Abstract
The arsH gene or its homologs are a frequent part of the arsenic resistance system in bacteria and eukaryotes. Although a specific biological function of the gene product is unknown, the ArsH protein was annotated as a member of the NADPH-dependent FMN reductase family based on a conserved (T/S)XRXXSX(T/S) fingerprint motif common for FMN binding proteins. Presented here are the first crystal structure of an ArsH protein from Shigella flexneri refined at 1.7 Å resolution and results of enzymatic activity assays that revealed a strong NADPH-dependent FMN reductase and low azoreductase activities. The ArsH apo protein has an α/β/α−fold typical for FMN binding proteins. The asymmetric unit consists of four monomers, which form a tetramer. Buried surface analysis suggests that this tetramer is likely to be the relevant biological assembly. Dynamic light scattering experiments are consistent with this hypothesis and show that ArsH in solution at room temperature does exist predominantly in the tetrameric form.
Keywords: crystal structure, flavin binding, arsenic resistance, ArsH, NADPH-dependant FMN reductase
Arsenic is a carcinogenic and toxic metalloid, and its trivalent (arsenite) and pentavalent (arsenate) oxyanions can inhibit a variety of biological pathways (Shi et al. 2004). The toxicity of arsenicals is determined by the ability of arsenate (As(V)O4 3−) to mimic phosphate ion, by the reaction of As(III) oxides and anions with protein thiol groups, and by the cross-linking of DNA with another DNA or protein (Norman 1998). An arsenic and antimony (III) resistance mechanism was revealed in a wide range of microorganisms and was found to be encoded in specific ars operons located in either plasmids or chromosomes. Arsenic/antimony (III) detoxication genes and pathways were studied in a variety of species, including a eukaryote (Bobrowicz et al. 1997) and a number of prokaryotes (Ji et al. 1994; Carlin et al. 1995; Diorio et al. 1995; Neyt et al. 1997; Butcher et al. 2000; Saltikov and Olson 2002).
All known arsenical resistance operons include at least three components:
An As(III)/Sb(III)-responsive repressor protein ArsR, which regulates the basal level of the arsenic resistance proteins expression. In some organisms, an additional ArsD protein that regulates the upper level of expression is also present (Chen and Rosen 1997).
An arsenic (III), and usually antimony (III), extrusion system. A few known families of arsenic transporters include (1) bacterial ArsB integral membrane protein functioning as a secondary arsenite carrier and (2) ArsB coupled with the ArsA ATPase and functioning as an ATP-driven pump, (3) eukaryotic Arr3p (which homolog in prokaryotes is called YqclL) membrane carrier protein and (4) Ycf1p transport ATPase (a member of multidrug resistance-associated protein, MRP, family) that sequesters As-glutathione complexes into a vacuole, and, finally, (5) AqpS aquaglyceroporin that generates concentration gradient flows of arsenite out of the cell (Rosen 1999; Yang et al. 2005).
An arsenate reductase protein that establishes resistance to As(V) through its reduction to As(III) as a transport substrate. Arsenate reductases also belong to several unrelated families, including (1) ArsCec (named after Escherichia coli R773 plasmid product) Glutaredoxin-glutathione (Grx/GSH) linked arsenate reductase, (2) ArsCsa (named after Staphylococcus aureus plasmid pI258) Thioredoxin (Trx) linked arsenate reductase, and (3) the Saccharomyces cerevisiae Arr2p arsenate reductase, related to the tyrosine phosphatase family (Mukhopadhyay et al. 2002).
The observed diversity of arsenic/antimony resistance operon expression products has been explained by their convergent evolution from proteins of different origins (Rosen 2002; Jackson and Dugas 2003; Messens and Silver 2006). This makes a boundary between “typical” for bacteria ArsRBC and eukaryote Arr123 constructs very blurred. There are frequently present other products of the arsenic resistance operons in addition to those mentioned above.
Two such additional coexpressed proteins are ArsH, found in Yersinia enterocolitica (Neyt et al. 1997), Thiobacillus ferrooxidans (Butcher et al. 2000), Pseudomonas putida (Cánovas et al. 2003) and the cyanobacterium Synechocystis (López-Maury et al. 2003; Yang et al. 2005), and ArsO protein from Streptomyces (Wang et al. 2006). The function of ArsH in the context of the arsenic resistance pathway is annotated as unknown, and its importance for arsenic detoxication is ambiguous. It was shown to be important for As(III,V) resistance in Y. enterocolitica and As(III,V)/Sb(III) resistance in Sinorhizobium meliloti. At the same time, in cyanobacterium Synechocystis and proteobacteria T. ferrooxidans ArsH showed no arsenic resistance phenotype under the conditions tested; the phenotype in P. putida was not investigated.
Bioinformatic classification of the ArsH protein based on sequence homology has shed some light on this issue. Generally, the ArsH protein belongs to the family of NADPH-dependent FMN reductases (ArsO protein shows similarity to FAD binding reductases), which have representatives in many species, including all cited in Supplemental Table S1 (see Supplemental materials). Members of the FMN reductase family are not always a part of arsenic resistance operons. Microorganisms usually have a few flavin reductases, for example, the well-known flavoenzymes Fre and Wrba in E. coli (Fieschi et al. 1995; Grandori et al. 1998), which may provide a similar function. Because of this functional duplication by other proteins, an ars operon with deleted arsH gene may not affect arsenic resistance. At the same time, there were no direct biochemical studies or structural evidence for the ArsH protein to confirm its assignment as an NADPH-dependent FMN reductase.
The arsenic resistance operon in the genome of Shigella flexneri consists of four genes arsR, arsH, arsC, and arsB (Wei et al. 2003). The ArsR protein shows more than 48% sequence identity to transcriptional regulators from S. meliloti and T. ferrooxidans; the ArsC and ArsB proteins have 35% and 23% identity to ArsCsa from S. aureus and Arr3p from S. cerevisiae, respectively. The ArsH protein from S. flexneri has more than 60% sequence identity to ArsH proteins in other proteobacteria and cyanobacteria (Supplemental Table S1). Based on sequence homology, the protein under study here could be considered a typical representative of bacterial ArsH proteins.
In this study, we present the structure of the S. flexneri (2a strain 2457T) ArsH protein, which proves its relation to FMN binding proteins. Preliminary biochemical data support its activity as an NADPH-dependent FMN reductase.
Results and Discussion
Identification of FMN as a prosthetic group
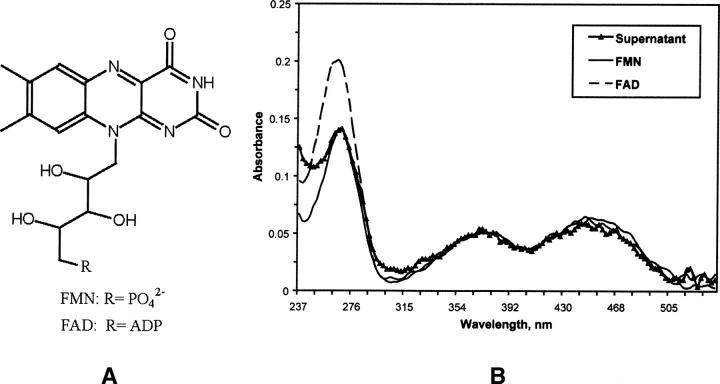
The purified ArsH in solution has a yellow color. In addition to the usual “protein” peak at 280 nm, its UV-Vis absorption spectrum had two peaks at 373 and 455 nm, which are typical for flavin molecules, with FMN and FAD as the most commonly encountered representatives (Fig. 1A). Although solutions of pure oxidized FMN and FAD show similar UV/Vis spectra in the area greater than 320 nm, the FAD in solution shows additional absorption at 267 nm in comparison with the FMN (Massey 2000). The ratio of absorption peaks at 267 and 373 nm, 3.93 for FAD, and 2.75 for FMN, allows one to distinguish between these prosthetic groups. After the protein was denatured by boiling, the yellow-colored compound was released, indicating a noncovalent nature of the protein–prosthetic group interaction (Lostao et al. 2003). The ratio of absorbance of the supernatant solution at 267 and 373 nm was equal to 2.7, clearly identifying the prosthetic group as FMN (Fig. 1B). The concentration of FMN in the supernatant solution was 47 μM, calculated using ɛM = 11,300 cm−1M−1 for pure FMN at 373 nm. Taking into account the concentration of protein (576 μM) and the concentration of FMN, the enzyme–prosthetic group ratio after protein purification was 12:1. This pronounced deficiency of prosthetic group might be a reason why FMN was not detected in the crystal structure. Alternatively, the apo form may have preferentially crystallized.
Figure 1.
Identification of FMN as a prosthetic group. (A) chemical sketch for FMN and FAD molecules. (B) Comparison of UV-Vis absorption spectrum (1-mm path length) of the supernatant after protein denaturation and centrifugation with spectra of FMN and FAD solutions in 25 mM Tris-HCl (pH 7.5). FMN/FAD absorption spectra were normalized to the supernatant absorption peak at 373 nm.
Structure description and putative FMN binding site
An overall view of the ArsH protein is presented in Figure 2A. The structure of the ArsH protein has a flavodoxin-like α/β/α−core. A central β-sheet, which consists of five parallel strands in the order β2, β1, β3, β4, β5, is sandwiched between helixes α1 and α5 on one side and three helixes α2, α3, α4 on the other. In addition, the ArsH protein has a 34-residue N-terminal extension and a 28-residue extension at the C terminus.
Figure 2.
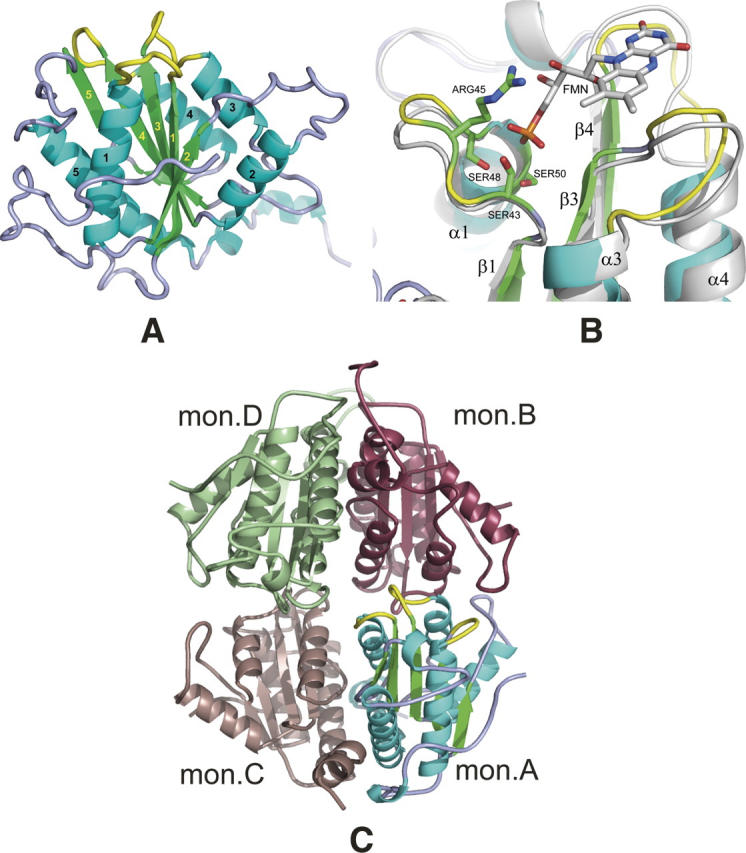
Structural overview. (A) General view of the S. flexneri ArsH protein. Secondary structures are colored in cyan (helixes), green (strands), and light blue/yellow (loops). Loops forming a binding site are shown in yellow. Labeling for β-strands and α-helixes is shown in yellow and black, respectively. (B) “Magic Fit” from Swiss-PDBViewer of the ArsH protein (colored) from S. flexneri with 1X77 FMN binding holoprotein (gray with colored FMN cofactor). Side chains of ArsH serine residues 43, 48, and 50 and arginine 45 are considered to participate in direct interaction with FMN phosphate group. Yellow-colored loops are also interacting with FMN and, probably, NADPH second cofactor. (C) General view of S. flexneri ArsH tetramer in the crystal, which is formed by two dimers A and C and B and D. Monomer A is colored as on the panel A, color codes for monomers B, C, and D are raspberry, dark salmon, and pale green, respectively.
A VAST search (Gibrat et al. 1996) was carried out to find structural homologs of the ArsH protein. The 11 closest structures from the VAST search had Z-scores more then 15.5 and included the following proteins: Azobenzene NADPH-dependent FMN reductases (Protein Data Bank [PDB] IDs 2GSW, 1X77, 1NNI), Wrba tryptophan repressor binding proteins (1ZWL, 1RLI, 1YDG, 2A5L, 1T0I), and Flavodoxins (2ARK, 5NUL, 1OBO). This search revealed a conserved flavin binding domain that spans the central part of ArsH, residues 37–200. These proteins are generally shorter than ArsH and are similar to the central conserved domain. Although their sequence identity to the ArsH protein is very low, 7.5%–24.9%, they show high structural similarity in terms of root mean square deviation for Cα positions (1.6–2.8 Å over 141–169 residues). Eight out of these 11 proteins have been shown to form complexes with FMN in the crystal. Structural alignment of ArsH from S. flexneri with the highest VAST-score structures 2GSW and 1X77 is presented in Supplemental Figure S1 (see Supplemental materials), which also shows the ClustalX sequence alignment of ArsH proteins and their homologs from proteobacteria cited earlier.
Figure 2B presents the putative FMN binding site of ArsH superimposed with the complex of Pseudomonas aeruginosa azobenzene reductase (1X77) with FMN (Agarwal et al. 2006). The FMN binding site is formed by three loops connecting strand β1 with helix α1, β3 with α3 and β4 with α4. Highly conserved residues Ser43, Arg45, Ser48, and Ser50 (ASN residue in case of 1X77) were shown to form a novel fingerprint motif (T/S)XRXXSX(T/S) directly interacting through hydrogen bonds with the phosphate group of FMN (Agarwal et al. 2006). Residues of two other loops (yellow in Fig. 2B) were shown to interact with the isoalloxazine ring directly through oxygen and nitrogen atoms of the main chain or mediated by water molecules. Additionally, the ArsH protein loop between β4 and α4 has a highly conserved V(S/C)GGSQS sequence similar to TTGGSGS suggested to participate in interaction with the other cofactor, NADPH (Liu et al. 1989; Ma et al. 1992). Although in the crystal ArsH protein is its apo form, this structural comparison allowed us to suggest that ArsH has the ability to bind an FMN molecule and, probably, an NADPH as a second cofactor.
Biological assembly: Buried surface analysis
The asymmetric unit of the crystal structure of the S. flexneri ArsH protein consists of four monomers (Fig. 2C). These monomers are likely to represent a biological assembly which is a tetramer, or more precisely, a dimer of dimers A and C and B and D. Buried surface areas between molecules A and C (2136 Å2) and molecules B and D (2387 Å2) per monomer exceed the surface buried between molecules A and B (1236 Å2) by almost a factor of two and the surface buried between molecules C and D (697 Å2) by three times. The shape of a dimer roughly resembles a letter V with the active centers at the ends of the arms. The tetramer is formed by two dimers and has approximate internal symmetry 222, such that all four active sites are exposed to the solvent. Interaction of the tetramer with the neighboring oligomers is mediated by direct contact of the C-terminal region of monomer B (residues 236–244) and monomer D (residues 232–242) with monomers A and C of neighboring tetramers. This results in distortion of the tetramer from the ideal 222 symmetry (Fig. 2C).
The same type of biological unit, a dimer of dimers, was observed earlier for the structural analogs of ArsH, the Wrba proteins (Gorman and Shapiro 2005). Interestingly, the buried areas for Wrba proteins (PDB ID 1YRH and 1ZWL) from Deinococcus radiodurans and Pseudomonas aeruginosa (1212 and 800 Å2 per monomer, respectively) formed upon the dimerization are half of that in ArsH dimers (2262 Å2 on average). This is due to the sequence feature mentioned above, that all ArsH proteins and their homologs with sequence identity greater than 60% have an extended C terminus and most of them an extended N terminus (Supplemental Fig. S1). These termini participate in dimerization. Their removal significantly reduces the contact surface in dimers A and C and B and D by 53% and 63%, respectively, but does not affect the surface between dimers in the tetrameric unit. The ArsH dimeric substructure is probably more stable than that of Wrba proteins.
Oligomerization state in solution
A dynamic light scattering (DLS) experiment on a solution of the ArsH protein at 25°C revealed a single peak in the particle size volume distribution with a hydrodynamic diameter of 8.9 and width of 2.2 nm (averaged on four measurements between 0.25 and 1.0 mg/mL concentration). The molecular weight of the oligomer estimated using Mark-Houwink-Kuhn-Sakurada (MHKS) relations for globular proteins was 111 kDa. Given the 28.4-kDa molecular weight of the ArsH monomer, the oligomer observed corresponded to the tetramer in good agreement with the theoretically calculated value of 113.6 kDa. The estimated contribution of the dimer in this case was 10% by mass. As it was shown above, the purified ArsH was essentially in its apo form. Conversion to the holo form after an addition of a 10-fold excess of FMN to the protein solution showed the tetrameric form as well.
The temperature dependence of the results of DLS from a 0.5 mg/mL solution revealed a shift of the hydrodynamic diameter peak maximum from 8.9 to 7.6 nm between 20°C and 48°C (Supplemental Table S2). The estimated average molecular weight of 77 kDa in this case corresponds neither to the dimer nor to tetramer, but to a tetramer–dimer mixture with prevalence of the dimeric form. Contribution of the dimer at 48°C in this case was estimated to be 80% by mass. These results showed that at room temperature, the ArsH protein in solution preferentially forms a tetrameric assembly in both apo and holo forms and indicates a tetramer–dimer mixture at higher temperature. This is in good agreement with the buried surface analysis of the tetramer found in the crystal structure and described above. Similar to ArsH, the Wrba protein from E. coli forms a tetramer in the crystal and was shown to be a dimer–tetramer mixture in solution (Grandori et al. 1998).
NADPH-dependent FMN reductase activity
In assaying the NAD(P)H-dependent FMN reductase activity, the following two half-reactions were tracked:
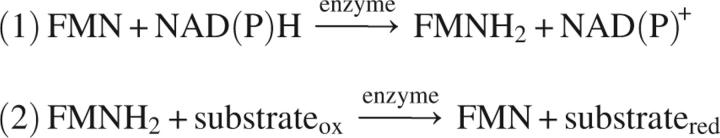 |
During step 2, FMNH2 was recycled through proton transfer to substrate or oxygen, naturally dissolved in the reaction mixture, since all reactions were performed in aerobic conditions. The reaction stops with the full utilization of NAD(P)H.
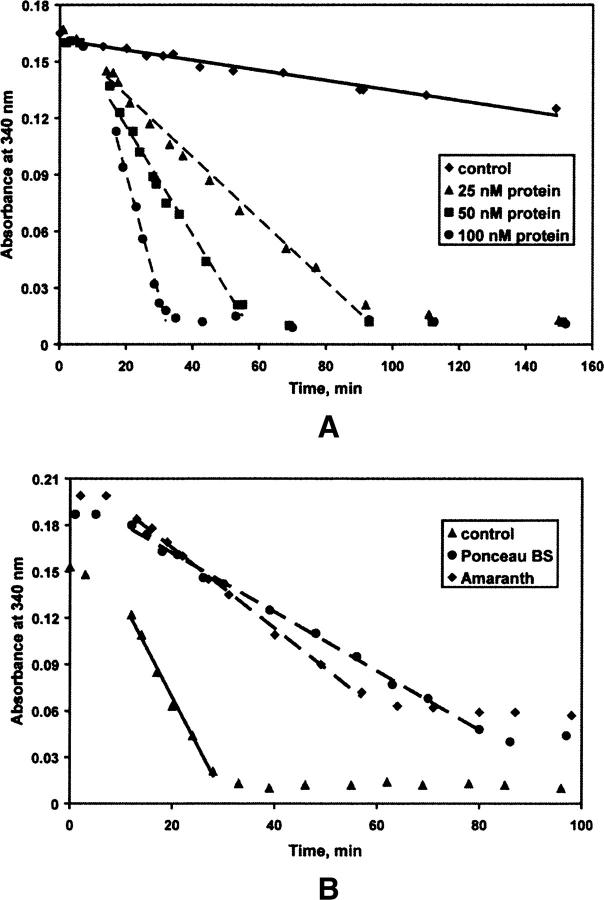
First, ArsH was assayed for NAD(P)H-dependent FMN reductase activity. No catalytic activity was detected with NADH, while reaction in the presence of NADPH was observed (Fig. 3A). An average specific activity of 2.5(1) μmol/(mg min) for NADPH oxidation was measured in the 25–100 nM range of protein concentrations. This value is 2.5 times the specific activity of 0.9 μmol/(mg min) found for YLR011wp reductase from yeast (ArsH structural homolog, PDB ID 1T0I) and by a factor of 17, the specific activity of 0.15 μmol/(mg min) for Azo1 reductase from S. aureus (Liger et al. 2004; Chen et al. 2005). Thus, ArsH revealed efficient FMN- and NADPH-dependent activity.
Figure 3.
ArsH enzymatic activity. (A) FMN- and NADPH-dependent activity. Reaction mixtures contained 300 μM NADPH, 25 mM Tris-HCl (pH 7.5), 10 μM FMN, and 25, 50, or 100 nM of protein. Control reaction did not contain the protein. NADPH oxidation rates (ɛM = 6,220 M−1cm−1 at 340 nm) were 3.5, 6.4, and 12.5 μM/min for reaction mixtures containing 25, 50, and 100 nM of the protein, respectively. (B) Azoreductase activity: competition with NADPH oxidation. Reaction mixtures contained 300 μM NADPH, 25 mM Tris-HCl (pH 7.5), 10 μM FMN, 100 nM protein, and 50 μM of dye (Ponceau BS or amaranth). Reactions were initiated by addition of the enzyme at 8 min after mixing of the other ingredients. Control reaction did not contain any dye. NADPH oxidation rates were 15.0 μM/min for control and 4.1 and 5.7 μM/min for reaction mixtures with Ponceau BS and amaranth, respectively. Path length for absorption measurements is 1 mm.
Next, ArsH was tested for reductase activity on three commercially available azo dyes (Ponceau BS, amaranth, and methyl red) and GSSG, as a model compound for the disulfide ArsC arsenate reductase products (Fig. 3B). No change in the NADPH oxidation rate was observed for methyl red and GSSG potential substrates. At the same time, the presence of 50 μM Ponceau BS or amaranth in the reaction mixture (100 nM protein, 10 μM FMN, 300 μM NADPH, 25 mM Tris-HCl at pH 7.5) reduces the NADPH oxidation rate by factors of 3.7 and 2.6 for Ponceau BS or amaranth, respectively. This inhibition, very probably, means that these azo dyes compete with NADPH for the substrate binding site. At the same time, in the course of the reaction 62% and 35% reduction was observed in the absorbance at 502 nm and 520 nm, specific for Ponceau BS or amaranth, respectively (Supplemental Fig. 2S). Interestingly, after all the NADPH is consumed, a gradual increase of the absorbance for Ponceau BS was detected, while for amaranth the reduction was irreversible. The azoreductase-specific activity of 0.07 and 0.05 μmol/(mg min) with Ponceau BS and amaranth, correspondingly, is low compared to the few thousands of μmol/(mg min) activity for the most efficient bacterial azoreductases (Liger et al. 2004). Therefore, by its ability to reduce amaranth and Ponceau BS, ArsH appears to have a very low azoreductase activity.
In summary, the S. flexneri ArsH protein has been shown to be structurally homologous to NADPH-dependent FMN reductases with unique N- and C-terminal extensions that are involved in oligomerization. The enzyme has been demonstrated to have NADPH-dependent FMN reductase activity, but the role of this activity in arsenic resistance remains to be elucidated.
Materials and Methods
ArsH expression and purification
The cloning, expression, and purification of the S. flexneri ArsH protein were carried out mostly as described for other protein targets at the Midwest Center for Structural Genomics (Dieckman et al. 2002; Stols et al. 2002; Godsey et al. 2007). The only modification from the standard protocol was that an N-terminal His-tag with a Tobacco Etch Virus protease recognition site (MHHHHHHSSGVDLGTENLYFQ↓SNA) was not removed from the protein.
ArsH crystallization and X-ray data collection
For crystallization experiments, protein was used at a concentration of 8 mg/mL in buffer containing 0.5 M NaCl, 5 mM β-mercaptoethanol, and 10 mM Tris-HCl (pH 8.3). Crystal screening was done in 96-well Corning crystallization plates using a Hydra II Plus 1 liquid handling system (MATRIX Technology Inc.). Diffraction quality crystals were grown in 2-μL sitting-drops at 22°C by mixing the protein solution with the well solution containing 0.2 M calcium chloride, 20% w/v polyethylene glycol 3350 (Nextal, Qiagen screen the PEGs) in 1:1 ratio. A platinum derivative was prepared by a 30-min soak of a native crystal at 22°C in well solution containing 5 mM of platinum nitrate. Crystals were rapidly cooled by immersion in liquid nitrogen. Native and derivative diffraction data sets were collected at 100 K at the DND-CAT (5ID-B) and NE-CAT (8-BM) beamlines at the Advanced Photon Source, Argonne National Laboratory. The crystals belonged to the tetragonal space group P43212 and initially diffracted to 1.7 and 2.3 Å for native and derivative, respectively. The data were processed with HKL2000 and XDS packages (Kabsch 1993; Otwinowski and Minor 1997).
Structure determination
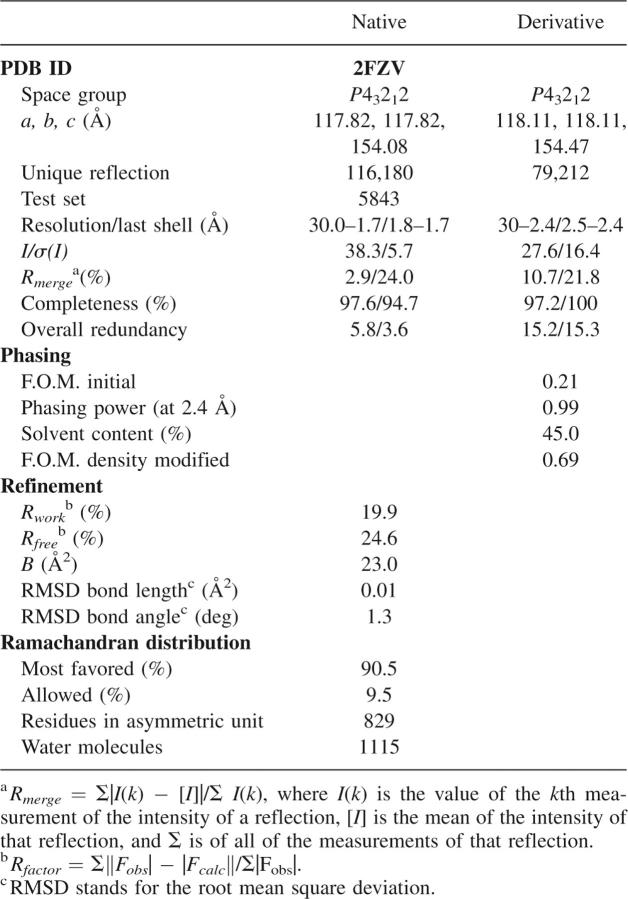
Phases were determined using the SAD method. Three consistent platinum sites were found from anomalous difference electron density maps using both SHELXD and Phenix-HYSS software packages (Sheldrick and Schneider 1997; Adams et al. 2004). Positions, occupancies, and B-factors of these three sites were refined with the program SHARP (Fortelle and Bricogne 1997) and gave initial phases with FOM 0.21 at 2.4 Å. The phases were improved using SOLOMON with an assumption of 45% solvent content in the crystal (Cowtan 1994). Modified phases (FOM 0.69) were used to calculate electron density maps, which clearly showed elements of secondary structure. An initial model consisting of 506 residues in 27 chains (45% of the total number of residues) was autotraced with the ARP/wARP program (Perrakis et al. 1999). The model was completed using iterative cycles of model building with TURBO-FRODO and refinement with the REFMAC programs (Roussel and Cambillau 1991; Murshudov et al. 1997). Crystallographic data and results of the refinement are summarized in Table 1.
Table 1.
Data and refinement statistics for the ArsH protein from S. flexneri
Attempts to make a complex of ArsH with FMN by soaking the crystal were not successful. Analyses of the crystal structure showed that access to all active sites is blocked by side chains of Arg45 and Arg110 residues and, additionally for monomers A and D, by C-terminal loops of neighboring monomers from symmetry related tetramers.
Sequence and structure alignments
A preliminary search for sequence and structural homology was performed by the ProFunc Web server (Laskowski et al. 2005). A more extensive search was done using Basic Local Alignment Search Tool (BLAST) and Vector Alignment Search Tool (VAST) on the National Center for Biotechnology Information (NCBI) Web server (Gibrat et al. 1996; Altschul et al. 1997). Structure and sequence alignments were performed using VAST and ClustalX 1.8 programs, respectively (Thompson et al. 1997). Graphical presentations of the alignments and protein structure were performed using ALSCRIPT (Barton 1993), Swiss-PDBViewer 3.7 (Guex and Peitsch 1997), and PyMOL (De Lano 2002) program packages. Buried molecular surface analysis was done using GRASP (Nicholls et al. 1991).
Identification of the prosthetic group
The concentration of the protein was measured through its absorbance at 280 nm using an extinction coefficient of 22,460 M−1cm−1 calculated from its sequence. One hundred microliters of the 576 μM of purified protein solution were heated to 100°C and kept for 10 min. After being cooled down to room temperature, the denatured protein was removed by 5-min centrifugation at 14,000 rpm. A UV/Vis absorption spectrum of the resulting yellow supernatant was analyzed and compared with the standard spectra of 100 μM solutions of pure FMN and FAD in 25 mM Tris-HCl (pH 7.5) using a NanoDrop ND-1000 UV/Vis Spectrophotometer (NanoDrop Technologies, Inc.).
Enzymatic assays
ArsH was assayed for FMN reductase activity in a reaction mixture (500 μL of final volume) consisting of 150 μM NADH or NADPH, 100 μM FMN, and 250 nM protein in 25 mM Tris-HCl (pH 7.5). NAD(P)H oxidation was monitored by the decrease in absorbance at 340 nm (ɛM = 6,220 M−1cm−1). Specific activity of the protein was measured in reaction mixtures (500 μL each) consisting of 300 μM NADPH, 10 μM FMN, and 25, 50, and 100 nM of protein in 25 mM Tris-HCl (pH 7.5).
Azoreductase assay was performed in a reaction mixture (500 μL) containing 25 μM azo dye, 10 μM FMN, 300 μM NADPH, and 0.05 μM of protein in 25 mM Tris-HCl at 22°C. The azo dye oxidation was determined by monitoring the decrease in absorbance at optimal wavelength for each azo dye tested as a substrate: 502 nm for Ponceau BS, 520 nm for amaranth, and 430 nm for methyl red. The rate of reaction was determined through NADPH oxidation by measuring the absorbance at 340 nm. Specific activity of the protein in the presence of Ponceau BS or amaranth was measured in a reaction mixture (500 μL) containing 50 μM azo dye, 10 μM FMN, 300 μM NADPH, and 100 nM of protein in 25 mM Tris-HCl (pH 7.5).
ArsH was also assayed for GSSG (L-glutathione oxidized) reductase activity. The reaction mixture and protocol were identical to that for the azoreductase assay except that 100 μM of GSSG was added.
Reactions were initiated by addition of the protein. All spectra were recorded on a NanoDrop ND-1000 UV/Vis Spectrophotometer (NanoDrop Technologies, Inc.).
Dynamic light scattering
Experiments were done on Zetasizer Nano ZEN1600 DLS photometer using Dispersion Technology Software 4.10 (Malvern Instruments Ltd.). Measurements were performed with samples containing 0.25, 0.50, 0.75, and 1.00 mg/mL of protein solution. Temperature-dependence trend measurements were done on the 0.50 mg/mL protein solution in the temperature range of 20°C–48°C with 7°C step. Molecular weight of aggregates was estimated using the Protein Utility component of Malvern Software through Mark-Houwink-Kuhn-Sakurada (MHKS) relations for globular proteins (Harding 1997).
Electronic supplemental material
Table S1 shows arsenic resistance components in studied microorganisms; Table S2, oligomerization state in solution (DLS experiment); Figure S1, sequence alignment; and Figure S2, azoreductase activity.
Acknowledgments
This work is supported by grant NIH-GM-62414 for the Midwest Center for Structural Genomics. Portions of this research were carried out at the Advanced Photon Source (APS), Argonne National Laboratory. DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) at the APS is supported by the E.I. DuPont de Nemours & Co., The Dow Chemical Company, the U.S. National Science Foundation through grant DMR-9304725, and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. This work is also based on research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines of the APS, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the APS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38. We thank Dr. Ulf Nobbmann from Malvern Instruments, Malvern, United Kingdom, for providing additional materials for oligomerization analyses from DLS experiments.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Wayne F. Anderson, Department of Molecular Pharmacology and Biological Chemistry, Ward 8-264, Feinberg School of Medicine, Northwestern University, 303 East Chicago Avenue, Chicago, IL 60611, USA; e-mail: wf-anderson@northwestern.edu; fax: (312) 503-5349.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073029607.
References
- Adams P.D., Gopal, K., Grosse-Kunstleve, R.W., Hung, L.-W., Ioerger, T.R., McCoy, A.J., Moriarty, N.W., Pai, R.K., Read, R.J., Romo, T.D., et al. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Agarwal R., Bonanno, J.B., Burley, S.K., and Swaminathan, S. 2006. Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal. Acta Crystallogr. D Biol. Crystallogr. 62: 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6: 37–40. [DOI] [PubMed] [Google Scholar]
- Bobrowicz P., Wysocki, R., Owsianik, G., Goffeau, A., and Ulaszewski, S. 1997. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae . Yeast 13: 819–828. [DOI] [PubMed] [Google Scholar]
- Butcher B.G., Deane, S.M., and Rawlings, D.E. 2000. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli . Appl. Environ. Microbiol. 66: 1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas D., Cases, I., and de Lorenzo, V. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5: 1242–1256. [DOI] [PubMed] [Google Scholar]
- Carlin A., Shi, W., Dey, S., and Rosen, B.P. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. and Rosen, B.P. 1997. Metalloregulatory properties of the ArsD repressor. J. Biol. Chem. 272: 14257–14262. [DOI] [PubMed] [Google Scholar]
- Chen H., Hopper, S.L., and Cerniglia, C.E. 2005. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiol. 151: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. 1994. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31: 34–38. [Google Scholar]
- De Lano W.L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- Dieckman L., Gu, M., Stols, L., Donnelly, M.I., and Collart, F.R. 2002. High-throughput methods for gene cloning and expression. Protein Expr. Purif. 25: 1–7. [DOI] [PubMed] [Google Scholar]
- Diorio C., Cai, J., Marmor, J., Shinder, R., and DuBow, M.S. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in Gram-negative bacteria. J. Bacteriol. 177: 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieschi F., Niviere, V., Frier, C., Decout, J.L., and Fontecave, M. 1995. The mechanism and substrate specificity of the NADPH:flavin oxidoreductase from Escherichia coli . J. Biol. Chem. 270: 30392–30400. [DOI] [PubMed] [Google Scholar]
- Fortelle E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In Methods in enzymology: Macromolecular crystallography, part A (eds. C.W. Carter and R.M. Sweet), pp. 472–494. Academic Press, San Diego, CA. [DOI] [PubMed]
- Gibrat J.F., Madej, T., and Bryant, S.H. 1996. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6: 377–385. [DOI] [PubMed] [Google Scholar]
- Godsey M.H., Minasov, G., Shuvalova, L., Brunzelle, J.S., Vorontsov, I.I., Collart, F.R., and Anderson, W.F. 2007. The 2.2 Å resolution crystal structure of Bacillus cereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain. Protein Sci. 16: 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J. and Shapiro, L. 2005. Crystal structures of the tryptophan repressor binding protein WrbA and complexes with flavin mononucleotide. Protein Sci. 14: 3004–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori R., Khalifah, P., Boice, J.A., Fairman, R., Giovanielli, K., and Carey, J. 1998. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins. J. Biol. Chem. 273: 20960–20966. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Harding S.E. 1997. The intrinsic viscosity of biological macromolecules. Progress in measurement, interpretation and application to structure in dilute solution. Prog. Biophys. Mol. Biol. 68: 207–262. [DOI] [PubMed] [Google Scholar]
- Jackson C.R. and Dugas, S.L. 2003. Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 3: 18. doi: 10.1186/1471-2148-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Garber, E.A., Armes, L.G., Chen, C.M., Fuchs, J.A., and Silver, S. 1994. Arsenate reductase of Staphylococcus aureus plasmid pI258 . Biochemistry 33: 7294–7299. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26: 795–800. [Google Scholar]
- Laskowski R.A., Watson, J.D., and Thornton, J.M. 2005. ProFunc: A server for predicting protein function from 3D structure. Nucleic Acids Res. 33: W89–W93. doi: 10.11093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liger D., Graille, M., Zhou, C.Z., Leulliot, N., Quevillon-Cheruel, S., Blondeau, K., Janin, J., and van Tilbeurgh, H. 2004. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J. Biol. Chem. 279: 34890–34897. [DOI] [PubMed] [Google Scholar]
- Liu X.F., Yuan, H., Haniu, M., Iyanagi, T., Shively, J.E., and Chen, S.A. 1989. Reaction of rat liver DT-diaphorase (NAD(P)H:quinone acceptor reductase) with 5′-[p-(fluorosulfonyl)benzoyl]-adenosine. Mol. Pharmacol. 35: 818–822. [PubMed] [Google Scholar]
- López-Maury L., Florencio, F.J., and Reyes, J.C. 2003. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185: 5363–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostao A., Daoudi, F., Irun, M.P., Ramon, A., Fernandez-Cabrera, C., Romero, A., and Sancho, J. 2003. How FMN binds to Anabaena apoflavodoxin: A hydrophobic encounter at an open binding site. J. Biol. Chem. 278: 24053–24061. [DOI] [PubMed] [Google Scholar]
- Ma Q., Cui, K., Xiao, F., Lu, A.Y., and Yang, C.S. 1992. Identification of a glycine-rich sequence as an NAD(P)H-binding site and tyrosine 128 as a dicumarol-binding site in rat liver NAD(P)H:quinone oxidoreductase by site-directed mutagenesis. J. Biol. Chem. 267: 22298–22304. [PubMed] [Google Scholar]
- Massey V. 2000. The chemical and biological versatility of riboflavin. Biochem. Soc. Trans. 28: 283–296. [PubMed] [Google Scholar]
- Messens J. and Silver, S. 2006. Arsenate reduction: Thiol cascade chemistry with convergent evolution. J. Mol. Biol. 362: 1–17. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R., Rosen, B.P., Phung, L.T., and Silver, S. 2002. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 26: 311–325. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Neyt C., Iriarte, M., Thi, V.H., and Cornelis, G.R. 1997. Virulence and arsenic resistance in Yersiniae . J. Bacteriol. 179: 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Norman N.C. 1998. Chemistry of arsenic, antimony, and bismuth. Blackie Academic and Professional, London, UK.
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. In Methods in enzymology: Macromolecular crystallography, part A (eds. C.W. Carter Jr and R.M. Sweet), pp. 307–326. Academic Press, New York. [DOI] [PubMed]
- Perrakis A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6: 458–463. [DOI] [PubMed] [Google Scholar]
- Rosen B.P. 1999. Families of arsenic transporters. Trends Microbiol. 7: 207–212. [DOI] [PubMed] [Google Scholar]
- Rosen B.P. 2002. Biochemistry of arsenic detoxification. FEBS Lett. 529: 86–92. [DOI] [PubMed] [Google Scholar]
- Roussel A. and Cambillau, C. 1991. TURBO-FRODO, silicon graphics applications directory. Silicon Graphics, Mountain View, CA.
- Saltikov C.W. and Olson, B.H. 2002. Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl. Environ. Microbiol. 68: 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. and Schneider, T. 1997. SHELXL: High resolution refinement. In Methods in enzymology (eds. R.M. Sweet et al.), pp. 319–343. Academic Press, New York. [PubMed]
- Shi H., Shi, X., and Liu, K.J. 2004. Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol. Cell. Biochem. 255: 67–78. [DOI] [PubMed] [Google Scholar]
- Stols L., Gu, M., Dieckman, L., Raffen, R., Collart, F.R., and Donnelly, M.I. 2002. A new vector for high-throughput, ligation-independent cloning encoding a Tobacco Etch Virus protease cleavage site. Protein Expr. Purif. 25: 8–15. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen, S., Xiao, X., Huang, X., You, D., Zhou, X., and Deng, Z. 2006. arsRBOCT arsenic resistance system encoded by linear plasmid pHZ227 in Streptomyces sp. strain FR-008. Appl. Environ. Microbiol. 72: 3738–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Goldberg, M.B., Burland, V., Venkatesan, M.M., Deng, W., Fournier, G., Mayhew, G.F., Plunkett III, G., Rose, D.J., Darling, A., et al. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.C., Cheng, J., Finan, T.M., Rosen, B.P., and Bhattacharjee, H. 2005. Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti . J. Bacteriol. 187: 6991–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]