Abstract
Calsenilin is a member of the recoverin branch of the EF-hand superfamily that is reported to interact with presenilins, regulate prodynorphin gene expression, modulate voltage-gated Kv4 potassium channel function, and bind to neurotoxins. Calsenilin is a Ca+2-binding protein and plays an important role in calcium signaling. Despite its importance in numerous neurological functions, the structure of this protein has not been reported. In the absence of Ca+2, the protein has limited spectral resolution that increases upon the addition of Ca+2. Here, we describe the three-dimensional solution structure of EF-hands 3 and 4 of calsenilin in the Ca+2-bound form. The Ca+2-bound structure consists of five α-helices and one two-stranded antiparallel β-sheet. The long loop that connects EF hands 3 and 4 is highly disordered in solution. In addition to its structural effects, Ca+2 binding also increases the protein's propensity to dimerize. These changes in structure and oligomerization state induced upon Ca+2 binding may play important roles in molecular recognition during calcium signaling.
Keywords: calsenilin, DREAM, KChIP3, calcium-binding protein, EF-hands, NMR structure
Calcium (Ca+2), a fundamental signal in many biological processes, exerts its effect primarily through Ca+2-binding proteins. Calsenilin, DREAM, and KChIP3 were independently discovered and named (Buxbaum et al. 1998; Carrion et al. 1999; An et al. 2000; Cheng and Penninger 2003), but they all contain the same four putative Ca+2-binding EF-hand domains. Calsenilin was first identified to interact with presenilins and regulate the levels of their proteolytic product (Buxbaum et al. 1998), and thus it may play a role in Alzheimer disease (Zaidi et al. 2002; Buxbaum 2004; Dong-Gyu et al. 2004) and epilepsy (Hong et al. 2003). Subsequently, in the search for transcriptional factors for the regulation of the prodynorphin gene involved in memory acquisition and pain, a Ca+2-binding protein named DREAM was discovered to bind the downstream regulatory element (DRE) (Carrion et al. 1999). DREAM constitutively suppresses prodynorphin expression, and knocking out DREAM results in an increase of prodynorphin expression and decrease in pain behavior (Cheng et al. 2002; Costigan and Woolf 2002; Vogt 2002). DREAM is therefore also a potential drug target for pain modulation (Reisch et al. 2006). More recently, a class of Ca+2-binding proteins named KChIP (Kv-channel-interacting protein) was discovered to bind the C-terminal fragment of the Kv4 channels (An et al. 2000). KChIP3, identical in sequence to calsenilin, is a member of the KChIP family and has been reported to rescue the function of tetramerization mutants of Kv4 channel by promoting tetrameric channel assembly (Kunjilwar et al. 2004) and play a key role in pacemaker control (Liss et al. 2001). KChIP3 has also been reported to bind to cAMP response element binding protein (CREB) and cAMP response element modulator (αCREM) in pain modulation (Ledo et al. 2000, 2002; Cheng and Penninger 2003) and to neurotoxin β-bungarotoxin (Lin et al. 2006). Hereafter, we will refer to calsenilin/DREAM/KChIP3 using the single name calsenilin.
Many of calsenilin's biological activities are Ca+2 regulated (Osawa et al. 2001; Craig et al. 2002). For example, the binding of a single Ca+2 ion at either EF-hand 3 or 4 has been suggested to be sufficient to drive the protein conformational changes that abolish DNA binding (Osawa et al. 2005). In the presence of Ca+2, calsenilin forms a dimer at low concentrations (Osawa et al. 2001, 2005). Calsenilin binds two calcium ions with high affinity and another with lower affinity. The two tightest Ca+2-binding sites are in EF-3 and EF-4 (Osawa et al. 2005).
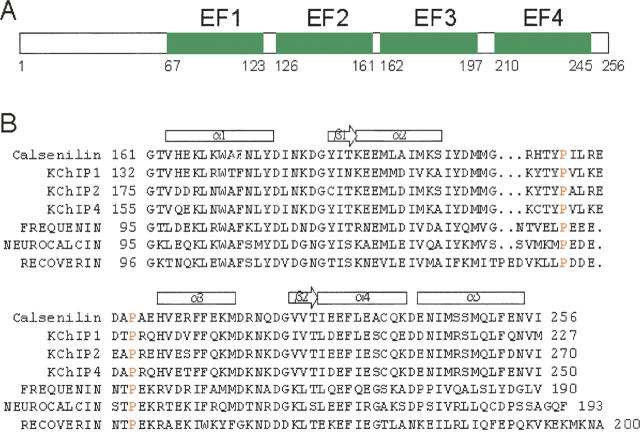
The three-dimensional structure of calsenilin is currently not known. The protein has a domain structure as shown in Figure 1A and consists of an N-terminal fragment and four EF-hands (labeled as EF1–EF4). The high-affinity Ca+2-binding site is thought to reside in EF3 or EF4 (Osawa et al. 2005). Here, we report on the three-dimensional solution structure of EF-hands 3 and 4 (hereafter denoted as EF3–4) of calsenilin in the Ca+2-bound form as determined by heteronuclear multidimensional NMR spectroscopy. The structure of this protein was compared with previously determined structures of Ca+2-binding proteins to provide clues about the binding sites involved in a variety of protein–protein and protein–DNA interactions.
Figure 1.
Domain structure of calsenilin (A) and sequence alignment of calsenilin homolog proteins in the EF3–4 region (B). Calsenilin is also known as DREAM or KChIP3. The α-helices (rectangles) and β-strands (arrows) for calsenilin EF3–4 are indicated above the sequence. (Red) The two conserved proline residues located in a loop between α2- and α3-helices. The accession numbers are Q9Y2W7 for human calsenilin, Q9NZI2 for human KChIP1, Q9NS61 for human KChIP2, Q6PIL6 for human KChIP4, AAF01804 for human frequenin, AAK34951 for human neurocalcin, and NP_002894 for human recoverin.
Results and Discussion
Ca+2-binding studies
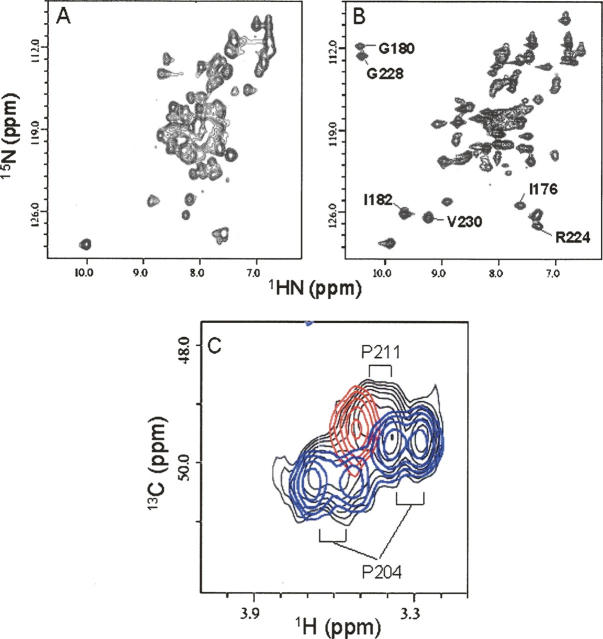
The protein sequences of calsenilin EF3–4 and its homologous Ca+2-binding proteins are shown in Figure 1B. 15N/1H HSQC spectra of the uniformly 15N-labeled calsenilin EF3–4 are shown in Figure 2. The 15N/1H cross peaks of the HSQC spectrum in the absence of Ca+2 ions are broad and poorly dispersed (Fig. 2A). This is consistent with a previous analysis that suggested a flexible molten globule-like structure in the metal-free form (Osawa et al. 2005). However, upon the addition of Ca+2 ions, the cross peaks become narrower and more dispersed (Fig. 2B). A careful Ca+2 titration by NMR indicated that saturation of the bound form occurred at a calcium to protein ratio of 2:1, suggesting that two Ca+2 ions were bound to each molecule of calsenilin EF3–4. Since the Ca+2-bound and Ca+2-free proteins are in slow exchange on the NMR timescale, the affinities for Ca+2 binding at these two sites are <1 μM.
Figure 2.
15N/1H HSQC spectra recorded on a DRX500 NMR spectrometer of the uniformly 15N-labeled calsenilin EF3–4 in the absence (A) and presence (B) of 15 mM CaCl2. The well-shifted cross peaks upon Ca+2 binding are labeled. (C) HMQC spectra of the Pro CδH2 region of the uniformly 13C-labeled calsenilin EF3–4 of the wild type (black), P204A mutant (red), and P211A mutant (blue). The assignments of the proline residues are indicated. The Pro to Ala mutation data clearly shows that P204 exits in two conformations, while P211 is present in a single conformation in calsenilin EF3–4.
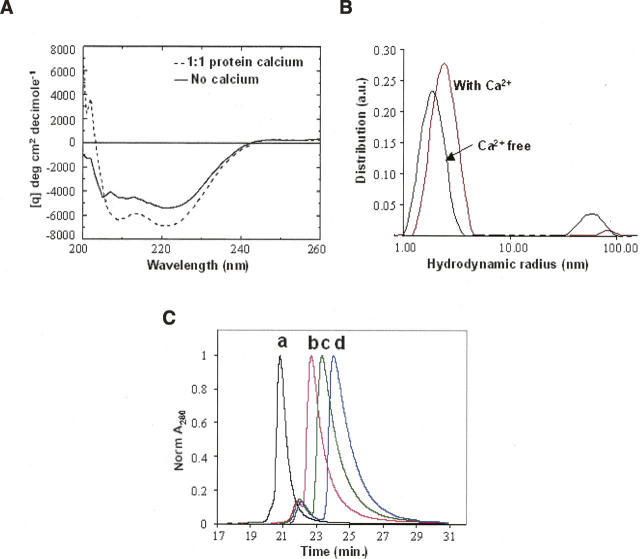
The dramatic changes observed in the 15N/1H HSQC spectra upon the addition of calcium suggest changes in the protein's structural state. To investigate these changes further we used CD to monitor secondary structural changes, and Dynamic Light Scattering (DLS) to monitor changes in protein oligomerization that occur upon binding to Ca+2 ions. The CD data shown in Figure 3A indicate that more α-helical structure is formed in the presence of calcium based on the decreased ellipticity at 208 and 222 nm. Using the simplest model for interpreting CD spectra (Chen et al. 1972), the Ca+2-bound protein is estimated to contain ∼7% more α-helical structure (corresponding to approximately one turn) than the Ca+2-free protein. This change in overall secondary structure is modest and is unlikely to account for all of the changes observed in the NMR spectra. However, it is likely that Ca+2 binding causes the reorganization of these secondary structures that are already formed in the Ca+2-free state. From DLS studies, we found that the oligomerization state of EF3–4 is also influenced by Ca+2 binding. In the Ca+2-free state, EF3–4 has a hydrodynamic radius of 1.9 nm (Fig. 3B) that increases to 2.4 nm at saturating amounts of Ca+2. The observed change in hydrodynamic radius is consistent with a spherical monomer (Ca+2-free state) versus dimer (at saturating amounts of Ca+2 and high protein concentration). A series of dilution studies of HPLC-SEC (Fig. 3C) and SLS indicates that EF3–4 is under monomer–dimer equilibrium in the presence of saturating calcium concentrations. The average molecular weight changed from 15.1 kDa at an injected protein concentration of 0.4 mg/mL to 20.3 kDa at an injected protein concentration of 15 mg/mL. The fully dimerized EF3–4 is expected to have a molecular weight of 23.6 kDa. The CD spectra collected at various Ca+2 ion/protein ratios for EF3–4 protein indicate that single Ca+2-ion binding is sufficient to induce the maximal increase in the α-helical content of EF3–4 protein (Fig. 3A), while our light-scattering data indicate a concomitant change in oligomerization state. Together, these data suggest that Ca+2 binding to the EF3–4 domain of calsenilin is sufficient to cause dimerization and likely reflect the same changes contributing to the oligomerization of the full-length protein. We did not observe any significant shift changes in the well-resolved resonances in our NMR spectra at 50 μM or 1 mM protein concentration. Mapping the dimer interface using chemical shift changes as a function of protein concentration was therefore not pursued.
Figure 3.
(A) Circular dichroism (CD) spectra of calsenilin EF3–4 in the absence (solid line) and presence (dashed line) of 10 μM Ca+2 ions. The protein concentration used in these measurements was 10 μM. Increasing Ca+2 concentrations resulted in similar CD spectra as the one obtained at 10 μM Ca+2 ions. (B) DLS analysis of 15 mg/mL calsenilin EF3–4 in the presence vs. absence of 15 mM CaCl2. The curves represent the relative mass-weighted molecular size distributions. The average hydrodynamic radii are 2.4 nm (with Ca2+) and 1.9 nm (Ca2+-free). A minor large aggregate component (50–100 nm) is also present. (C) Normalized A280 HPLC-SEC chromatograms of calsenilin EF3–4 at four injected concentrations: (a) 15 mg/mL; (b) 3 mg/mL; (c) 1 mg/mL; (d) 0.4 mg/mL. The monotonic shift of the SEC peaks to later elution times as a function of decreasing protein concentration is consistent with the decreasing absolute molecular weights determined by SLS measurements in each case (see Electronic supplemental Table).
NMR structure determination
NMR assignments and structure determination were carried out on the calcium-bound calsenilin EF3–4 protein. Three-dimensional heteronuclear multidimensional NMR experiments were used to assign backbone resonances using a uniformly 15N-, 13C-, and 2H-labeled sample with selectively protonated methyl groups of Val, Leu, and Ile (δ1). The side-chain resonances of the protein were assigned from an analysis of several heteronuclear multidimensional NMR spectra using a variety of labeled samples, as described in the Materials and Methods section.
The three-dimensional structure of calsenilin EF3–4 was determined using a simulated annealing protocol (Nilges et al. 1988) with the CNX program (Accelrys) from a total of 1294 NMR-derived distance and torsional angle restraints (Table 1). No intermonomer NOEs were observed. It is likely that the additional broadening of resonances at the interface as a result of the dimer–monomer equilibrium (Electronic supplemental material) precluded the observation of any intermonomer NOEs. A superposition of the 20 lowest-energy NMR structures is shown in Figure 4. Except for the N-terminal three residues and residues 195–213 (located in a long loop between α2- and α3-helices) (Fig. 5), the protein structure is well-defined by the NMR data with an RMSD about the mean coordinate positions of 0.56 ± 0.13 Å for the backbone atoms and 1.08 ± 0.10 Å for all heavy atoms for the residues 163–194 and 214–255 (Table 1).
Table 1.
Structural statistics and root-mean-square deviation for 20 structures of calsenilin EF3–4
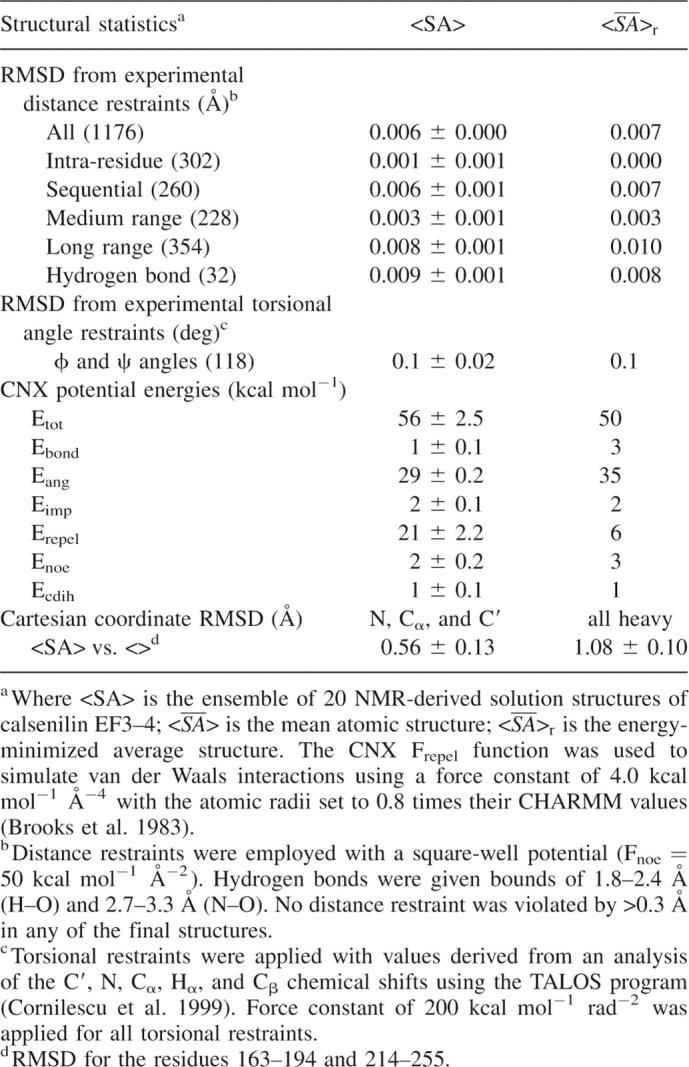
Figure 4.
Stereoview of the backbone (N, Cα, C′) of 20 superimposed NMR-derived structures of calsenilin EF3–4 (residues 161–256). The residues 163–194 and 214–255 in the well-defined regions were used here for the superimposition. (N) N terminus, (C) C terminus.
Figure 5.
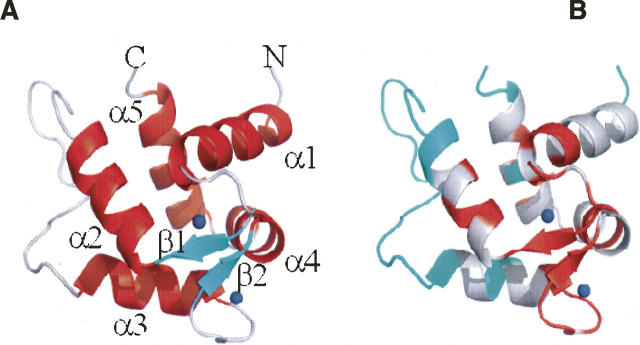
Ribbon plot (The PyMOL Molecular Graphics System, DeLano Scientific) depicting the averaged minimized NMR structure of calsenilin EF3–4 (residues 161–256). (Blue dots) Ca+2 ions present in the structure. (A) (Red) α-helices, (green) β-strands, and (gray) loops. (B) The ribbons are colored red for those residues whose backbone amides are observed after 30 min in D2O. The ribbon corresponding to residues with unassigned amides are colored blue.
As shown in Figure 5, the structure of calsenilin EF3–4 consists of a short two-stranded antiparallel β-sheet and five α-helices. The α1-helix, β1-strand, and α2-helix make up EF-hand 3, while the α3-helix, β2-strand, and α4-helix make up EF-hand 4. The two Ca+2 ions bind in the Ca+2-binding loops and β-strand regions as indicated by the two blue spheres where highly negatively charged protein surfaces are located in the electrostatic surface display (Fig. 6A). The two EF-hands are packed in such a way that the two β-strands from each EF-hand form a short antiparallel β-sheet. This β-sheet structure together with the two Ca+2-binding loops may be described as the palm of a left hand, while the helices α1 and α4 and helices α2 and α3 serve as a thumb and fingers, respectively, that grab the C-terminal α5-helix in its hand. Interestingly, the backbone amide exchange rates are much faster in the fingers and thumb than in the palm, where the slowest backbone amide exchange rates were observed (Fig. 5B). There is a cluster of hydrophobic surfaces near the junction of the α1-, α2-, and α5-helices (Figs. 5A, 6C) where EF-hands 1 and 2 could form an interface with EF3–4. The hydrophobic patch located in the back of the structure between α5-helix and the long-flexible loop (Fig. 6D) could serve as an anchorage point for interacting with functional binding partners of calsenilin during calcium signaling. The sequences of calsenilin and members of the KChIP family (Fig. 1B) are conserved for these hydrophobic patches.
Figure 6.
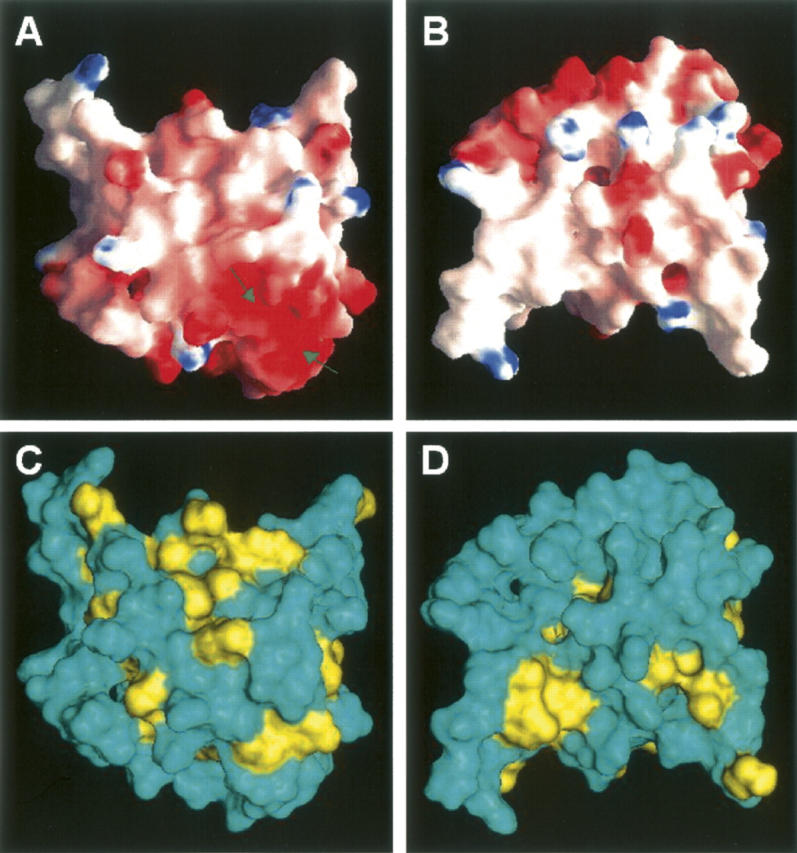
Molecular surfaces of calsenilin EF3–4 colored by local electrostatic potential (A,B) and by hydrophobicity (C,D). The electrostatic potential surface was prepared with the GRASP program (Nicholls et al. 1991), and the hydrophobicity surface was prepared with the Insight II program (Accelrys). (Blue in A,B) Positively charged areas, (red in A,B) negatively charged areas. (Yellow in C,D) The surfaces of the hydrophobic residues (Phe, Tyr, Trp, Leu, Ile, Met, Val). Panels A and C are displayed in the same orientation as Figure 5A. Panels B and D are rotated 180° along the X-axis in the paper plane relative to panels A and C. The two arrows in the panel A indicate locations where the two Ca+2 ions were bound.
While nearly all (93%) of the backbone amide peaks that were present in the NMR spectra were assigned, these assignments accounted for only 71% of the backbone amides. Interestingly, the majority of residues for which backbone amide resonances could not be detected are located in a long loop between α2- and α3-helices (Fig. 5A). In contrast to the backbone amide resonances, many of the side chain resonances for those residues that lack backbone amide peaks were observable and assigned. It is interesting to note that three sets of proline CδH2 peaks were observed in the analysis of the NMR data of the wild-type calsenilin EF3–4 (Fig. 2C). However, there are only two proline residues present in calsenilin EF3–4 (P204 and P211). Since the inter-residue NOEs in this region of the protein were very weak, we resorted to making proline to alanine mutations to confirm the assignments of these residues. The P204A mutant exhibited only one cross peak in the CδH2 region of the HMQC spectrum in Figure 2C (red), which therefore corresponds to P211 CδH2. The P211A mutant exhibited two pairs of doublets (colored blue, Fig. 2C), suggesting that P204, which is conserved in this family of calcium-binding proteins (Fig. 1B), exists in both cis and trans conformations at approximately a 1:1 ratio. These data as well as the line broadening observed for the resonances in the loop suggest conformational mobility that may play an important role in regulating molecular recognition with other biomolecules during calcium signaling.
Comparison with other Ca+2-binding proteins
Our structure of calsenilin EF3–4 has a similar fold to other Ca+2-binding proteins such as KChIP1 (Scannevin et al. 2004; Zhou et al. 2004), neurocalcin (Vijay-Kumar and Kumar 1999), frequenin (Bourne et al. 2001), and recoverin (Flaherty et al. 1993). The structure of KChIP1, a homologous protein of calsenilin, has been solved by crystallography (Scannevin et al. 2004; Zhou et al. 2004). It contains four EF-hands, with EF-hands 1 and 2 forming a pair while the other EF-hands 3 and 4 form the second pair. Among the four EF-hands, only two Ca+2 ions were bound in EF-hands 3 and 4 in KChIP1. This result is similar to our NMR data on calsenilin EF3–4 in which two Ca+2 ions were bound in both EF-hands 3 and 4 (Figs. 2, 5).
The structures of calsenilin and KChIP1 EF3–4 domains are compared in Figure 7. Our solution structure of calsenilin EF3–4 has shorter α2- and α3-helices than the crystal structures of the corresponding parts of EF3–4 domains of KChIP1 (Fig. 7; Scannevin et al. 2004; Zhou et al. 2004). The backbone amides in the α3-helix are broadened in calsenilin EF3–4, while no backbone amide exchange data are currently available for KChIP1. P204 in calsenilin EF3–4 exists in both cis and trans conformations in solution. However, the status of the isomerization for the corresponding residue P175 of KChIP1 in solution is not reported. The corresponding long loop that is disordered in our solution structure of calsenilin EF3–4 (Figs. 4, 5) was not observed in the crystal structure of KChIP1 that contains a fused N-terminal peptide from Kv4.2 (Fig. 7; Zhou et al. 2004), presumably due to conformational heterogeneity as we observed for calsenilin EF3–4. Similarly, in the crystal structure of another homologous protein, frequenin, the same corresponding long loop was not observed in some of the structural subunits (Bourne et al. 2001). Therefore, this long loop that connects the EF-hands 3 and 4 may be inherently flexible to function as fingers during calcium signaling for the binding and recognition to other molecules such as presenilins in Alzheimer disease (Buxbaum et al. 1998), DRE, CREB, and αCREM in pain modulation (Ledo et al. 2000, 2002), and the N-terminal domain of the Kv4 channels for modulating potassium channel functions (An et al. 2000).
Figure 7.
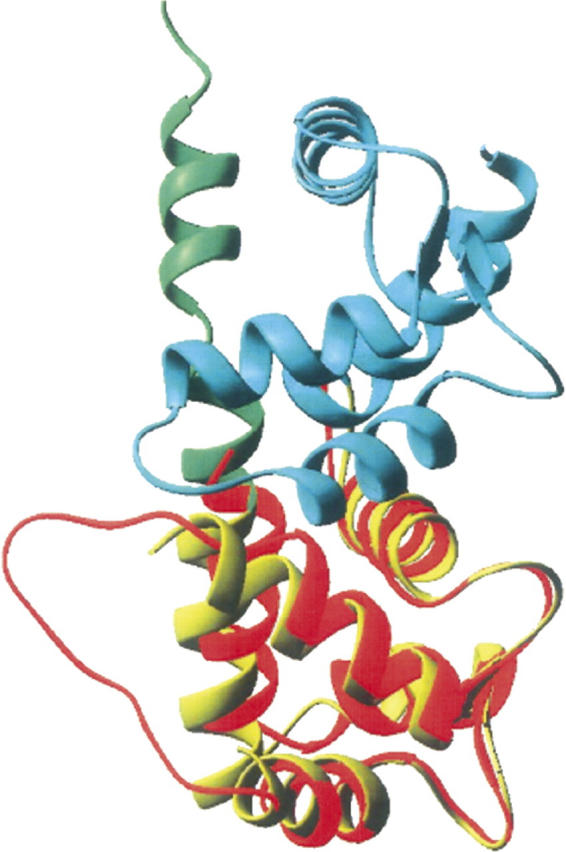
Ribbon plots comparing the structures of calsenilin EF3–4 (red) and KChIP1 that contains a fused Kv4.2 N-terminal peptide at its C terminus (Zhou et al. 2004). (Cyan) EF1–2, (yellow) EF3–4, (green) the fused peptide. The long loop in the EF-hands 3 and 4 is not determined for this structure of KChIP1. The structures were superimposed by using 58 pairs of Cα, C′, and N atoms in the EF3 and EF4 domains.
Conclusions
We have determined the three-dimensional solution structure of the Ca+2-bound form of calsenilin EF3–4 by NMR spectroscopy. The EF-hand 3 consists of α1-helix, a Ca+2-binding loop, β1-strand, and α2-helix, while the EF-hand 4 is composed of α3-helix, a Ca+2-binding loop, β2-strand, and α4-helix. These two EF-hands are packed tightly in such a way that the two β-strands contributed from each EF-hand form a short two-stranded antiparallel β-sheet where some of the slowest exchangeable backbone amides were located. The C-terminal α5-helix packed against the helices of the EF-hands. The long loop that links the EF-hands 3 and 4 is disordered in solution and may play an important role in interacting with other cellular molecules for relaying the biological signals during calcium signaling processes.
Materials and Methods
NMR sample preparation
Calsenilin EF3–4 was cloned into a pET28a vector with an N-terminal His tag (MGSSHHHHHHSSGLVPRGSHM) and expressed in Escherichia coli BL21 (DE3) cells. Uniformly 15N- and 15N, 13C-labeled proteins were prepared for the NMR experiments by growing bacteria in a medium containing 15NH4Cl with or without [U-13C]-glucose. A uniformly 15N-, 13C-, and ∼70% 2H-labeled sample was prepared by growing cells in a medium containing in 80% 2H2O, 15NH4Cl (1 g/L), and [U-13C]-glucose (3 g/L). A uniformly 15N-, 13C-labeled sample with selectively protonated methyl groups of Val, Leu, and Ile (δ1) was also prepared by growing cells in 100% 2H2O and by supplementing media with [U-13C] α-ketobutyrate (50 mg/L), [U-13C] α-ketoisovalerate (100 mg/L), [U-13C, 2H] glucose (3 g/L), and 15NH4Cl (1 g/L) (Goto et al. 1999; Hajduk et al. 2000). To confirm the Pro assignments, site-directed Pro to Ala mutants were prepared by using the QuikChange mutagenesis kit (Strategene) for the two proline residues (P204 and P211) present in calsenilin EF3–4.
The labeled recombinant proteins were purified by affinity chromatography on a nickel-IDA column (Invitrogen). The N-terminal His-tag of the purified protein was then cleaved with thrombin and purified further by size-exclusion chromatography. The purified protein was concentrated to 1.0 mM for NMR studies in a buffer containing 25 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, with or without 15 mM CaCl2.
Far UV circular dichroism (CD) spectra of the purified samples were collected on a JASCO J-715 spectropolarimeter from 195–260 nm using three accumulated scans, 5-nm bandwidth, and a 4-sec time constant. Sample temperature was maintained at 25°C by using the built-in Peltier heating/cooling unit. The protein concentration used in these measurements was 10 μM in the absence or presence of various concentrations of Ca+2 ions.
Evaluation of the molecular weight and molecular size of calsenilin EF3–4 at different protein concentrations and in the presence or absence of Ca+2 ions was performed by static and dynamic light scattering experiments (SLS, DLS). SLS studies were carried out with a Hewlett-Packard Series 1100 HPLC system (Agilent Technologies) and a Tosoh TSKgel G2000SWXL 7.8 mm × 30 cm SEC column (Tosoh Biosep LLC) connected with three chromatographic detectors in series: the H-P 1100s diode array absorbance detector, a Wyatt DAWN HELEOS multi-angle light scattering instrument, and a Wyatt Optilab rEX differential refractive index detector (Wyatt Technology Corporation). Absolute molecular weights were calculated at 1-sec intervals across chromatographic peaks using the Wyatt Astra V software, utilizing light scattering intensities typically at 10–15 angles in conjunction with UV 280 nm absorbance and refractive index values as independent measurements of protein concentration at each respective time point. DLS experiments were conducted using an ALV model DLS/SLS-5000F Compact Goniometer System (ALV-Laser Vertriebsgesellschaft m.b.H.) equipped with an ALV multi-τ digital correlator, APD detector, and 647 nm input from an Innova 400 Krypton ion laser (Coherent Inc.). Dust and microparticulates were removed from samples either by microcentrifugation at 14,000g, or filtration with 0.22-μm filters. Samples were measured in 8-mm cylindrical quartz tubes and placed in a toluene bath thermostated at 20 ± 0.1°C. DLS data were acquired at three angles (40, 90, and 140 degrees) and the autocorrelation functions analyzed for multiple diffusion components by a CONTIN-based in-house program. The molecular radius (equivalent spherical hydrodynamic radius) was calculated by averaging the results obtained at the three angles.
NMR spectroscopy and structure determination
The NMR spectra were collected at 30°C on a Bruker DRX500 or DRX800 NMR spectrometer. The 1H, 15N, and 13C resonances of the backbone were assigned using triple resonance experiments (HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, and HN(CA)CO) (Yamazaki et al. 1994) using the uniformly 15N-, 13C-, and 2H-labeled sample with selectively protonated methyl groups of Val, Leu, and Ile (δ1). 1Hα resonances were assigned from a 15N-edited TOCSY spectrum using a uniformly 15N-labeled protein (Clore and Gronenborn 1994). The side-chain signals were assigned from 3D H(CCO)NH-TOCSY, C(CO)NH-TOCSY, HCCH-TOCSY, 15N-edited TOCSY experiments, and 3D 15N- or 13C-edited NOESY (Fesik and Zuiderweg 1988; Clore and Gronenborn 1994). Many of the resonances in the loop were assigned to residue type by identifying their side-chain resonances. Ambiguities in sequence position were resolved from short-range NOEs.
Structures of calsenilin EF3–4 were generated using a simulated annealing protocol (Nilges et al. 1988) with the CNX program (Accelrys). Structure calculations for calsenilin EF3–4 employed a total of 1144 NMR-derived distance restraints from the analysis of 3D 15N- and 13C-resolved NOESY spectra (Fesik and Zuiderweg 1988) collected with mixing times of 100–150 ms. The NOE-derived distance restraints were given upper bounds of 3.0, 4.0, 5.0, and 6.0 Å based upon the measured NOE intensities. From an analysis of the amide exchange rates measured from a series of 1H/15N HSQC spectra recorded after the addition of 2H2O, 32 hydrogen bonds from the α-helices, β-sheets, and Ca+2-binding loops were included in the structural calculations. In addition, 118 φ and ψ angular restraints derived from an analysis of the C′, N, Cα, Hα, and Cβ chemical shifts using the TALOS program (Cornilescu et al. 1999) were included in the structural calculations.
Protein Data Bank accession number
The coordinates of calsenilin EF3–4 have been deposited into the Protein Data Bank under the identifier code 2E6W.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Edward T. Olejniczak, Pharmaceutical Discovery Division, GPRD, Abbott Laboratories, Abbott Park, IL 60064-6098, USA; e-mail: Edward.olejniczak@abbott.com; fax: (847) 938-2478.
Abbreviations: NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; HSQC, heteronuclear single-quantum coherence; RMSD, root-mean-square deviation; DRE, down-stream regulatory element; CREB, cAMP response element binding protein; CREM, cAMP response element modulator; DREAM, downstream regulatory element antagonistic modulator; KChIP, Kv-channel-interacting protein; DLS, Dynamic Light Scattering; SLS, Static Light Scattering.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072928007.
References
- An W.F., Bowlby, M.R., Betty, M., Cao, J., Ling, H.-P., Mendoza, G., Hinson, J.W., Mattsson, K.I., Strassle, B.W., Trimmer, J.S., et al. 2000. Modulation of A-type potassium channels by a family of calcium sensors. Nature 403: 553–556. [DOI] [PubMed] [Google Scholar]
- Bourne Y., Dannenberg, J., Pollmann, V., Marchot, P., and Pongs, O. 2001. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1). J. Biol. Chem. 276: 11949–11955. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. 1983. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem. 4: 187–217. [Google Scholar]
- Buxbaum J.D. 2004. A role for calsenilin and related proteins in multiple aspects of neuronal function. Biochem. Biophys. Res. Commun. 322: 1140–1144. [DOI] [PubMed] [Google Scholar]
- Buxbaum J.D., Choi, E.-K., Luo, Y., Lilliehook, C., Crowley, A.C., Merriam, D.E., and Wasco, W. 1998. Calsenilin: A calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 4: 1177–1181. [DOI] [PubMed] [Google Scholar]
- Carrion A.M., Link, W.A., Ledo, F., Mellstrom, B., and Naranjo, J.R. 1999. DREAM is a Ca2+-regulated transcriptional repressor. Nature 398: 80–84. [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Yang, J.T., and Martinez, H.M. 1972. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11: 4120–4131. [DOI] [PubMed] [Google Scholar]
- Cheng H.-Y.M., Pitcher, G.M., Laviolette, S.R., Whishaw, I.Q., Tong, K.I., Kockeritz, L.K., Wada, T., Joza, N.A., Crackower, M., Goncalves, J., et al. 2002. DREAM is a critical transcriptional repressor for pain modulation. Cell 108: 31–43. [DOI] [PubMed] [Google Scholar]
- Cheng H.-Y.M. and Penninger, J.M. 2003. When the DREAM is gone: From basic science to future prospectives in pain management and beyond. Expert Opin. Ther. Targets 7: 249–263. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn, A.M. 1994. Multidimensional heteronuclear magnetic resonance of proteins. Meth. Enzymol. 239: 349–363. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Costigan M. and Woolf, C.J. 2002. No DREAM, no pain: Closing the spinal gate. Cell 108: 297–300. [DOI] [PubMed] [Google Scholar]
- Craig T.A., Benson, L.M., Venyaminov, S.Y., Klimtchuk, E.S., Bajzer, Z., Prendergast, F.G., Naylor, S., and Kumar, R. 2002. The metal-binding properties of DREAM. Evidence for calcium-mediated changes in DREAM structure. J. Biol. Chem. 277: 10955–10966. [DOI] [PubMed] [Google Scholar]
- Dong-Gyu J., Joo-Yong, L., Yeon-Mi, H., Sungmin, S., Inhee, M.-J., Jae-Young, K., and Yong-Keun, J. 2004. Induction of pro-apoptotic calsenilin/DREAM/KChIP3 in Alzheimer's disease and cultured neurons after amyloid-β exposure. J. Neurochem. 88: 604–611. [DOI] [PubMed] [Google Scholar]
- Fesik S.W. and Zuiderweg, E.R.P. 1988. Heteronuclear three-dimensional NMR spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson. 78: 588–593. [Google Scholar]
- Flaherty K.M., Zozulya, S., Stryer, L., and McKay, D.B. 1993. Three-dimensional structure of recoverin, a calcium sensor in vision. Cell 75: 709–716. [DOI] [PubMed] [Google Scholar]
- Goto N.K., Gardner, K.H., Mueller, G.A., Willis, R.C., and Kay, L.E. 1999. A robust and cost-effective method for the production of Val, Leu, Ile (d1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J. Biomol. NMR 13: 369–374. [DOI] [PubMed] [Google Scholar]
- Hajduk P.J., Augeri, D.J., Mack, J., Mendoza, R., Yang, J., Betz, S.F., and Fesik, S.W. 2000. NMR-based screening of proteins containing 13C-labeled methyl groups. J. Am. Chem. Soc. 122: 7898–7904. [Google Scholar]
- Hong Y.-M., Jo, D.-G., Lee, M.-C., Kim, S.-Y., and Jung, Y.-K. 2003. Reduced expression of calsenilin/DREAM/KChIP3 in the brains of kainic acid-induced seizure and epilepsy patients. Neurosci. Lett. 340: 33–36. [DOI] [PubMed] [Google Scholar]
- Kunjilwar K., Strang, C., DeRubeis, D., and Pfaffinger, P.J. 2004. KChIP3 rescues the functional expression of Shal channel tetramerization mutants. J. Biol. Chem. 279: 54542–54551. [DOI] [PubMed] [Google Scholar]
- Ledo F., Carrion, A.M., Link, W.A., Mellstrom, B., and Naranjo, J.R. 2000. DREAM-α-CREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Mol. Cell. Biol. 20: 9120–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo F., Kremer, L., Mellström, B., and Naranjo, J.R. 2002. CA2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 21: 4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-L., Wu, P.-F., Wu, T.T., and Chang, L.-S. 2006. KChIP3: A binding protein for Taiwan banded krait β-bungarotoxin. Toxicon 47: 265–270. [DOI] [PubMed] [Google Scholar]
- Liss B., Franz, O., Sewing, S., Bruns, R., Neuhoff, H., and Roeper, J. 2001. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChIP3.1 transcription. EMBO J. 20: 5715–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A.J., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Nilges M., Clore, G.M., and Gronenborn, A.M. 1988. Determination of three-dimensional structures of proteins from interproton distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 229: 317–324. [DOI] [PubMed] [Google Scholar]
- Osawa M., Tong, K.I., Lilliehook, C., Wasco, W., Buxbaum, J.D., Cheng, H.-Y.M., Penninger, J.M., Ikura, M., and Ames, J.B. 2001. Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 276: 41005–41013. [DOI] [PubMed] [Google Scholar]
- Osawa M., Dace, A., Tong, K.I., Valiveti, A., Ikura, M., and Ames, J.B. 2005. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J. Biol. Chem. 280: 18008–18014. [DOI] [PubMed] [Google Scholar]
- Reisch N., Aeschlimann, A., Gay, S., and Sprott, H. 2006. The DREAM of pain relief. Curr. Rheumatol. Rev. 2: 69–82. [Google Scholar]
- Scannevin R.H., Wang, K., Jow, F., Megules, J., Kopsco, D.C., Edris, W., Carroll, K.C., Lu, Q., Xu, W., Xu, Z., et al. 2004. Two N-terminal domains of Kv4 K+ channels regulate binding to and modulation by KChIP1. Neuron 41: 587–598. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S. and Kumar, V.D. 1999. Crystal structure of recombinant bovine neurocalcin. Nat. Struct. Biol. 6: 80–88. [DOI] [PubMed] [Google Scholar]
- Vogt B.A. 2002. Knocking out the DREAM to study pain. N. Engl. J. Med. 347: 362–364. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Lee, W., Arrowsmith, C.H., Muhandiram, D.R., and Kay, L.E. 1994. A suite of triple-resonance NMR experiments for the backbone assignment of 15N-, 13C-, 2H-labeled proteins with high sensitivity. J. Am. Chem. Soc. 116: 11655–11666. [Google Scholar]
- Zaidi N.F., Berezovska, O., Choi, E.K., Miller, J.S., Chan, H., Lilliehook, C., Hyman, B.T., Buxbaum, J.D., and Wasco, W. 2002. Biochemical and immunocytochemical characterization of calsenilin in mouse brain. Neuroscience 114: 247–263. [DOI] [PubMed] [Google Scholar]
- Zhou W., Qian, Y., Kunjilwar, K., Pfaffinger, P.J., and Choe, S. 2004. Structural insights into the functional interaction of KChIP1 with Shal-type K+ channels. Neuron 41: 573–586. [DOI] [PubMed] [Google Scholar]