibril, generally believed to be an ordered β-sheet structure. The aggregation of such peptide/protein systems, which give rise to these structured fibrils, has been the subject of intense investigation (e.g., Ferrone 1999; Tycko 2004; Urbanc et al. 2004; Dobson 2006; Dusa et al. 2006; Fink 2006; Powers and Powers 2006; Wetzel 2006a), and has been recently reviewed (Kheterpal and Wetzel 2006b,a; Wetzel 2006b). In spite of the extensive literature, however, the mechanism of aggregation is poorly understood. Delineating mechanisms for processes that start from unstructured states and results in structured fibrils, however, is critical to understanding the role of such aggregates in diseases, and is essential if one is to develop drugs that may interfere with the process.
Of particular relevance to the mechanism of the aggregation process is that these systems typically exhibit a very long lag period, in which nothing appears to be happening, followed by a rapid formation of the fibril. This review deals primarily with an exploration of what actually does happen during the early steps that delineate the lag period which may define, in large part, the mechanism of aggregation.
The terms to describe amyloid formation, polymerization and aggregation, are fundamentally the same, although aggregation implies a nonspecific process while polymerization implies a more defined process. This distinction between polymerization and aggregation becomes unclear when dealing with the formation of amyloids because both specific and nonspecific processes may be occurring at the same time. I start with a discussion of polymerization mechanisms, in which the monomers are structured and every oligomeric species is on the pathway for forming a polymeric species. I will then turn to the condition where the monomers may be an ensemble of poorly structured states frequently characterized as intrinsically disordered. Having such structures complicates the ability to understand or delineate the mechanism of polymerization. Furthermore, as discussed later, the term IDPs, which is commonly used to describe peptides/proteins that aggregate, does not fully describe the nature of these proteins. Finally, I discuss some experimental methods that can be used to help delineate the mechanism of aggregation by investigating early steps in the process.
In general, polymerization from structured monomers has been considered to be of two types that reflect extreme situations: isodesmic (or linear) polymerization, and nucleation–elongation polymerization. In isodesmic polymerization, the dissociation constants for monomer addition to any protein species are considered to be identical, independent of the size of the polymer. In a nucleation–elongation process, several monomers form a nucleus that serves as the structure for adding more monomers with the same rate constants controlling each step for monomer addition and dissociation. Nucleation–elongation processes are generally differentiated from isodesmic processes on the basis of three criteria: (1) There is a time-dependent lag in the formation of the polymer, (2) the lag can be abolished by the addition of a preformed nucleus (seeding), and (3) there is a critical concentration representing the monomer in equilibrium with the polymer. A process cannot be considered to be a nucleation–elongation polymerization unless all three criteria hold, since at least two of the three can be observed in the isodesmic case.
Polymerization: The isodesmic mechanism
In this process, the total concentration of monomer M, Mt, is defined as
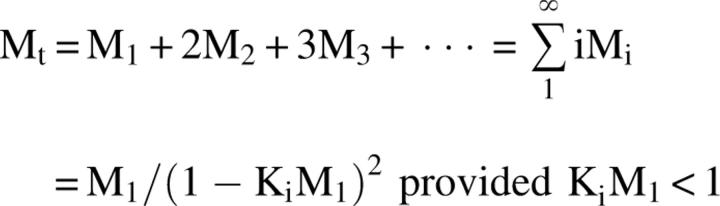 |
where Ki is the equilibrium constant for monomer addition. At equilibrium, the concentration of the monomeric species, M 1, is
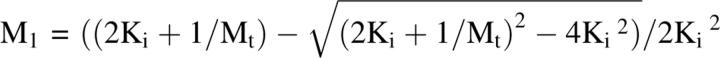 |
and at high protein concentrations under polymerizing conditions M 1 will asymptotically approach 1/Ki (Oosawa and Asakura 1975). That is, some monomer will remain after polymerization but there is not a specific critical concentration (see below).
Using the previously described (Barshop et al. 1983) numerical integration program, KINSIM, it is possible to simulate the polymerization process for isodesmic polymerization.
The simplest mechanism is
Scheme 1.
We have shown elsewhere that n = 6 is appropriate for polymerizing systems (Frieden 1983). This mechanism assumes that all oligomeric species are productive with respect to monomer addition. Presumably this will be true when Mn and Mn +1 are equivalent structures. Interestingly, if the rate constants are such that oligomeric species are not favored (kr > kf), one can observe a lag in polymer formation. In this case, the concentration of monomer after polymerization might be appreciable relative to the initial monomer concentration. The appearance of a lag in the time-dependent formation of the polymer is therefore not limited to nucleation–elongation mechanisms. When the equilibria favor the higher molecular weight oligomers, any lag in polymer formation disappears.
Polymerization: The nucleation–elongation mechanism
A generic definition of a nucleation–elongation process is one in which a set of monomers forms a multimonomeric species, an oligomer, or nucleus, and subsequent monomer addition (the elongation step) occurs with the same forward and reverse rate constants independent of the size of the polymer. As described elsewhere (Frieden and Goddette 1983) the kinetics of polymerization were developed in polymer chemistry by Flory (1953) and first applied to protein systems by Oosawa and Asakura (1975) and later, with different assumptions by Wegner and coworkers (1975). As noted by Frieden and Goddette (1983), assumptions made by Oosawa and Asakura (1975) could lead to an incorrect value of the nucleus size. In order to develop analytical expressions to define the time course of polymerization, early studies assumed that nucleus formation, whatever its size, could be defined by a single overall dissociation constant, and that all elongation steps occurred with the same rate constant. Using a steady-state assumption, the analytic expressions given by Wegner and coworkers (1975) are
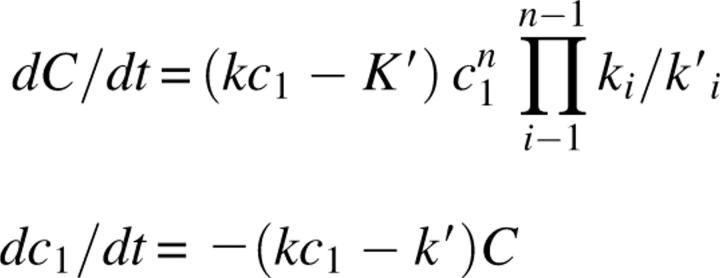 |
where C is the molar concentration of filaments, ki and k′ I are rate constants related to nuclei formation and dissociation, n is the size of the nucleus, c 1 is the monomer concentration, and k and k′ are rate constants for addition and dissociation of monomer to and from the filament. At equilibrium, dc 1 /dt = 0 and the ratio k′/k is defined as the critical concentration.
Actin and microtubule polymerization have served as model systems for this type of polymerization (Frieden 1985). The assumption that there is a single dissociation constant for nucleus formation, however, provides no information on the mechanism for forming the nucleus.
With the introduction of systems for numerical integration of the differential equations that describe polymerization (i.e., KINSIM) (Barshop et al. 1983), it was not necessary to make any simplifying assumptions as discussed elsewhere (Frieden 1985).
Polymerization: The middle ground
For nucleation–elongation systems as discussed above, the question becomes at what point does the association of the monomer become independent of the length of the polymer. In other words, the distinction between an isodesmic mechanism and a nucleation–elongation mechanism rests solely on the nucleus size and the associated rate constants for formation and dissociation of oligomers. Analysis of the time dependence of actin polymerization as a function of actin concentration showed that dimer formation was poor, trimer formation better, and tetramer formation even better (Frieden 1983). The consequence of this observation was that it was not possible to define a specific nucleus size because the concentration of any given small oligomeric species depended on the rate of its formation and the rate of monomer dissociation. The rate of formation is a second-order process and thus dependent on monomer concentration, while the rate of dissociation of a monomer is a first-order process independent of monomer concentration. If one assumes that the rate of elongation is independent of the number of monomers in the polymer, then the apparent nucleus size for actin polymerization could range from 3 to 4, depending on the monomer concentration and on the rate constant for monomer addition to the monomer, dimer, or trimer. Thus, at high monomer concentrations, the apparent nucleus size tends to be small because monomer addition is more favored. This has important consequences for the assembly of intrinsically disordered systems discussed later.
As suggested from the above discussion, however, depending upon the rate constants chosen for the first few steps, a mechanism could be either an isodesmic or a nucleation–elongation process. Since these mechanisms are extreme cases, a general mechanism might be that rate (or equilibrium) constants change depending on the size or structure of each intermediate oligomer. Thus, it is truly necessary to be able to measure intermediate steps in the polymerization.
There is no a priori reason to assume that all association rate constants be the same in an aggregating system. They could vary with the number of subunits to be added or the nature of the oligomeric intermediate. The extreme case would be the formation of relatively small and defined nuclei which then elongate by monomer addition all adding with the same second-order rate constant. That would formally become a nucleation–elongation mechanism. The mechanism for polymerization of IDPs discussed below is guaranteed to be complex, not falling into either of the two extreme cases, but rather somewhere in between. To explore the mechanism, we need the appropriate kinetic studies, and any attempt to determine the mechanism will depend on what is being measured (see Experimental Approaches).
The case of IDPs
As noted earlier, IDPs are those that lack a defined stably structured state. It is usually assumed that the monomeric species of proteins associated with the formation of aggregates in neurodegenerative diseases exist in a disordered state. For example, Aβ is a small disordered peptide but the final structure of the aggregate is of stacked β-sheets (Luhrs et al. 2005). Similarly, α-synuclein is a disordered protein that may be incorporated into Lewy bodies (Shults 2006) and CsgA is a secreted bacterial protein that forms amyloid-type fibers on the surface of bacteria and is predicted to form fibrils consisting of β-sheets. There have been extensive efforts to understand why some peptides/proteins are intrinsically disordered (Romero et al. 2004; Weathers et al. 2004; Oldfield et al. 2005). Radivojac et al. have determined disorder biases from a large database of proteins (Radivojac et al. 2007; Sickmeier et al. 2007). These investigators also discuss in detail computational methods for determining disorder. Databases of IDPs and regions of proteins that are disordered are available (Sim et al. 2001; Vucetic et al. 2005).
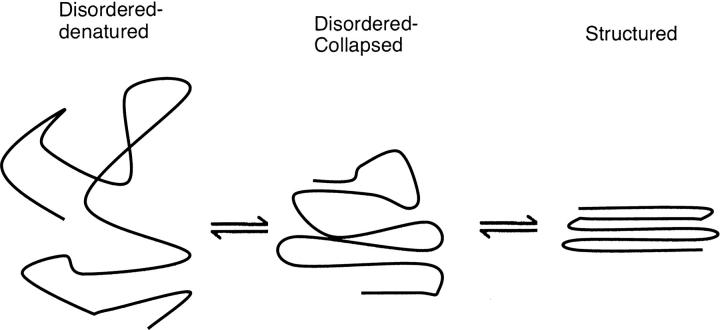
These IDPs probably do not, however, represent denatured states, that is, random coils. Polyglutamine exists as an ensemble of collapsed structures (Crick et al. 2006; Tran and Pappu 2006) as does Aβ (Zhang et al. 2000), α-synuclein (Lee et al. 2007), and regions of the yeast prion protein (Mukhopadhyay et al. 2007). Furthermore, the addition of denaturant unfolds the collapsed state (Mukhopadhyay et al. 2007; S.L. Crick, R. Pappu, and C. Frieden, unpubl.). Collapsed states may also exist at low denaturant (i.e., guanidine) concentrations (Sherman and Haran 2006; Merchant et al. 2007). Based on these and similar results, we now define two states of an intrinsically disordered system, both of which are ensembles of states: disordered–collapsed and disordered–denatured. This is illustrated by Figure 1, which indicates the various states of the IDP monomer. It should be noted, however, that a single large protein, such as a prion, may contain both collapsed and denatured ensembles (Mukhopadhyay et al. 2007). Presumably it is only the disordered–collapsed ensemble which are viable in forming oligomers. In the discussion below, this ensemble will be represented by the symbol U.
Figure 1.
A representation of the possible states of a monomer of intrinsically disordered proteins. It should be recognized that both the disordered–denatured and the disordered–collapsed states represent ensembles of different conformational forms.
The polymerization process of intrinsically disordered proteins as observed in vitro is frequently very slow, on the order of hours or even days. The usual explanation appears to be that nucleation is a slow process. Similar to defining at least two states of the intrinsically disordered protein, we now need to understand the term “nucleation,” and the term that needs to be clarified is “nucleus.” In the above discussion, I have referred to the nucleus as a collection of monomers. If, as Wetzel (Chen et al. 2002) proposes for polyglutamine polymerization, the nucleus is a monomer, then the remaining polymerization would be essentially isodesmic. Wetzel (Chen et al. 2002) considers the conversion of a disordered form (see below) to a structured form as a nucleation step and, therefore, that polymerization is a nucleation–elongation process with the nucleus as the monomer. This definition tends to confuse the distinction between isodesmic and nucleation–elongation mechanisms.
In describing mechanisms for protein folding, the term nucleus refers to that region, or those regions, that form transiently, and that are essential for folding to occur rapidly. For example, Daggett and Fersht (2003) conclude that a unifying mechanism for protein folding is a nucleation–condensation process. It is important to distinguish between a nucleus within a denatured monomer from a nucleus which is an assembly of monomers. Thus, one should describe the nuclei forming within a peptide or protein as the “folding nucleus” and define the term nuclei to be assemblies of monomers. While the aggregation of these disordered peptides and proteins may be presumed to proceed through a nucleation–elongation process, the slow step is almost certainly buried in the fact that the disordered collapsed state is an ensemble of states. Thus, the folding nucleus could be an essential factor in the formation of the assembly of monomeric structures.
How does the above discussion relate to the assembled nucleus formation? Ferrone (1999) describes the initial part of the polymerization reaction using a perturbation method showing that the lag may not be just due to an assembly step. We (Frieden and Goddette 1983) showed in an earlier paper that the lag time could be influenced by a conformational transition. Thus, some portion of the lag period may reflect the conversion of the disordered–collapsed state to a structured monomer or the assembly of a rare form of monomers or both.
The structure of the assembly of monomers
The formation of an assembly of intrinsically disordered monomers brings up a number of important mechanistic questions. These include: What species of the disordered–collapsed monomer forms an oligomer? Are all oligomers productive species on the pathway to protofibril or fibril formation? Are small oligomers composed solely of disordered–collapsed monomers? At what oligomeric size will the subsequent monomer addition be characterized as simple elongation? When does the conversion from the disordered–collapse form to a β-strand or β-sheet structure occur (see Fig. 2 as an example for a dimer)? What species binds to the oligomer? The number of possible variations of structures of small oligomers generated from collapsed and structured forms can be quite large.
Figure 2.
A simplified representation of different dimeric forms. As discussed in the text, the formation of a dimer would be a second-order process with the rate depending on the concentration of a monomeric species. The conversion from a disordered–collapsed state to a structured state is a first-order process independent of concentration.
It would seem likely that dimer and higher oligomers form by diffusion of the disordered–collapsed species. Thus, for some disordered–collapsed species Ui
Scheme 2.
where n represents the number of monomers in the oligomer.
At some point, however, one or more of the Ui species must be converted to a structured (presumably β-sheet) form:
where M represents a monomer with stable structure. This conversion could occur stepwise or cooperatively.
It is instructive to consider the rates of oligomer formation versus the rate of this conversion and whether it be stepwise or cooperative. Oligomer formation is a second or multiorder process and dependent on the concentration of Ui. The conversion of Ui to M, however, is a unimolecular process, and therefore independent of monomer concentration. Thus, the nature and perhaps the size of the oligomer depends on the relative rate constants of these two processes. For example, at high concentrations of Ui the formation of oligomer might be faster than the conversion of (Ui)n to M and the intermediate may be composed of monomers in the disordered–collapsed form. On the other hand, at low Ui concentrations, the conversion of disordered–collapsed to structured monomer might be faster than formation of oligomer and the oligomer may consist of structured monomers. By the same argument, at high monomer concentrations the nucleus size may be relatively small because the high monomer concentration tends to drive the elongation process. Thus, it is not possible, in this mechanism, to define the nature of the oligomer without detailed knowledge of the appropriate rate constants. For this reason, the rate of polymerization may not directly relate to the concentration of the disordered protein because the nature of the intermediate is undefined. As indicated above, a related issue is how the U to M conversion occurs within the assembled nucleus. Do all U molecules deep within the nucleus convert to M faster than those close to the surface? Andrews and Roberts (2007) have recently examined the aggregation of α-chymotrypsinogen and suggest that formation of irreversible aggregates and conversion to β-sheet secondary structures occur simultaneously suggesting that the conversion to a β-sheet structure is a necessary step during nucleation and further elongation.
Productive, nonproductive oligomers
The possibility that small oligomers are the toxic species in some diseases suggests that such intermediates may not be productive in forming fibrils. Whether an intermediate oligomer is productive or nonproductive depends on a number of factors that include its structure and its ability to dissociate to monomers. Mechanistically, a nonproductive oligomer decreases the concentration of monomer that eventually forms the fibril. If the nonproductive oligomer cannot dissociate or dissociates very slowly and cannot serve as an nucleus for elongation, the lag time may be extended relative to the productive case. If, on the other hand, the nonproductive oligomer were to form and dissociate rapidly, it would be equivalent to lowering the concentration of monomer that can form the productive oligomer.
Fibril formation
The focus of this review is not on fibril formation, but rather what occurs prior to the aggregation step. Once productive nuclei are formed, however, elongation is assumed to occur with the same rate constants for every monomeric species added. For defined systems, such as actin or tubulin polymerization where the monomer has structure, the process is relatively simple even if the two ends of the polymer differ in on and off rate constants. For systems involving IDPs, the situation is more complex. First, what species of monomer is actually adding to the polymer? Second, are there side-to-side interactions that occur as well as the elongation steps? Third, do oligomers interact with other oligomers and, if so, how should those interactions be described? None of these questions have been adequately answered. Moore et al. (2007) have evidence for fibril formation from large assemblies of monomers. Finally, what is the meaning of the critical concentration? Methods for examining amyloid fibril formation have been reviewed by Nilsson (2004). It should be noted, however, that methods for investigating this part of the process are generally not revealing.
The critical concentration
The critical concentration for a polymerizing system is frequently considered to be the concentration of monomer in equilibrium with polymer and, as mentioned earlier, is equal to the ratio of rate constants,
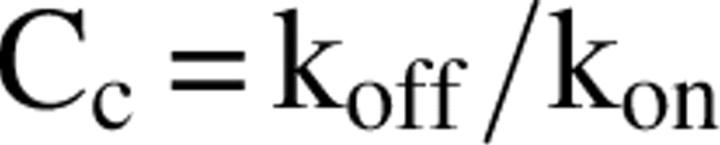 |
k off being the rate constant for dissociation of the monomer from the polymer and k on, the rate constant for monomer addition to the polymer. In an ideal system, no polymerization should occur below the critical concentration. Two implicit assumptions are that all monomeric species are capable of interacting with polymer or that, if there are several different monomeric species, they are in rapid equilibrium relative to the monomer–polymer equilibrium. An additional assumption is that the appropriate sites on the polymer (e.g., the ends) are all equally accessible to the polymer. None of these assumptions may be true for aggregating systems that make fibrils from intrinsically disordered structures. First, the monomer that dissociates from a fibril is probably structured, and may or may not be the structure that binds to the fibril. That is, the fibril may be elongated by the addition of disordered–collapsed monomers, but that may not be the form that dissociates from the polymer. Second, if it is an ensemble of disordered collapsed forms that bind to the polymer it is not clear what the rate constants for interconversion to different disordered–collapsed structures are or how they differ from the rate constants for monomer addition to the polymer. Again, this is an issue of rate constants for a unimolecular interconversion of monomers versus rate constants for a second-order process of monomer addition. Therefore, we must redefine what happens when the concentration of polymer does not change with time, i.e., at equilibrium.
Under these conditions, the monomer may consist of several forms
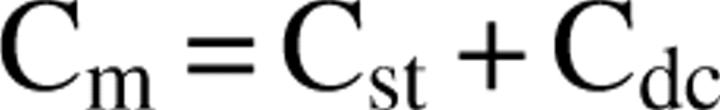 |
where C st and C dc are concentrations of structured and disordered collapsed monomer, respectively. The meaning of the rate constant k on then becomes more complex reflecting the addition of different species. Certainly, the critical concentration no longer represents a simple equilibrium system. One can imagine a number of different scenarios. All this means is that the determination of a critical concentration, if indeed there is one, may not be a simple matter. Finally, there is a technical problem in that fibrils become insoluble that can form large intertwined aggregates. Accessibility to sites to which monomers bind may be restricted. These problems may give rise to large variations in reported critical concentrations. Some of these caveats have been discussed by Williams et al. (2006) when discussing alanine or proline scanning experiments (Williams et al. 2004).
Experimental approaches
In the following discussion, I ignore issues with respect to purity of starting material. Clearly, these are important and every effort needs to be made to ensure that the peptide/protein is as pure as possible, but that is not the focus of the approaches discussed here. Now that we have laid out many, but probably not all, of the issues we need to find ways to disentangle the polymerization process. I start with methods that may be useful for examining what occurs during the long lag phase of the aggregation process.
Using low protein concentrations: Fluorescence
Fluorescence methods may have the ability to detect the formation of the collapsed monomer, the formation of a structured monomer, and the formation of dimer and higher oligomeric species during the polymerization. Monomer incorporation into oligomers might best be followed by changes in the properties of a covalently attached fluorophore or other spectroscopic reporter. There are, or course, some caveats that need to be considered. Most importantly, the attached moiety should not affect the polymerization. One can test this by changing the ratio of labeled to unlabeled protein. Steady-state fluorescence measurements are most convenient, but other fluorescence methods could be considered. For example, Förster Resonance Energy Transfer (FRET) measurements, where monomers are labeled with an appropriate pair of FRET dyes could be used to measure monomer–monomer or monomer–oligomer interactions. A clever experiment would be to use a fluorescent dye that monitors the disordered to structured monomeric form. A tryptophan in the monomer might report on such a transition as well as incorporation of monomer into oligomer. The most useful method may be fluorescence correlation spectroscopy (FCS) using either fluorescence quenching or FRET.
The experimental and theoretical basis for FCS was developed in the early 1970s (Elson and Magde 1974; Magde et al. 1974). Initially, the technique was used mostly to measure diffusion coefficients of fluorescently labeled molecules. As pointed out early on, however, one can also use FCS to make kinetic measurements under certain conditions. For freely diffusing molecule, measurements of dynamic motions are accessible if (1) the dynamics are faster than the diffusion time of the molecule and (2) if they are associated with some change in fluorescence. There are at least two experimental approaches that may be complementary. In the first, one can use a FRET pair. Using a FRET measurement only, one might be able to measure the distance between monomers within an assembled nucleus after mixing monomers with different dyes that would give a FRET signal. FRET can also be used to measure dynamic motions within the monomer. Nettels et al. (2007) have used FRET to determine dynamics within unfolded Cold Shock Protein (62 residues) after attaching dyes to the C and N termini. They determined a reconfigure time of ∼50 ns. Similarly, Mukhopadhyay et al. (2007) observed fast (20–300 ns) conformational fluctuations of a region of the yeast prion protein. These rates might be considered as the dynamics of a small motion within a disordered–collapsed protein.
FCS measurements can also use dye quenching. In this case, the dye moieties have to be close enough to quench, and it would be expected that the time might be considerably longer. Chattopadhyay et al. (2005) used rhodamine-labeled intestinal fatty acid binding protein with labeled residues 48 amino acids apart. When the protein was denatured by guanidine, they observed that the fluorescence decreased and interpreted that as rhodamine self-quenching. With that observation they could measure the apparent rate of loop closure in the unfolded protein. Their results gave dynamics of 1.6 μs. As expected, no such dynamics were observed in the native folded protein. FCS can also be used, as mentioned above, to measure diffusion times. Crick et al. (2006) measured the diffusion times of different chain lengths of polyglutamine and showed that from 20–55 residues, the diffusion time changed as expected for a collapsed state rather than a random coil. Urea unfolds this collapsed state similar to results with Aβ (S.L. Crick, R. Pappu, and C. Frieden, unpubl.).
One of the great advantages of FCS is that the concentrations used in these experiments are quite low—on the order of a nanomolar. In principle, this should slow down or stop any assembly or polymerization process. Thus, one could measure diffusional or possibly conformational changes within the monomer. With the addition of unlabeled protein, it should be possible to measure the change in diffusion with time. The disadvantage is that the diffusion coefficient, for a sphere, for example, changes with the cube root of the molecular weight. Thus, it may be difficult to deconvolute the concentrations of lower molecular weight oligomers. Photon counting histograms, that measure the brightness of the molecule and are directly proportional to the number of monomers in an oligomer, may circumvent this problem, but such an analysis for molecules in solution (i.e., in three dimensions) is difficult.
FCS cross-correlation
With different labels on different monomers, and using dual-color cross-correlation, it might be possible to obtain information about dimers in equilibrium with monomer.
Dual-color cross-correlation is an extremely powerful tool to probe interactions between different molecular species, and a number of experiments have been carried out applying this technique to different kinds of reactions (Schwille et al. 1997; Dittrich et al. 2001).
Using high protein concentrations
Fluorine NMR
In recent years, we have incorporated fluorine-labeled amino acids into proteins and then doing either equilibrium or stopped flow NMR (e.g., (Hoeltzli and Frieden 1996; Bann and Frieden 2004; Shu and Frieden 2004; Li and Frieden 2005). Examining the NMR spectrum of a protein that contains a fluorine-labeled amino acid has not been used for the study of the polymerization of natively disordered proteins. Yet the method has considerable promise when used in conjunction with a fluorine cryoprobe allowing relatively low protein concentrations to be used. As discussed elsewhere (Frieden et al. 2004), the incorporation of fluorine-labeled amino acid usually has minimal effects on protein structure yet serves as an excellent reporter group because the fluorine chemical shift, although poorly understood, is very sensitive to its environment. It is possible that the fluorine label will appear solvent exposed in the disordered–collapsed state as observed by Winkler et al. (2006) for α-synuclein but, if properly placed, chemical shifts could report on the disordered–collapsed state or, more likely, on the formation of lower molecular weight oligomers. Currently, the most common substitutions include fluorotryptophan, fluorophenylalanine, and fluorotyrosine. Of these, the most useful would be fluorophenylalanine, because it is possible to incorporate that amino acid site specifically (Furter 1998; Frieden et al. 2004). Stopped-flow NMR, which allows real-time measurements of the fluorine chemical shift, may be particularly useful for measuring time-dependent processes occurring, for example, in the lag time of the polymerization. NMR experiments, of course, require much higher concentrations of material than do the fluorescence experiments discussed above, thus raising the issue of solubility. Furthermore, the time regimes are quite different.
Light scattering
Light-scattering techniques, either static or dynamic, have the advantage that the protein does not have to be labeled. The disadvantage is that at the current time the concentrations required are much greater than those needed for FCS or other fluorescent methods. Static scattering methods can yield bimodal molecular weight distributions, but the scattering profile is biased toward high molecular weight species. Furthermore, the scattering envelope will change if the particle size approaches or exceeds the wavelength used for scattering. Dynamic scattering can be used to measure diffusion times like FCS, but again, is limited to high protein concentrations and is particularly good at sensing the presence of very small amounts of aggregated protein (<0.01% by weight). If more sensitive scattering techniques become available, dynamic light scattering could provide results similar to that obtained by FCS with respect to diffusion times. The difference between conformational forms, however, depends on the difference in refractive index gradients, and these may be too small to measure compared to fluorescence changes that can be detected by FCS.
Methods currently used for fibril formation
Fibril formation, as mentioned above, is frequently measured based on changes in ThioflavinT or Congo Red fluorescence, but it is not clear at what point in the polymerization process these dyes add to the polymer. Thus, this assay is qualitative at best, and gives no information as to the mechanism of the process. Indeed, Necula et al. (2007) point out that many compounds interfere with ThioflavinT fluorescence or absorbance.
The best method for kinetic analysis would be one that measures both the disordered–collapsed to structured monomer transition and the incorporation of the monomer into higher molecular weight species. This might be done by specific fluorescence labeling of the monomer with the caveat that labeling does not affect the polymerization. The early steps of a polymerization process (except when starting with a disordered denatured monomer) are the incorporation of the monomer into dimer. This step could be quite fast, and being able to measure it would give useful information concerning the initial step(s) in the polymerization mechanism.
The most extensively studied system is probably that using Aβ. Solid-state NMR experiments have been conducted on amyloid fibrils primarily by Tycko and coworkers (Balbach et al. 2002; Petkova et al. 2004; Sharpe et al. 2004). Teplow et al. (2006) have surveyed several methods for elucidating folding and assembly of Aβ including solution NMR, mass spectrometry, and cross-linking studies in conjunction with molecular dynamics simulations to obtain information on the monomer as well as oligomer size distribution. The multidisciplinary approach emphasized the role of turns in nucleating monomer folding and helped to identify specific oligomeric types. Cannon et al. (2004) attached fibrils to a chip surface and used surface plasmon resonance to measure kinetics. They proposed a three-step model in which peptide bound to the growing fibril is stabilized after binding.
These studies provide valuable information on the structure of the fibrils but only partial information on the mechanism of fibril formation, emphasizing the importance of hydrophobic interactions. Solution NMR studies have provided some information on the nature of the monomeric state, but again, little information of the mechanism of polymerization (Panchal et al. 2001; Whittemore et al. 2005), although some structural motifs do appear (i.e., Lazo et al. 2005). These solution NMR studies suggest that Aβ, for example, may exist as a collapsed state (Zhang et al. 2000; Hou et al. 2004). Chen and Glabe (2006) have shown that the far UV CD of Aβ1–40 changes with increasing urea concentration consistent with the idea that the monomer is a collapsed state. According to these investigators, Aβ1–40 exists as an unstable monomer with a large random coil population, mostly collapsed, while Aβ1–42 tends to form small oligomers.
In studies with α-synuclein Fink and coworkers and others (Kaylor et al. 2005; Dusa et al. 2006) have used a variety of techniques to characterize oligomers of prior to fibril formation. These included FTIR, FRET (between tyrosine and tryptophan), tryptophan fluorescence, dynamic light scattering, ANS binding, and fluorescence anisotropy. The results clearly show the existence oligomers prior to formation of fibrils and the possibility of multiple forms of those oligomeric states. The studies, using FRET between tyrosine and tryptophan in conjunction with more standard techniques, revealed two classes of oligomeric intermediates, one formed during the lag period and the other after fibril formation. The appearance of multiple species clearly complicates mechanistic studies. In contrast to Aβ, NMR studies of α-synuclein also indicate transient helical structure in the monomer (Eliezer et al. 2001). Hoyer et al. (2004) have used atomic force microscopy in a tapping mode to follow the self-assembly of α-synuclein, and have shown different aggregation modes depending on the experimental conditions, including pH and the presence of polyamines.
Myers et al. (2006) have used limited proteolysis with pepsin combined with mass spectrometry to determine the conformational properties of β2-microglobulin fibrils formed under different conditions. They showed different cleavage sites for fibrils of different morphologies that were formed under different conditions. The experiments show the importance of controlling the conditions used for fibril formation in order to obtain reproducible results.
The rational use of inhibitors
Inhibition of polymerization may occur at different places along the pathway: the disordered–collapsed form to structured monomer, the formation of assembled nuclei, or the elongation. Each step may involve using different approaches. Perhaps the best way to approach the formation of structured monomer would be site-directed mutagenesis. It is well known, for example, that the folding rate of proteins from denaturant can be affected by altering specific residues. For proteins that are mostly composed of β-strands, mutations in the predicted turns appear quite effective (Kim and Frieden 1998; Frieden et al. 2001). To inhibit dimer formation, an inhibitor that blocks addition to both the top and bottom of the putative structured monomer would be effective This might require the addition of two inhibitors with different specificities. One such inhibitor might be effective in blocking elongation. Rzepecki et al. (2004), for example, have found aminopyrazole derivatives to be effective in blocking the formation of high molecular weight aggregates of Aβ. There is, however, a large literature on inhibitors of Aβ fibril formation (not reviewed here). Necula et al. (2007), using an antibody specific for Aβ oligomers, have postulated three classes of small molecule inhibitors: those that inhibit oligomerization but not fibrillization, those that inhibit fibrillization but not oligomerization, and those that inhibit both. The approach provides useful information on the mechanism of the polymerization process.
Site-directed mutagenesis
Site-directed mutagenesis, when coupled with fluorescent studies, should be a powerful technique to understand the mechanism of aggregation. Carefully chosen mutations could interfere with the folding nucleus formation, with monomer–monomer interactions, with the formation of a structured form and with formation of an assembled nucleus.
Turns
Of particular interest are mutations in putative turns between β-strands which is especially relevant to structure in the fibril. Frieden et al. (2001) have coined the term “turn scanning,” and outlined the importance of turn formation in the folding process. While there are no general rules of mutations in turns, those of most interest might contain glycine. For example, the intestinal fatty acid binding protein consists primarily of two β-sheets with each sheet containing five antiparallel β-strands. Folding studies with several proteins (Hoeltzli and Frieden 1996; Bann and Frieden 2004; Li and Frieden 2007) have shown that the final step in folding is the stabilization of side chains, and that the stabilization is a highly cooperative process. For the intestinal fatty acid binding protein many of the turns between strands contain glycine, and Kim and Frieden (1998) have mutated each of these glycine residues to valine showing that replacing glycine with valine (or alanine) results not only in destabilization of the protein but much slower folding as well. In particular, a glycine to valine mutation between the last two strands slows the folding dramatically (Kim and Frieden 1998; Li and Frieden 2007). Folding is not complete until all turns are formed. While these mutations are relatively drastic, less draconian changes can be made. For example, substitutions between hydrophobic and hydrophilic residues may change solvent accessibility of the turn. Residues directly adjacent to the turn may also influence the turn type. These results suggest that perhaps the reason that the polymerization of some peptides is so slow is because some turn or turns do not form easily. Therefore, a test of this prediction would be to mutate one or more glycines that occur in turns to see the effect on polymer formation.
Strands
Mutations in putative strands should also provide information on the ability of strands to form or their structure in oligomers or fibrils. For Aβ, Wetzel et al. (2007) have replaced almost every residue with cysteine, alanine, or proline.
Using cysteine mutants, for example, Shivaprasad and Wetzel (2006) examined the structure of the fibril by assessing the ability of the cysteine in the fibril to react with an alkylating reagent.
Conclusions
For peptides/proteins that fall into the category of intrinsically disordered, the polymerization process is complex. Indeed, there may not be a single mechanism that is appropriate to describe the process for different systems. There have been many attempts to understand these mechanisms, and I have discussed some of the simple possible mechanisms. Yet, important questions remain to be addressed, and include the nature of the ensemble of states in aqueous solvent, the dynamic motions within the monomers, the rate of formation of oligomers occurring prior to the polymerization step, the nature of oligomers and whether such oligomers are on or off the polymerization pathway, the rate of formation of structured monomers either before or after oligomer formation, and the appropriate measurement of the polymer. I have suggested some experimental procedures to examine these issues.
Acknowledgments
I thank Dr. Rohit Pappu and Scott Crick for reading and commenting on the manscript. This work was supported by NIH Grant DK 13332.
Footnotes
Editor's Note: Dr. Frieden is the 2007 recipient of the Protein Society Christian B. Anfinsen Award for significant technical achievements in the field of protein science. This review develops themes from his Award Lecture.
Reprint requests to: Carl Frieden, Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, USA; e-mail: frieden@biochem.wustl.edu; fax: (314) 362-7183.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073164107.
References
- Andrews J.M. and Roberts, C.J. 2007. Non-native aggregation of α-chymotrypsinogen occurs through nucleation and growth with competing nucleus sizes and negative activation energies. Biochemistry 46: 7558–7571. doi: 10.1021/bi700296f. [DOI] [PubMed] [Google Scholar]
- Balbach J.J., Petkova, A.T., Oyler, N.A., Antzutkin, O.N., Gordon, D.J., Meredith, S.C., and Tycko, R. 2002. Supramolecular structure in full-length Alzheimer's β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 83: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bann J.G. and Frieden, C. 2004. Folding and domain–domain interactions of the chaperone PapD measured by 19F NMR. Biochemistry 43: 13775–13786. [DOI] [PubMed] [Google Scholar]
- Barshop B.A., Wrenn, R.F., and Frieden, C. 1983. Analysis of numerical methods for computer simulation of kinetic processes: Development of KINSIM—A flexible, portable system. Anal. Biochem. 130: 134–145. [DOI] [PubMed] [Google Scholar]
- Cannon M.J., Williams, A.D., Wetzel, R., and Myszka, D.G. 2004. Kinetic analysis of β-amyloid fibril elongation. Anal. Biochem. 328: 67–75. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay K., Elson, E.L., and Frieden, C. 2005. The kinetics of conformational fluctuations in an unfolded protein measured by fluorescence methods. Proc. Natl. Acad. Sci. 102: 2385–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Berthelier, V., Hamilton, J.B., O'Nuallain, B., and Wetzel, R. 2002. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 41: 7391–7399. [DOI] [PubMed] [Google Scholar]
- Chen Y.R. and Glabe, C.G. 2006. Distinct early folding and aggregation properties of Alzheimer amyloid-β peptides Aβ40 and Aβ42: Stable trimer or tetramer formation by Aβ42. J. Biol. Chem. 281: 24414–24422. [DOI] [PubMed] [Google Scholar]
- Crick S.L., Jayaraman, M., Frieden, C., Wetzel, R., and Pappu, R.V. 2006. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc. Natl. Acad. Sci. 103: 16764–16769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V. and Fersht, A.R. 2003. Is there a unifying mechanism for protein folding? Trends Biochem. Sci. 28: 18–25. [DOI] [PubMed] [Google Scholar]
- Dittrich P., Malvezzi-Campeggi, F., Jahnz, M., and Schwille, P. 2001. Accessing molecular dynamics in cells by fluorescence correlation spectroscopy. Biol. Chem. 382: 491–494. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. 2006. Protein aggregation and its consequences for human disease. Protein Pept. Lett. 13: 219–227. [DOI] [PubMed] [Google Scholar]
- Dusa A., Kaylor, J., Edridge, S., Bodner, N., Hong, D.P., and Fink, A.L. 2006. Characterization of oligomers during α-synuclein aggregation using intrinsic tryptophan fluorescence. Biochemistry 45: 2752–2760. [DOI] [PubMed] [Google Scholar]
- Eliezer D., Kutluay, E., Bussell Jr, R., and Browne, G. 2001. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307: 1061–1073. [DOI] [PubMed] [Google Scholar]
- Elson E.L. and Magde, D. 1974. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers 13: 1–27. [DOI] [PubMed] [Google Scholar]
- Ferrone F. 1999. Analysis of protein aggregation kinetics. Methods Enzymol. 309: 256–274. [DOI] [PubMed] [Google Scholar]
- Fink A.L. 2006. The aggregation and fibrillation of α-synuclein. Acc. Chem. Res. 39: 628–634. [DOI] [PubMed] [Google Scholar]
- Flory P.J. 1953. Principles of polymer chemistry. Cornell University Press, Ithaca, NY.
- Frieden C. 1983. Polymerization of actin: Mechanism of the Mg2+-induced process at pH 8 and 20°C. Proc. Natl. Acad. Sci. 80: 6513–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. 1985. Actin and tubulin polymerization: The use of kinetic methods to determine mechanism. Annu. Rev. Biophys. Biophys. Chem. 14: 189–210 [Review]. [DOI] [PubMed] [Google Scholar]
- Frieden C. and Goddette, D.W. 1983. Polymerization of actin and actin-like systems: Evaluation of the time course of polymerization in relation to the mechanism. Biochemistry 22: 5836–5843. [DOI] [PubMed] [Google Scholar]
- Frieden C., Huang, E.S., and Ponder, J.W. 2001. Experimental and theoretical approaches to the role of turns. In Protein structure, stability and folding (ed. K.P. Murphy), pp. 133–158. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Frieden C., Hoeltzli, S.D., and Bann, J.G. 2004. The preparation of 19F-labeled proteins for NMR studies. Methods Enzymol. 380: 400–415. [DOI] [PubMed] [Google Scholar]
- Furter R. 1998. Expansion of the genetic code: Site-directed p-fluoro-phenylalanine incorporation in Escherichia coli . Protein Sci. 7: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeltzli S.D. and Frieden, C. 1996. Real-time refolding studies of 6-19F-tryptophan labeled Escherichia coli dihydrofolate reductase using stopped-flow NMR spectroscopy. Biochemistry 35: 16843–16851. [DOI] [PubMed] [Google Scholar]
- Hou L., Shao, H., Zhang, Y., Li, H., Menon, N.K., Neuhaus, E.B., Brewer, J.M., Byeon, I.J., Ray, D.G., Vitek, M.P., et al. 2004. Solution NMR studies of the A β(1–40) and A β(1–42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 126: 1992–2005. [DOI] [PubMed] [Google Scholar]
- Hoyer W., Cherny, D., Subramaniam, V., and Jovin, T.M. 2004. Rapid self-assembly of α-synuclein observed by in situ atomic force microscopy. J. Mol. Biol. 340: 127–139. [DOI] [PubMed] [Google Scholar]
- Kaylor J., Bodner, N., Edridge, S., Yamin, G., Hong, D.P., and Fink, A.L. 2005. Characterization of oligomeric intermediates in α-synuclein fibrillation: FRET studies of Y125W/Y133F/Y136F α-synuclein. J. Mol. Biol. 353: 357–372. [DOI] [PubMed] [Google Scholar]
- Kheterpal I. and Wetzel, R. 2006a. Amyloid, prions and other protein aggregates, Part B. Academic Press, New York.
- Kheterpal I. and Wetzel, R. 2006b. Amyloid, prions, and other protein aggregates, Part C. p. 375. Academic Press, New York.
- Kim K. and Frieden, C. 1998. Turn scanning by site-directed mutagenesis: Application to the protein folding problem using the intestinal fatty acid binding protein. Protein Sci. 7: 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo N.D., Grant, M.A., Condron, M.C., Rigby, A.C., and Teplow, D.B. 2005. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 14: 1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C., Lai, B.T., Kozak, J.J., Gray, H.B., and Winkler, J.R. 2007. α-synuclein tertiary contact dynamics. J. Phys. Chem. B 111: 2107–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Frieden, C. 2005. NMR studies of 4-19F-phenylalanine-labeled intestinal fatty acid binding protein: Evidence for conformational heterogeneity in the native state. Biochemistry 44: 2369–2377. [DOI] [PubMed] [Google Scholar]
- Li H. and Frieden, C. 2007. Observation of sequential steps in the folding of intestinal fatty acid binding protein using a slow folding mutant and 19F NMR. Proc. Natl. Acad. Sci. 104: 11993–11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrs T., Ritter, C., Adrian, M., Riek-Loher, D., Bohrmann, B., Dobeli, H., Schubert, D., and Riek, R. 2005. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. 102: 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D., Elson, E.L., and Webb, W.W. 1974. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 13: 29–61. [DOI] [PubMed] [Google Scholar]
- Merchant K.A., Best, R.B., Louis, J.M., Gopich, I.V., and Eaton, W.A. 2007. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. 104: 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.A., Hayes, S.F., Fischer, E.R., and Priola, S.A. 2007. Amyloid formation via supramolecular peptide assemblies. Biochemistry 46: 7079–7087. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Krishnan, R., Lemke, E.A., Lindquist, S., and Deniz, A.A. 2007. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. 104: 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S.L., Thomson, N.H., Radford, S.E., and Ashcroft, A.E. 2006. Investigating the structural properties of amyloid-like fibrils formed in vitro from β2-microglobulin using limited proteolysis and electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 20: 1628–1636. [DOI] [PubMed] [Google Scholar]
- Necula M., Kayed, R., Milton, S., and Glabe, C.G. 2007. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 282: 10311–10324. [DOI] [PubMed] [Google Scholar]
- Nettels D., Gopich, I.V., Hoffmann, A., and Schuler, B. 2007. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Natl. Acad. Sci. 104: 2655–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M.R. 2004. Techniques to study amyloid fibril formation in vitro. Methods 34: 151–160. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Cheng, Y., Cortese, M.S., Brown, C.J., Uversky, V.N., and Dunker, A.K. 2005. Comparing and combining predictors of mostly disordered proteins. Biochemistry 44: 1989–2000. [DOI] [PubMed] [Google Scholar]
- Oosawa F. and Asakura, S. 1975. Thermodynamics of the polymerization of protein, p. 204. Academic Press, London.
- Panchal S.C., Bhavesh, N.S., and Hosur, R.V. 2001. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: Application to unfolded proteins. J. Biomol. NMR 20: 135–147. [DOI] [PubMed] [Google Scholar]
- Petkova A.T., Buntkowsky, G., Dyda, F., Leapman, R.D., Yau, W.M., and Tycko, R. 2004. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J. Mol. Biol. 335: 247–260. [DOI] [PubMed] [Google Scholar]
- Powers E.T. and Powers, D.L. 2006. The kinetics of nucleated polymerizations at high concentrations: Amyloid fibril formation near and above the “supercritical concentration.” Biophys. J. 91: 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P., Iakoucheva, L.M., Oldfield, C.J., Obradovic, Z., Uversky, V.N., and Dunker, A.K. 2007. Intrinsic disorder and functional proteomics. Biophys. J. 92: 1439–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic, Z., and Dunker, A.K. 2004. Natively disordered proteins: Functions and predictions. Appl. Bioinformatics 3: 105–113. [DOI] [PubMed] [Google Scholar]
- Rzepecki P., Nagel-Steger, L., Feuerstein, S., Linne, U., Molt, O., Zadmard, R., Aschermann, K., Wehner, M., Schrader, T., and Riesner, D. 2004. Prevention of Alzheimer's disease-associated Aβ aggregation by rationally designed nonpeptidic β-sheet ligands. J. Biol. Chem. 279: 47497–47505. [DOI] [PubMed] [Google Scholar]
- Schwille P., Meyer-Almes, F.J., and Rigler, R. 1997. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 72: 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe S., Kessler, N., Anglister, J.A., Yau, W.M., and Tycko, R. 2004. Solid-state NMR yields structural constraints on the V3 loop from HIV-1 Gp120 bound to the 447-52D antibody Fv fragment. J. Am. Chem. Soc. 126: 4979–4990. [DOI] [PubMed] [Google Scholar]
- Sherman E. and Haran, G. 2006. Coil-globule transition in the denatured state of a small protein. Proc. Natl. Acad. Sci. 103: 11539–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad S. and Wetzel, R. 2006. Scanning cysteine mutagenesis analysis of Aβ-(1–40) amyloid fibrils. J. Biol. Chem. 281: 993–1000. [DOI] [PubMed] [Google Scholar]
- Shu Q. and Frieden, C. 2004. Urea-dependent unfolding of murine adenosine deaminase: Sequential destabilization as measured by 19F NMR. Biochemistry 43: 1432–1439. [DOI] [PubMed] [Google Scholar]
- Shults C.W. 2006. Lewy bodies. Proc. Natl. Acad. Sci. 103: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmeier M., Hamilton, J.A., LeGall, T., Vacic, V., Cortese, M.S., Tantos, A., Szabo, B., Tompa, P., Chen, J., Uversky, V.N., et al. 2007. DisProt: The database of disordered proteins. Nucleic Acids Res. 35: D786–D793. doi: 10.1093/nar/gk1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim K.L., Uchida, T., and Miyano, S. 2001. ProDDO: A database of disordered proteins from the Protein Data Bank (PDB). Bioinformatics 17: 379–380. [DOI] [PubMed] [Google Scholar]
- Teplow D.B., Lazo, N.D., Bitan, G., Bernstein, S., Wyttenbach, T., Bowers, M.T., Baumketner, A., Shea, J.E., Urbanc, B., Cruz, L., et al. 2006. Elucidating amyloid β-protein folding and assembly: A multidisciplinary approach. Acc. Chem. Res. 39: 635–645. [DOI] [PubMed] [Google Scholar]
- Tran H.T. and Pappu, R.V. 2006. Toward an accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys. J. 91: 1868–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R. 2004. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 14: 96–103. [DOI] [PubMed] [Google Scholar]
- Urbanc B., Cruz, L., Yun, S., Buldyrev, S.V., Bitan, G., Teplow, D.B., and Stanley, H.E. 2004. In silico study of amyloid β-protein folding and oligomerization. Proc. Natl. Acad. Sci. 101: 17345–17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic S., Obradovic, Z., Vacic, V., Radivojac, P., Peng, K., Iakoucheva, L.M., Cortese, M.S., Lawson, J.D., Brown, C.J., Sikes, J.G., et al. 2005. DisProt: A database of protein disorder. Bioinformatics 21: 137–140. [DOI] [PubMed] [Google Scholar]
- Weathers E.A., Paulaitis, M.E., Woolf, T.B., and Hoh, J.H. 2004. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 576: 348–352. [DOI] [PubMed] [Google Scholar]
- Wegner A. and Engel, J. 1975. Kinetics of the cooperative association of actin to actin filaments. Biophys. Chem. 3: 215–225. [DOI] [PubMed] [Google Scholar]
- Wetzel R. 2006a. Kinetics and thermodynamics of amyloid fibril assembly. Acc. Chem. Res. 39: 671–679. [DOI] [PubMed] [Google Scholar]
- Wetzel R. 2006b. On amyloid assemble and structure. Acc. Chem. Res. 39: 567–679. [DOI] [PubMed] [Google Scholar]
- Wetzel R., Shivaprasad, S., and Williams, A.D. 2007. Plasticity of amyloid fibrils. Biochemistry 46: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore N.A., Mishra, R., Kheterpal, I., Williams, A.D., Wetzel, R., and Serpersu, E.H. 2005. Hydrogen-deuterium (H/D) exchange mapping of Aβ1-40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry 44: 4434–4441. [DOI] [PubMed] [Google Scholar]
- Williams A.D., Portelius, E., Kheterpal, I., Guo, J.T., Cook, K.D., Xu, Y., and Wetzel, R. 2004. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 335: 833–842. [DOI] [PubMed] [Google Scholar]
- Williams A.D., Shivaprasad, S., and Wetzel, R. 2006. Alanine scanning mutagenesis of Aβ(1–40) amyloid fibril stability. J. Mol. Biol. 357: 1283–1294. [DOI] [PubMed] [Google Scholar]
- Winkler G.R., Harkins, S.B., Lee, J.C., and Gray, H.B. 2006. α-Synuclein structures probed by 5-fluorotryptophan fluorescence and 19F NMR spectroscopy. J. Phys. Chem. B 110: 7058–7061. [DOI] [PubMed] [Google Scholar]
- Zhang S., Iwata, K., Lachenmann, M.J., Peng, J.W., Li, S., Stimson, E.R., Lu, Y., Felix, A.M., Maggio, J.E., and Lee, J.P. 2000. The Alzheimer's peptide Aβ adopts a collapsed coil structure in water. J. Struct. Biol. 130: 130–141. [DOI] [PubMed] [Google Scholar]