Abstract
Francisella tularensis is a highly infectious Gram-negative intracellular pathogen that causes the fulminating disease tularemia and is considered to be a potential bioweapon. F. tularensis pathogenicity island proteins play a key role in modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm of macrophages. The 23 kDa pathogenicity island protein IglC is essential for the survival and proliferation of F. tularensis in macrophages. Seeking to gain some insight into its function, we determined the crystal structure of IglC at 1.65 Å resolution. IglC adopts a β-sandwich conformation that exhibits no similarity with any known protein structure.
Keywords: Francisella tularensis, IglC, crystal structure, bioterrorism
Francisella tularensis is an intracellular Gram-negative bacterial pathogen that is capable of infecting a variety of small mammals such as voles, rabbits, and muskrats, as well as humans. Tularemia, the severe disease caused by F. tularensis, is endemic in North America, parts of Europe, and Asia (Oyston et al. 2004). The disease has a very rapid onset, with flu-like symptoms such as headache, fatigue, dizziness, muscle pains, nausea, fever, and chills. The face and eyes redden and become inflamed. Inflammation spreads to the lymph nodes, which enlarge and may suppurate. Colonization of the lymph nodes is accompanied by a high fever. Other clinical manifestations may vary according to different routes of pathogen entry and the F. tularensis subspecies (Santic et al. 2006). Because of its ease of dissemination, multiple routes of infection, high infectivity, and lethality, F. tularensis has been classified a Category A bioterrorism agent by the CDC.
F. tularensis infects and rapidly multiplies in macrophages (Fortier et al. 1992), but the mechanism by which it does so is still poorly understood. Most intracellular pathogens have either a type III or a type IV secretion system, through which effector proteins are transported into the host cytosol. However, neither of these systems has been identified in the F. tularensis genome. Recently, a 30 kb Francisella pathogenicity island (FPI) was shown to encode several genes that are important for its intracellular growth (Santic et al. 2005). Bioinformatic analysis of the FPI suggests that IglA and IglB are components of a newly described type VI secretion system (de Bruin et al. 2007). The 23 kDa protein IglC has no sequence similarity to any known protein, but recent studies have shown that it and its regulator, MglA, play essential roles in the modulation of phagesome biogenesis and the escape into the cytoplasm (Lindgren et al. 2004; Santic et al. 2005).
Hoping to gain some insight into the function of IglC, we solved its crystal structure at a resolution of 1.65 Å. However, the three-dimensional structure of IglC is unique among known protein folds and possesses no recognizable functional motifs. Nevertheless, the availability of the IglC structure should facilitate further study of this important virulence factor.
Results and Discussion
General features of the IglC structure
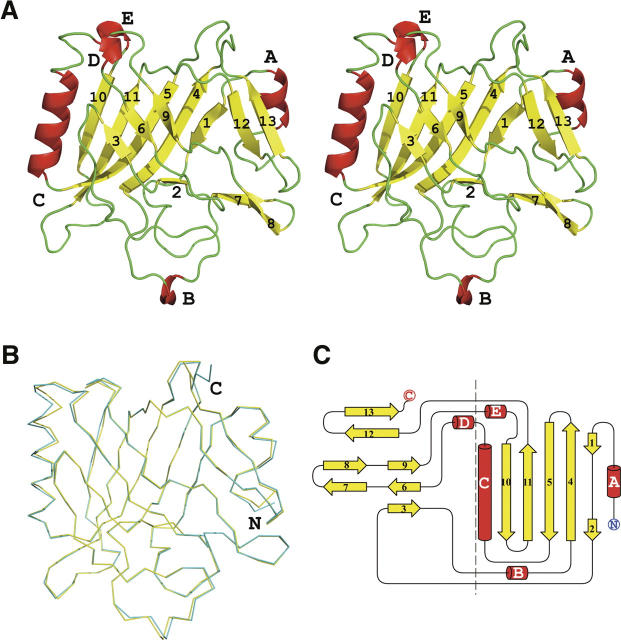
The crystal structure of IglC was determined to 1.65 Å resolution using the Single-wavelength Anomalous Dispersion (SAD) method. The Matthews' coefficient for the IglC crystals is 2.06 Å3/Da, and the estimated solvent content is 40.4%. The final model includes two molecules (residues 3–209 for both Chains A and B) and 431 water molecules in one asymmetric unit (Fig. 1A). No electron density was observed for residues 1–2 and 210–211. The two copies in the asymmetric unit are superimposable with an RMSD value of 0.56 Å2 between them (Fig. 1B). There is only a slight difference at the N and C termini. Different crystal packing arrangements observed in two other crystal forms (data not shown), which belong to space groups P32 and C2, respectively, suggest that IglC is a monomer in solution.
Figure 1.
(A) Stereoview of the crystal structure of IglC. The structure is colored according to its secondary structure elements: helices (red), strands (yellow), and loops (green). The 13 β-strands are numbered and the helices are labeled A–E (including α-helices A and C and 310-helices B, D, and E). (B) Superposition of the two monomers in one asymmetric unit. Chains A and B were colored yellow and cyan, respectively. (C) Topology diagram of IglC. Helices are represented as cylinders and β-strands as arrows. The two layers of the β-sandwich are separated by a dashed line. All images were generated with the graphics program PyMOL (DeLano Scientific).
The central feature of the IglC structure is a β-sandwich plate (β1–β13) with approximate dimensions of 50 × 50 × 25 Å3. The β-sandwich is augmented by two α-helices (A and C) and three 310-helices (B, D, and E) around its periphery. One layer of the β-sandwich is composed of six antiparallel β-strands with a strand order of β1/ β2–β4–β5–β11–β10 (Fig. 1C). The other layer is composed of three antiparallel β-strands: β3–β6–β9, β7–β8, and β12/β13. The two layers of the β-sandwich are packed together by virtue of extensive hydrophobic interactions at their interface.
Structure similarity search and bioinformatics analysis
A DALI (Holm and Sander 1998) search of the Protein Data Bank (PDB) database with the coordinates of IglC revealed a very low degree of structural similarity with other protein structures. The closest structural relative is pyridoxamine 5′-phosphate oxidase (PDB ID: 1T9M), with a Z-score of 2.4. The next three best matches are chain C of the largest subunit of RNA polymerase II (PDB ID: 2YU9, Z-score of 2.2), the sigma C capsid protein from avian reovirus (PDB ID: 2BSF, Z-score of 2.0), and gp27, a cell-puncturing device component of bacteriophage T4 (PDB ID: 1K28, Z-score of 2.0). In all of these cases, the structural similarity is limited to only a few β-strands from either or both layers of the IglC sandwich. Considering also that PSI-BLAST (Altschul et al. 1997) failed to identify any open reading frames with significant similarity to IglC, we believe that it represents a unique structure and a new protein fold.
In order to gain more insight into the possible function of IglC, the coordinates were submitted to the ProFunc server (Laskowski et al. 2005a,b) at EBL (http://www.ebi.ac.uk/thornton-srv/databases/ProFunc/) for further analysis. Some similarity was noted between the structure of IglC and a binary complex between a poxvirus-encoded viral CC Chemokine Inhibitor (vCCI) and human MIP-1β (PDB ID: 2FFK) (Zhang et al. 2006). Although these two structures cannot be aligned very well, vCCI does adopt a similar β-sandwich shape. However, this vague structural similarity is not enough to assign a function to IglC. Nevertheless, the availability of the three-dimensional structure of IglC should facilitate future efforts to investigate its role in virulence.
Materials and Methods
Protein expression and purification
The His6-MBP-IglC expression vector pKP1690 was constructed by Gateway recombinational cloning (Invitrogen) as described previously (Nallamsetty et al. 2005). To create additional sites for the incorporation of selenomethionine, L36M and L76M mutations were introduced into the IglC open reading frame in pKP1690 by site-directed mutagenesis, thereby generating pPS1986.
Escherichia coli B834(DE3) cells (EMD Biosciences) were transformed with pPS1986. A single ampicillin-resistant colony was used to inoculate Luria broth containing 100 μg/mL ampicillin, and the culture was grown to saturation overnight at 37°C and 250 rpm in an orbital shaker. The following morning, 50 mL of the saturated culture were used to inoculate each of four large shake flasks containing 1 L of selenomethionine medium (Molecular Dimensions) prepared according to the manufacturer's instructions and containing 100 μg/mL of ampicillin. The culture was incubated at 37°C and 250 rpm until the OD600 was ∼0.5, at which point IPTG was added to a final concentration of 1 mM and the temperature reduced to 30°C. The culture was allowed to incubate with shaking until the following morning. The cells were harvested by centrifugation at 4°C and ∼2500g for 10 min. The cell paste was frozen at −80°C until purification.
SeMet-labeled IglC was purified as described previously (Nallamsetty et al. 2005). The final product was judged to be >95% pure by SDS-PAGE. The molecular weight of SeMet IglC (L36M/L76M) was confirmed by electrospray mass spectrometry.
Crystallization, data collection, and structure determination
The purified IglC (L36M/L76M) protein (8.8 mg/mL) was subjected to crystallization trials with various kits from Hampton Research, Qiagen, and Emerald Biosystems. The Hydra II Plus One crystallization robot (Matrix Technologies) was used to setup the screens in a sitting-drop vapor diffusion format at 18°C. Hexagonal and orthorombic crystal forms were obtained from the initial screens. Further optimization, using the hanging-drop vapor diffusion method, focused on conditions that yielded the orthorombic crystals because they exhibited higher diffraction quality. The final crystallization condition consisted of 100 mM Tris-HCl (pH 8.5) and 25% PEG3350 with a ratio of protein to well solution of 1:3.
SeMet-labeled IglC (L36M/L76M) formed rod-shaped crystals in 100 mM Tris-HCl (pH 8.5) and 27%–30% PEG3350 that diffracted X-rays to 3 Å. Macro-seeding was used to obtain the orthorombic crystals. Before seeding with native IglC (L36M/L76M) crystals, SeMet-IglC (L36M/L76M) was mixed with 100 mM Tris-HCl (pH 8.5) and 25% PEG3350 at 1:3 volume ratio and incubated at 18°C for 2 h. Tiny native IglC (L36M/L76M) crystals were transferred into the incubated drops directly. Single orthorombic crystals appeared with 24 h.
The crystals were cryo-protected with 100 mM Tris (pH 8.5), 30% PEG3350, and 5% ethylene glycol and then flash-frozen in liquid nitrogen. The diffraction data were collected at the SER-CAT 22-ID beamline (Advanced Photon Source, Argonne National Laboratory). The wavelength was tuned to the selenium absorption edge after running a wavelength fluorescence scan. A 1.65 Å SAD data set was collected and then indexed and scaled with the HKL2000 program suite (Otwinowski and Minor 1997). The processed data were submitted into the SGXPRO program palette (Fu et al. 2005) for automatic structure determination. Fourteen of the 18 selenium sites in one asymmetric unit were found by SHELXD (Schneider and Sheldrick 2002). The handedness was subsequently determined by ISAS (Wang 1985). The phases were further improved by refinement of heavy atoms in SOLVE (Terwilliger and Berendzen 1999). After density modification, 84% of the residues were built as a polyalanine peptide chain by RESOLVE (Terwilliger 2000, 2003). The resulting model was manually corrected and finished in O (Jones et al. 1991). Refinement was carried out in CNS (Brunger et al. 1998). Five percent of the reflections were set aside for cross-validation (R free). Model quality was assessed with PROCHECK (Laskowski et al. 1993). Data collection, processing, and model refinement statistics are presented in Table 1. The atomic coordinates and structure factors for the IglC structure have been deposited in the Protein Data Bank (Berman et al. 2000) with accession code 2QWU.
Table 1.
Summary of the IglC crystallographic data

Acknowledgments
We thank Kerri Penrose for technical assistance and Zbignew Dauter for helpful discussions. Electrospray mass spectrometry experiments were conducted on the LC/ESMS instrument maintained by the Biophysics Resource in the Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute at Frederick. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38.
Footnotes
Reprint requests to: David S. Waugh, Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute at Frederick, P.O. Box B, Frederick, MD 21702-1201, USA; e-mail: waughd@ncifcrf.gov; fax: (301) 846-7148.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073177307.
References
- Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. doi: 10.1002/047001153X.g406303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- de Bruin O.M., Ludu, J.S., and Nano, F.E. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7: 1. doi: 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier A.H., Polsinelli, T., Green, S.J., and Nacy, C.A. 1992. Activation of macrophages for destruction of Francisella tularensis: Identification of cytokines, effector cells, and effector molecules. Infect. Immun. 60: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Rose, J., and Wang, B.C. 2005. SGXPro: A parallel workflow engine enabling optimization of program performance and automation of structure determination. Acta Crystallogr. D Biol. Crystallogr. 61: 951–959. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Moss, D.S., and Thornton, J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231: 1049–1067. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Watson, J.D., and Thornton, J.M. 2005a. ProFunc: A server for predicting protein function from 3D structure. Nucleic Acids Res. 33: W89–W93. doi: 10.1107/S002188982009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Watson, J.D., and Thornton, J.M. 2005b. Protein function prediction using local 3D templates. J. Mol. Biol. 351: 614–626. [DOI] [PubMed] [Google Scholar]
- Lindgren H., Golovliov, I., Baranov, V., Ernst, R.K., Telepnev, M., and Sjostedt, A. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53: 953–958. [DOI] [PubMed] [Google Scholar]
- Nallamsetty S., Austin, B.P., Penrose, K.J., and Waugh, D.S. 2005. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli . Protein Sci. 14: 2964–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Oyston P.C., Sjostedt, A., and Titball, R.W. 2004. Tularaemia: Bioterrorism defence renews interest in Francisella tularensis . Nat. Rev. Microbiol. 2: 967–978. [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret, M., Klose, K.E., Jones, S., and Kwaik, Y.A. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7: 969–979. [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret, M., Klose, K.E., and Abu Kwaik, Y. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 14: 37–44. [DOI] [PubMed] [Google Scholar]
- Schneider T.R. and Sheldrick, G.M. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58: 1772–1779. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. 2003. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59: 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.C. 1985. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 115: 90–112. [DOI] [PubMed] [Google Scholar]
- Zhang L., Derider, M., McCornack, M.A., Jao, S.C., Isern, N., Ness, T., Moyer, R., and LiWang, P.J. 2006. Solution structure of the complex between poxvirus-encoded CC chemokine inhibitor vCCI and human MIP-1beta. Proc. Natl. Acad. Sci. 103: 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]